Abstract
Osteosarcoma is known as a malignant tumour with a high mortality rate in orthopaedic settings; however, the factors associated with its degree of malignancy and the biological response remains to be elucidated. Although the essential role of the long noncoding RNA (lncRNA) maternally expressed 3 (MEG3) has been recently reported, its biological functions and regulatory mechanism in osteosarcoma cells have not yet been reported. In the present study, reverse transcription-quantitative polymerase chain reaction analysis revealed that the expression of MEG3 in MG63 cells was lower compared with in hFOB1.19 cells. Furthermore, it was observed that overexpressing MEG3 in MG63 cells resulted in a decline in the proliferation and invasion, and a marked increase in apoptosis. Additionally, western blotting was used to detect the changes in expression of p53 and MDM2 proto-oncogene, which may be regulated by MEG3, and proteins that associated with cell proliferation, invasion and apoptosis. It was demonstrated that the upregulation of MEG3 significantly increased the transactivation of p53 and induced downstream changes in protein expression. In conclusion, these experiments have demonstrated that MEG3 serves an essential regulatory role in the biological processes of human osteosarcoma cells, and imply that MEG3 may be a marker for predicting the occurrence and development of osteosarcoma.
Keywords: long non-coding RNA, MEG3, osteosarcoma, proliferation, invasion, apoptosis
Introduction
Osteosarcoma is one of the most common primary malignant bone tumours. It mainly affects children and adolescents, accounts for 5% of paediatric oncology cases (1) and has a tendency of high invasiveness and distant metastasis. With the development of adjuvant examination, operative therapy, chemotherapy and other treatment technologies, the five-year survival rate of osteosarcoma without metastasis has reached 70% in recent years. However, the survival rate of patients with multiple metastases has reached only 10 to 20% (2–4). Therefore, new and effective molecular markers need to be discovered in order to predict the occurrence and development of tumours and guide decisions regarding clinical treatment and prognosis. The functional mechanism of biological factors needs to be further studied to promote the development of clinical treatments.
lncRNA (long non-coding RNA) is non-protein-coding RNA that is longer than 200 nucleotides (5). Studies on the biological function and the coordination mechanism of lncRNA have drawn great attention. Many studies showed that lncRNA plays a key role in regulating gene expression, chromatin remodelling, transcription and post-transcriptional processing (6,7). Additionally, more and more studies have reported that disorder of lncRNA is closely related to many human diseases, especially tumours (8).
MEG3 (maternally expressed 3) is a maternally expressed, imprinted long non-coding RNA gene with a length of about 1.6 kb nucleotides. MEG3 is expressed in many human normal tissues, but has failed to express in a variety of human tumour cells, such as non-small cell lung cancer, bladder cancer, prostate cancer and other cancers (9–14). Additionally, many studies have articulated that MEG3 gene plays an inhibiting role in various tumour cells (15–20). However, the functional mechanism of MEG3 in MG63 has yet to be proven.
Tumour inhibition is a self-protective mechanism that prevents normal cells from undergoing neoplastic transformation (21). p53, as a tumour suppressor, inhibits tumours by regulating downstream gene expression (22,23). Some studies showed that MEG3 functions by activating p53 (17,18) while other studies indicated that the activation of p53 may be achieved by means of the inhibition of MEG3 on MDM2 (a major inhibitor of p53) (24–26). Our study found that p53 and MDM2 are important mediators through which MEG3 inhibits osteosarcoma.
In the present study, we found low-expression of MEG3 in MG63 cells. Over-expression of MEG3 inhibits proliferation and invasion and induces apoptosis. Further, our experiments showed that p53 was activated upon MEG3 over-expression and that the expression of MDM2 decreased. Based on these experimental data, MEG3 may inhibit the progression of osteosarcoma, and MEG3 has the potential to provide guidance for the diagnosis and treatment of osteosarcoma.
Materials and methods
Cell culture
The human osteosarcoma cell line MG63 and SaOS2 and the human foetal osteoblastic cell line hFOB1.19 were purchased from the Cell Bank of the Shanghai Institutes for Biological Sciences. MG63 cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Gibco, Gaithersburