Abstract
Malignant melanoma is a tumor with a high mortality rate. Previous studies have demonstrated that the oncogenesis of melanoma is associated with microRNA (miR)-150. However, the role of miR-150 in melanoma and its regulatory mechanisms are still unclear. In the present study, melanoma cancer tissues and adjacent normal tissues were obtained from 20 melanoma patients. The expression level of miR-150 in melanoma tissue and cell lines was detected by reverse transcription-quantitative polymerase chain reaction. miR-150 inhibitors/negative control were transfected into melanoma A375 cells in order to investigate the effects of miR-150 on cell proliferation, apoptosis, cell cycle migration and invasion using a Cell Counting Kit-8, colony formation, Hoechst 33528, flow cytometry, and Transwell assays. The association between miR-150 and programmed cell death protein-4 (PDCD4) was detected by a dual luciferase reporter assay. The functional role of PDCD4 in miR-150-affected melanoma cells was confirmed by small interfering (si)RNA knockdown. Results demonstrated that miR-150 was significantly upregulated and mRNA and protein expressions of PDCD4 were decreased in melanoma cancer tissues as compared with adjacent normal tissues. The level of PDCD4 was inversely associated with the level of miR-150. Transfection of miR-150 inhibitors suppressed cell proliferation, migration, and invasion, while the apoptosis of cells was promoted and G2/M cell arrest was induced. MiR-150 inhibitors enhanced the expression of caspase-8 and p21. The PDCD4 was identified as a direct target gene of miR-150. The effects of miR-150 inhibitors on apoptosis and apoptosis-associated proteins, including caspase-8 and p21, of A375 cells, were reversed following transfection of siRNA-PDCD4. Therefore, miR-150 inhibitors enhance cell apoptosis via upregulation of PDCD4-mediated activation of caspase-8 and p21. These findings demonstrate the potential for a promising therapeutic strategy in the management of melanoma.
Keywords: miR-150, PDCD4, melanoma, apoptosis, migration, invasion
Introduction
Malignant melanoma is a malignant tumor with a high mortality rate that originates from melanocytes (1). In the United States, the incidence of melanoma makes it the fifth-most-common tumor type, accounting for 5% of all cancers detected in 2013; and the number of newly diagnosed cases reached 73,870 in the year 2015 (2–4). In addition, in Queensland, Australia, the area where the highest incidence of malignant melanoma is seen, the annual incidence rate of this cancer in the female population was 55.8 per 10 million, and in the male population was 41.1 per 10 million (5). In China, despite the low incidence of malignant melanoma, the rate of incidence and death has increased over the years and this fact has attracted widespread concern (6). In the early stages of malignant melanoma, the five-year survival rate of patients is as high as 98% (7). However, malignant melanoma easily metastasize to regional lymph nodes, and this is associated with a poor prognosis due to a lack of effective interventions. The overall survival of patients thus affected is only 6–9 months, and the 5-year survival rate is only 16% (8,9). Because malignant melanoma that has metastasized is not amenable to conventional radiotherapy and chemotherapy, it is particularly important to identify relvant molecular markers for early diagnosis and treatment.
In recent years, researchers have come to realize the importance of microRNA in the treatment of cancer. MicroRNA (miRNA) is a short sequence of non-coding RNA that has a length of 21–25 nucleotides. It can regulate the expression of target genes involved in cell growth, proliferation, apoptosis, metastasis, cell cycle progression, and differentiation (10,11) by specific inhibition of translation or degradation of target mRNA (12). Evidence increasingly suggests that abnormal regulation of various microRNAs is closely related to the occurrence and development of tumors (11,13). miR-150 was reported to be associated with the progression of a variety of tumors. The reduction of miR-150 expression in non-small-cell carcinoma can inhibit tumor cell proliferation and induce cell apoptosis (14). miR-150 can indirectly activate the VEGF signaling pathway, and inhibition of the expression of miR-150 has an anti-tumor effect (15). miR-150 is highly expressed in gastric cancer and plays an important role in tumor progression via its effect on the tumor suppressor gene EGR2 (16).
It has been demonstrated that miR-150 is expressed at high levels in malignant melanoma and is associated with decreased long-term survival of metastatic melanoma patients (17–19). There is a positive association between miR-150 upregulation and a lowered risk of recurrence, and this suggests an important role for miR-150 in the treatment of melanoma (20–22). Although miR-150 is implicated in the oncogenesis of melanoma (23), the role of miR-150 in melanoma and the underlying mechanisms are still unclear.
The programmed cell death protein-4 (PDCD4) is a novel tumor suppressor protein involved in programmed cell death (24). Downregulation of PDCD4 is relevant with invasion, metastasis and poor prognosis of various types of cancers (24), including patients with melanoma (25). In this study, we investigated the functional significance of miR-150 in melanoma cancer and identified PDCD4 as miR-150-regulated novel cancer pathway, which could provide new insights into potential molecular mechanisms of melanoma carcinogenesis.
Materials and methods
Melanoma tissue
Twenty malignant melanoma tissues and adjacent normal tissues were collected during surgery at Department of Dermatology, the First Affiliated Hospital of Jinan University (Guangzhou, China). None of the patients received therapy including chemotherapy or radiotherapy before surgical resection. The study and protocol were approved by the Ethics Committee of the First Affiliated Hospital of Jinan University. A written informed consent was obtained from participants. All the samples were stored in liquid nitrogen.
Cell culture and transfection
The human malignant melanoma cell lines including M14, A357 and WM115 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). The adult melanocytes were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% FBS supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin, and maintained in a humidified incubator containing 5% CO2 at 37°C.
Subsequent analysis revealed that miR-150 was differentially expressed among the melanoma cell lines. The tested cell line which expressed the highest level of miR-150 was used for loss-of-function analysis by transfection miR-150 inhibitor or PDCD4 siRNA with Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA in cells and tissues were extracted by using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA concentration was measured and reverse transcription reaction was performed according to the manufacturer's instructions (Invitrogen; Thermo Fisher Scientific, Inc.). PCR analysis was performed on Applied Biosystems 7500 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Premix Ex Taq GC kit (Takara Bio, Inc., Otsu, Japan). The stem-loop primers used for the PCR amplification were synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The relative expression level of miR-150 was normalized against U6 expression level. The primers for miR-150: 5′-CTGCTTAGTGGCTCTACTCCTG-3′ (forward) and 5′-TCCCCTCTGGCTTATGTCC-3′ (reverse); for U6: 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-TGGTGTCGTGGAGTCG-3′ (reverse); for the PDCD4: 5′-AAGAAAGGTGGTGCAGGAGG-3′ (forward) and 5′-TGACTAGCCTTCCCCTCCAA-3′ (reverse). Gene expression of PDCD4 was normalized to the level of β-actin and analyzed by the relative 2−ΔΔCq method. Each experiment was performed in triplicate.
Western blotting
After transfection for 48 h, cells were treated to RIPA (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) to extract the total proteins. And the protein concentrations were determined using a BCA kit (Beyotime Institute of Biotechnology, Haimen, China). 30–50 µg of proteins were separated by 10% SDS-PAGE, and transferred to PVDF membranes (EMD Millipore, Billerica, MA, USA). Then, membranes were blocked with 5% non-fat milk and incubated overnight with primary antibody against PDCD4 (1:200; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), followed by an incubation of horseradish peroxidase-conjugated secondary antibody (1:1,000; Santa Cruz Biotechnology, Inc.). GAPDH was used as an internal control.
Cell proliferation
The cell proliferation was detected by Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology). The transfected A357 cells were added into 96-well plates. At 24, 48 and 72 h, the medium of each well was replaced with 100 µl fresh media contained 10% CCK-8 reaction solution and incubated for 2 h, and then the absorbance were measured using a microplate reader (Thermo Fisher Scientific, Inc.) at 450 nm. Also, the cell proliferation were detected by Edu staining according to the manufacture's instruction (Beyotime Institute of Biotechnology).
Colony formation assay
The transfected A357 cells were seeded at 4,000 cells/cm2 in 24-well plates in DMEM supplemented with 10% FBS, 100-U/ml PEST and 0.3% agarose with 0.6% agarose underlay. Dishes were maintained in a humidified incubator containing 5% CO2 at 37°C. After 10 days, colonies were stained with crystal violet (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and counted.
Cell apoptosis and cell cycle assay
After transfected 48 h, cell apoptosis was detected by Hoechst 33528 assay. For cell cycle detection, the PI/RNase staining kits (MultiSciences Biotech Co., Ltd., Hangzhou, China) was used and detected by a FACScan (BD Biosciences, Franklin Lakes, NJ, USA). The cells in G0-G1, S, and G2-M phases were counted.
Cell migration and invasion assay
Cell migration and invasion ability of melanoma cells were evaluated by transwell assay. Transfected A357 cells were digested and then resuspended in serum-free DMEM. For the transwell migration assays, cells were planted in the upper chambers of a 24-well plate and 500 µl DMEM containing 10% FBS was added into the lower chamber as a chemoattractant. Cells were then incubated for another 48 h. For the transwell invasion assays, cells were seeded on the top of the matrigel-coated transwell chambers (BD Biosciences) and proceeded the same as described above. Cotton swabs were used to remove the non-invasive cells after 24 h. The migrated or invaded cells were fixed and stained with 0.1% crystal violet, and counted using a microscope.
Immunostaining
After transfected for 48 h, cells were fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. After blocked by 2% serum goat for 30 min, the cells were incubated with the primary antibody against PDCD4 (1:200; Santa Cruz Biotechnology, Inc.) overnight and subsequent incubation with AF488 secondary antibody for 1 h. Cell nucleus were stained by DAPI. Then, images were acquired by confocal microscopy (Zeiss 510; Carl Zeiss, Jena, Germany).
Luciferase report assay
The 3′UTR fragment of PDCD4 containing the putative binding sequences of miR-150 was cloned into pMIR-REPORT vectors, and a mutated plasmid was used as a control. The H293T cells were co-transfected with a miR-150 mimics and related reporter plasmids. The luciferase activities were measured using a Dual Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) after transfection 48 h according to the manufacturer's instructions.
Statistically analysis
Data was presented as mean ± standard deviation and analyzed by SPSS 15.0 (SPSS, Inc., Chicago, IL, USA) by using student's t-test or one-way ANOVO. For detection the relationship between miR-150 and PDCD4 mRNA, the Pearson correlation analysis was performed. P<0.05 was considered to indicate a statistically significant difference.
Results
The upregulation of miR-150 is closely related to the downregulation of PDCD4 in melanoma
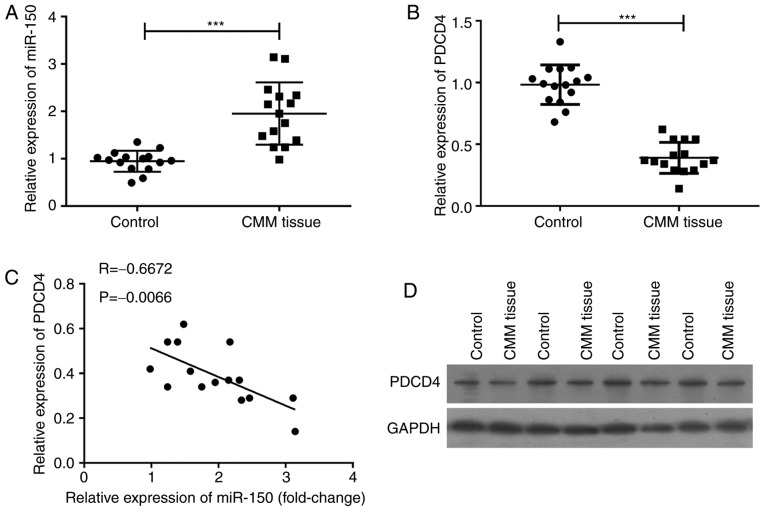
To investigate the relationship between miR-150 and PDCD4 in melanoma patients, cancer tissues and the adjacent normal tissues were collected from 20 melanoma patients. qPCR assay was used to detect the expression of miR-150 and PDCD4, and the results revealed that miR-150 was significantly upregulated (P<0.001) (Fig. 1A), and that PDCD4 mRNA was significantly downregulated (P<0.001) (Fig. 1B). The level of PDCD4 mRNA was inversely related to the expression of miR-150 (P=0.0066) (Fig. 1C). The levels of PDCD4 in cancer tissue and in its adjacent normal tissues were confirmed by western blot assay (Fig. 1D). PDCD4 protein expression in melanoma tissues was lower than that in adjacent normal tissues, suggesting that the expression of miR-150 was inversely related to the level of PDCD4 in melanoma.
Figure 1.
MicroRNA (miR)-150 was upregulated and programmed cell death protein-4 (PDCD4) was downregulated in melanoma cancer tissues compared with that in the adjacent normal tissues. 20 pairs of melanoma cancer and adjacent normal tissues were collected. (A) miR-150 and (B) PDCD4 mRNA were detected by RT-qPCR. (C) Relationship between miR-150 and PDCD4 mRNA. (D) Detection of PDCD4 by western blotting. ***P<0.001 vs. control.
Knockdown of miR-150 in melanoma cells by miR-150 inhibitors
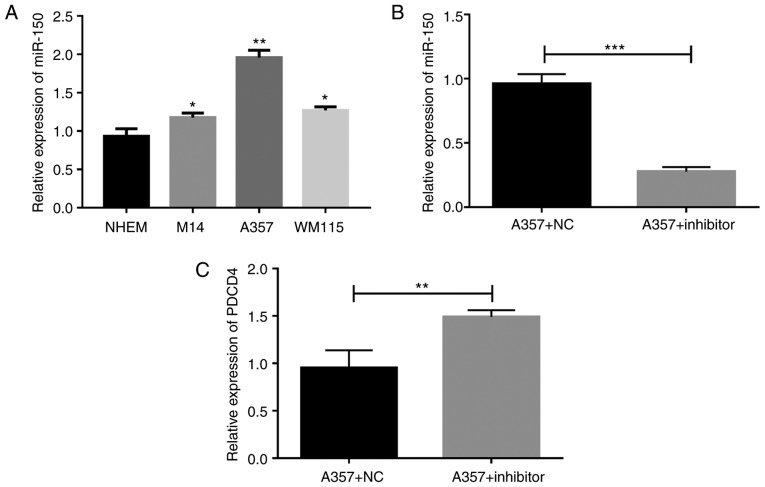
To detect the role of miR-150 in melanoma, the levels of miR-150 in three melanoma cells, including M14, A357, and WM115-and in NHEM cells, were detected (Fig. 2A). The levels of miR-150 in all the melanoma cells, including M14, A357, and WM115, were significantly upregulated as compared with levels in NHEM cells. As the levels in the A357 cells were the highest among those expressed within melanoma cells, we used A357 cells to detect the role of miR-150 in the subsequent experiments. A357 cells were transfected with miR-150 inhibitors, and our results showed that the level of miR-150 was significantly downregulated by miR-150 inhibitors transfection (Fig. 2B), and that the expression of PDCD4 was significantly upregulated (Fig. 2C), indicating that the PDCD4 was a target gene of miR-150.
Figure 2.
The levels of microRNA (miR)-150 in three melanoma cells and knockdown of miR-150. (A) The levels of miR-150 in three melanoma cells including M14, A357 and WM115 and in NHEM cells were detected by RT-qPCR. Then, A357 cells were transfected with miR-150 inhibitors. *P<0.05, **P<0.01 vs. NHEM. (B) The expression of miR-150 and (C) the expression of PDCD4 in miR-150 inhibitors-transfected cells. ***P<0.001.
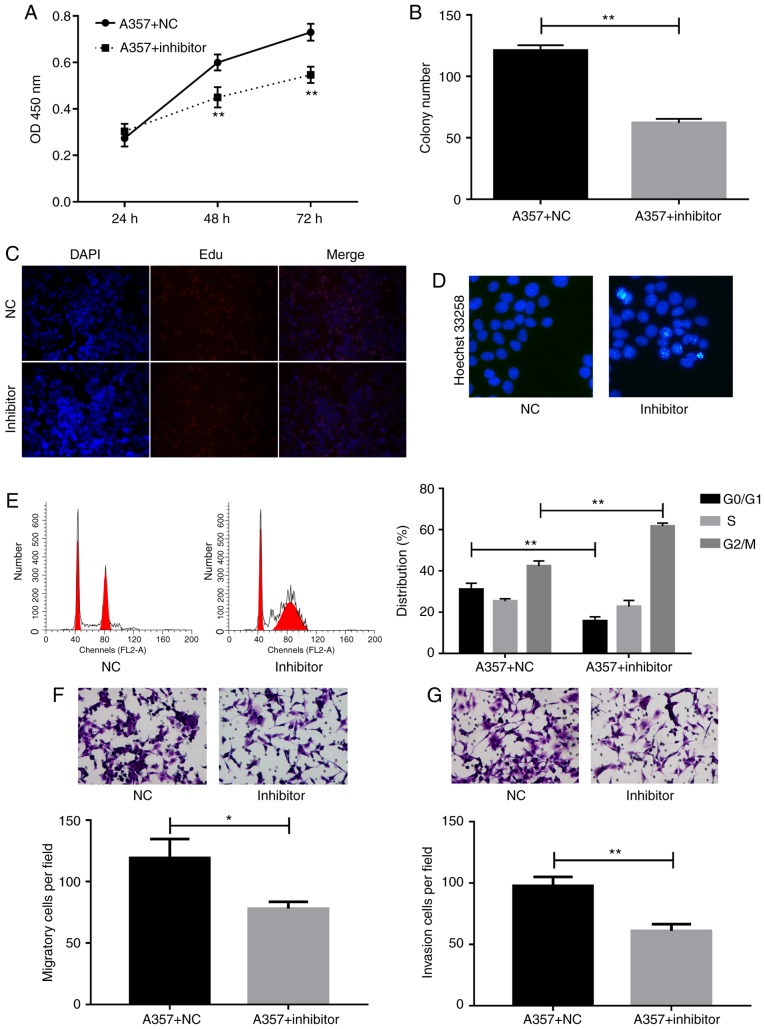
miR-150 inhibitor suppressed cell proliferation, migration, and invasion, whereas it enhanced cell aptoptosis and induced G2/M cell arrest. To detect the role of miR-150 in melanoma, cells were transfected with miR-150 inhibitors and then the cell proliferation were detected by using CCK-8, Edu, and colon formation assays. The CCK-8 assay showed that miR-150 inhibitors significantly suppressed cell proliferation at 48 h as compared with the NC group (Fig. 3A). Decreased colon formation was also observed after miR-150 inhibitors transfection in A357 cells (Fig. 3B). The inhibition of cell proliferation by miR-150 inhibitors was further confirmed by the Edu assay (Fig. 3C).
Figure 3.
Role of microRNA (miR)-150 in A357 cells. Cell proliferation was detected by (A) Cell Counting Kit-8, (B) colony formation and (C) Edu assays. (D) Cell apoptosis was detected by Hoechst 33258 assay. (E) Cell cycle was detected by flow cytometry and cell in G0/G1, S, G2/M phases were calculated. (F) Cell migration and (G) invasion were detected and calculated by Transwell assays. *P<0.05, **P<0.01.
To investigate the effect of miR-150 on cell apoptosis, the hoechst 33258 method was used to perform apoptosis assays. The data demonstrated that cell apoptosis rate was increased after the inhibition of miR-150 (Fig. 3D). Cell cycle assay provided more detailed information. As shown in Fig. 3E, accumulation of cells at G2/M phase and loss of cells at G0/G1 phase were detected after transfection of A357 cells with miR-150 inhibitors.
Moreover, migration Transwell assay indicated significant inhibition of cell migration in A357 cells transfected with miR-150 inhibitors compared to control group (Fig. 3F). Invasion capability of A357 cells transfected with NC or miR-150 inhibitors were evaluated by Matrigel Invasion assay. Indeed, the number of invading cells was decreased markedly in A357 cells as a result of knockdown of miR-150 (Fig. 3G).
Knockdown of miR-150 increased the expression of PDCD4
To detect the molecular mechanism underlying the miR-150-inhibitor-induced cell apoptosis, the expression of PDCD4 was detected by immunostaining and western blot assay (Fig. 4). The immunostaining results showed that miR-150 inhibitors increased the expression of PDCD4 in cell plasma (Fig. 4A). Western blotting confirmed the upregulation of PDCD4 in cells transfected with miR-150 inhibitors (Fig. 4B).
Figure 4.
Expression of programmed cell death protein-4 (PDCD4), caspase-8 and p21 in cells-transfected with miR-150 inhibitors. (A) Expression of PDCD4 was detected by immunostaining. (B) Western blotting detection of PDCD4, casepase-8 and p21.
The expressions of caspase-8 and p21 were also detected, and results showed that caspase-8 and p21 were upregulated by the miR-150 inhibitors. It indicated that knockdown of miR-150 might enhance cell apoptosis via upregulation of PDCD4-mediated activation of caspase-8 and p21.
Knockdown of miR-150-enhanced cell apoptosis via direct targeting of PDCD4
MiRNA mediates post-transcriptional regulation by binding to the 3′UTR of the downstream genes. To verify whether PDCD4 is a direct target of miR-150, the wild type or mutation of 3′UTR of PDCD4 was inserted into the downstream of luciferase reporter vector and transfected into H293T cells, together with the miR-150 mimics. Overexpression of miR-150 significantly suppressed the luciferase activity of reporter genes containing 3′UTR of PDCD4 compared with control group but partially rescued when the binding site was mutated (Fig. 5A). Thus, miR-150 directly targets PDCD4.
Figure 5.
Role of programmed cell death protein-4 (PDCD4) in microRNA (miR)-150 enhanced the cell apoptosis. (A) Dual-luciferase reporter system. The H293T cells were co-transfected with miR-150 mimics and a luciferase reporter containing a fragment of the PDCD4 3′UTR harboring either the miR-150 binding site (WT) or a mutant (MUT). After transfected with siPDCD4, (B) the expression of PDCD4 mRNA was detected by RT-qPCR, (C) cell apoptosis was detected by the Hoechst 33258 assay, and (D) the expression of PDCD4, caspase-8, p21 were detected by western blotting. **P<0.01.
The role of PDCD4 in miR-150-inhibitor-induced cell apoptosis was confirmed by siPDCD4 transfection (Fig. 5B-D). The A357 cells were co-transfected with NC or miR-150 inhibitors together with siPDCD4 for 48 h. As shown in Fig. 5B, the upregulation of PDCD4 induced by miR-150 inhibitors in A357 cells was abolished by siPDCD4 transfection. Hoechst 33258 assay revealed that cell apoptosis induced by miR-150 inhibitors was significantly inhibited by siPDCD4 transfection (Fig. 5C). The knockdown of PDCD4 significantly inhibited the expression of caspase-8 and p21 induced by miR-150 inhibitors (Fig. 5D). Thus, miR-150 inhibitors enhance cell apoptosis via upregulation of PDCD4-mediated activation of caspase-8 and p21.
Discussion
Melanoma is one of the most aggressive type of malignant skin cancers (26). In recent years, given the increase of incidence rate, it has become a common malignant tumor. Melanoma poses serious risks to human health, due to its rapid progression rate, ease of migration, and poor clinical prognosis. Although surgical resection is greatly facilitated the early treatment of melanoma, there is still no effective therapy for advanced melanoma. At present, it is very important to unveil the mechanisms of human melanoma cancer, and find effective therapeutic targets to improve the prognosis of melanoma cancer patients.
miRNA is a class of endogenous non-coding small RNA molecules that are involved in post-transcriptional regulation of single-stranded and conserved genes (12,13). Increasing evidence demonstrate that miRNA is involved in the development of many types of cancer, including melanoma cancer (27). Recent genome-wide miRNA expression technologies have clarified the alternation of miR-150 in several types of human cancers (28). Interestingly, divergent expression patterns of miR-150 among human cancers have been reported. miR-150 down-regulation was described in malignant lymphoma (29), osteosarcoma (30), and colorectal cancer (31), whereas its up-regulation was shown in lung cancer (28) and breast cancer (32). These controversial results suggest that the role of miR-150 is possibly tumor specific and highly dependent on its targets in different cancers. Since the role of miR-150 as a tumor suppressor or as an oncogene of tumor cell growth and metastasis in various cancers has been extensively investigated, we focused on its potential role in melanoma cancer. In this study, we observed that the expression level of miR-150 was increased in melanoma cancer specimens. In agreement with our results, Friedman et al have demonstrated that miR-150 is up-regulated in metastatic melanoma specimens and the patients with higher circulating expression of miR-150 have a high recurrence risk (33). In the in vitro study, we found that the loss of miR-150 suppresses proliferation, migration and invasion of melanoma cancer cell line, and we also showed that knockdown of miR-150 induces cell cycle arrest and apoptosis in A357 cells. Given that invasion and metastasis are two leading attributes of malignant cancer, these results suggest that miR-150 is a potent tumor suppressor in melanoma cancer. Additionally, it has been reported that the inhibition of miR-150 accelerates apoptosis in cancer cells and renders them more sensitive to various chemotherapy drugs, including gemcitabine and 5-fluorouracil, suggesting the association between miR-150 and apoptosis-related proteins during tumorigenesis.
PDCD4 is a key protein involved in programmed cell death (24). It has been reported that PDCD4 inhibits the translation of proteins and accelerates apoptosis by binding to the translation initiation factor eIF4A (34). In addition, by activating and regulating the transcription activator protein AP-1 and matrix metalloproteinase 2 (MMP-2), PDCD4 also inhibits tumor growth, invasion, and metastasis (35). Downexpression of PDCD4 is significantly associated with short overall survival of various types of cancer patients (24), including those suffering from melanoma (25). POLINA N found that PDCD4 loss is not a common event in melanoma progression, and PDCD4 can be used for molecular typing of melanoma (36). Loss of PDCD4 increases the proliferative activity, promotes tumor cell invasion, and contributes to apoptosis resistance of cancer cells, revealing the significance of PDCD4 loss in tumorigenesis. In the present study, we showed that miR-150 enhanced the cell apoptosis and elicited cell cycle arrest at G2/M phases in A357 cells. However, miR-150-inhibitor-induced cell apoptosis was reversed by knockdown of PDCD4 gene, suggesting that PDCD4 is an important mediator of cell apoptosis regulation by miR-150 in A357 cells. Our results also show that the increased levels of caspase-8 and p21, which were hallmarks of apoptosis induction and cell cycle arrest, was observed after miR-150 inhibitors transfection. Moreover, the induction of apoptosis by miR-150 inhibitors was attenuated by PDCD4 knockdown in A357 cells. Collectively, our results suggest that miR-150 induces cell proliferation and invasion via a mechanism dependent on PDCD4. Also, clinical melanoma cancer samples were used to confirm the relationship between the endogenous expression levels of PDCD4 and miR-150. We further confirmed through luciferase reporter gene assays that miR-150 directly targets PDCD4 by binding the 3′-UTR of PDCD4 mRNA, which is consistent with Lei et al (37). In essence, this provided the evidence that the loss of miR-150 may lead to PDCD4-mediated cell apoptosis and cell cycle in melanoma cancer, which would constitute a promising target for melanoma cancer therapy.
In conclusion, our study suggests that miR-150 is an anti-apoptotic factor in melanoma cancer that maintains tumor cell growth via regulation of PDCD4, and thus miR-150 may play an important role in melanoma carcinogenesis. The newly identified miR-150/PDCD4 link provides novel insights into the metastasis of melanoma cancer, especially with respect to cell apoptosis and cell cycle in vitro; and sheds new lights on therapeutic strategy for melanoma cancer.
References
- 1.Kosary CL, Altekruse SF, Ruhl J, Lee R, Dickie L. Clinical and prognostic factors for melanoma of the skin using SEER registries: Collaborative stage data collection system, version 1 and version 2. Cancer. 2014;23(Suppl 120):S3807–S3814. doi: 10.1002/cncr.29050. [DOI] [PubMed] [Google Scholar]
- 2.Coit DG, Thompson JA, Andtbacka R, Anker CJ, Bichakjian CK, Carson WE, III, Daniels GA, Daud A, Dimaio D, Fleming MD, et al. Melanoma, version 4.2014. J Natl Compr Canc Netw. 2014;12:621–629. doi: 10.6004/jnccn.2014.0066. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005–1011. [PubMed] [Google Scholar]
- 6.Hao M, Zhao G, Du X, Yang Y, Yang J. Clinical characteristics and prognostic indicators for metastatic melanoma: Data from 446 patients in north China. Tumour Biol. 2016;37:10339–10348. doi: 10.1007/s13277-016-4914-4. [DOI] [PubMed] [Google Scholar]
- 7.Song X, Zhao Z, Barber B, Farr AM, Ivanov B, Novich M. Overall survival in patients with metastatic melanoma. Curr Med Res Opin. 2015;31:987–991. doi: 10.1185/03007995.2015.1021904. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson H, Frohm-Nilsson M, Järås J, Kanter-Lewensohn L, Kjellman P, Månsson-Brahme E, Vassilaki I, Hansson J. Prognostic factors in localized invasive primary cutaneous malignant melanoma: Results of a large population-based study. Br J Dermatol. 2015;172:175–186. doi: 10.1111/bjd.13171. [DOI] [PubMed] [Google Scholar]
- 9.Pan Y, Haydon AM, McLean CA, McDonald PB, Kelly JW. Prognosis associated with cutaneous melanoma metastases. Australas J Dermatol. 2015;56:25–28. doi: 10.1111/ajd.12293. [DOI] [PubMed] [Google Scholar]
- 10.Lin X, Khalid S, Qureshi MZ, Attar R, Yaylim I, Ucak I, Yaqub A, Fayyaz S, Farooqi AA, Ismail M. VEGF mediated signaling in oral cancer. Cell Mol Biol (Noisy-le-grand) 2016;62:64–68. doi: 10.14715/cmb/2016.62.14.11. [DOI] [PubMed] [Google Scholar]
- 11.Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8:45–56. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Stein M, Ruggiero P, Rappuoli R, Bagnoli F. Helicobacter pylori CagA: From pathogenic mechanisms to its use as an anti-cancer vaccine. Front Immunol. 2013;4:328. doi: 10.3389/fimmu.2013.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 16.Zhang R, Peng Y, Wang W, Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunz M. MicroRNAs in melanoma biology. Adv Exp Med Biol. 2013;774:103–120. doi: 10.1007/978-94-007-5590-1_6. [DOI] [PubMed] [Google Scholar]
- 18.Shiiyama R, Fukushima S, Jinnin M, Yamashita J, Miyashita A, Nakahara S, Kogi A, Aoi J, Masuguchi S, Inoue Y, Ihn H. Sensitive detection of melanoma metastasis using circulating microRNA expression profiles. Melanoma Res. 2013;23:366–372. doi: 10.1097/CMR.0b013e328363e485. [DOI] [PubMed] [Google Scholar]
- 19.Tembe V, Schramm SJ, Stark MS, Patrick E, Jayaswal V, Tang YH, Barbour A, Hayward NK, Thompson JF, Scolyer RA, et al. MicroRNA and mRNA expression profiling in metastatic melanoma reveal associations with BRAF mutation and patient prognosis. Pigment Cell Melanoma Res. 2015;28:254–266. doi: 10.1111/pcmr.12343. [DOI] [PubMed] [Google Scholar]
- 20.Fleming NH, Zhong J, da Silva IP, de Miera Vega-Saenz E, Brady B, Han SW, Hanniford D, Wang J, Shapiro RL, Hernando E, Osman I. Serum-based miRNAs in the prediction and detection of recurrence in melanoma patients. Cancer. 2015;121:51–59. doi: 10.1002/cncr.28981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thanarajasingam U, Sanz L, Diaz R, Qiao J, Sanchez-Perez L, Kottke T, Thompson J, Chester J, Vile RG. Delivery of CCL21 to metastatic disease improves the efficacy of adoptive T-cell therapy. Cancer Res. 2007;67:300–308. doi: 10.1158/0008-5472.CAN-06-1017. [DOI] [PubMed] [Google Scholar]
- 22.Mullins IM, Slingluff CL, Lee JK, Garbee CF, Shu J, Anderson SG, Mayer ME, Knaus WA, Mullins DW. CXC chemokine receptor 3 expression by activated CD8+ T cells is associated with survival in melanoma patients with stage III disease. Cancer Res. 2004;64:7697–7701. doi: 10.1158/0008-5472.CAN-04-2059. [DOI] [PubMed] [Google Scholar]
- 23.Latchana N, Ganju A, Howard JH, Carson WE., III MicroRNA dysregulation in melanoma. Surg Oncol. 2016;25:184–189. doi: 10.1016/j.suronc.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Li JZ, Gao W, Ho WK, Lei WB, Wei WI, Chan JY, Wong TS. The clinical association of programmed cell death protein 4 (PDCD4) with solid tumors and its prognostic significance: A meta-analysis. Chin J Cancer. 2016;35:95. doi: 10.1186/s40880-016-0158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao XH, Chen M, Wang Y, Cui PG, Liu SB, Xu ZY. MicroRNA-21 regulates the ERK/NF-κB signaling pathway to affect the proliferation, migration, and apoptosis of human melanoma A375 cells by targeting SPRY1, PDCD4, and PTEN. Mol Carcinog. 2017;56:886–894. doi: 10.1002/mc.22542. [DOI] [PubMed] [Google Scholar]
- 26.Naves LB, Dhand C, Venugopal JR, Rajamani L, Ramakrishna S, Almeida L. Nanotechnology for the treatment of melanoma skin cancer. Prog Biomater. 2017;6:13–26. doi: 10.1007/s40204-017-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101:551–556. doi: 10.1038/sj.bjc.6605204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N, Wei X, Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013;587:2346–2351. doi: 10.1016/j.febslet.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Tagawa H, Watanabe A, Sawada K. Abstract 146: The role of Mir-150 as a tumor suppressor in malignant lymphoma. Cancer Res. 2011;71(8 Suppl):S146. doi: 10.1158/1538-7445.AM2011-146. [DOI] [Google Scholar]
- 30.Li CH, Yu TB, Qiu HW, Zhao X, Zhou CL, Qi C. miR-150 is downregulated in osteosarcoma and suppresses cell proliferation, migration and invasion by targeting ROCK1. Oncol Lett. 2017;13:2191–2197. doi: 10.3892/ol.2017.5709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, Liu W, Qin H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 32.Huang S, Chen Y, Wu W, Ouyang N, Chen J, Li H, Liu X, Su F, Lin L, Yao Y. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8:e80707. doi: 10.1371/journal.pone.0080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, Berman RS, Shapiro RL, Pavlick AC, Hernando E, et al. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med. 2012;10:155. doi: 10.1186/1479-5876-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colburn NH, Yang HS, Jansen A. Pdcd4 targets eIF4A to inhibit translation, transcription, tumorigenesis and invasion. EJC Suppl. 2006;4:23. doi: 10.1016/j.ejcsup.2006.04.053. [DOI] [Google Scholar]
- 35.Li JZ, Gao W, Lei WB, Zhao J, Chan JY, Wei WI, Ho WK, Wong TS. MicroRNA 744-3p promotes MMP-9-mediated metastasis by simultaneously suppressing PDCD4 and PTEN in laryngeal squamous cell carcinoma. Oncotarget. 2016;7:58218–58233. doi: 10.18632/oncotarget.11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vikhreva PN, Korobko IV. Expression of Pdcd4 tumor suppressor in human melanoma cells. Anticancer Res. 2014;34:2315–2318. [PubMed] [Google Scholar]
- 37.Lei Y, Hu X, Li B, Peng M, Tong S, Zu X, Wang Z, Qi L, Chen M. miR-150 modulates cisplatin chemosensitivity and invasiveness of muscle-invasive bladder cancer cells via targeting PDCD4 in vitro. Med Sci Monit. 2014;20:1850–1857. doi: 10.12659/MSM.891340. [DOI] [PMC free article] [PubMed] [Google Scholar]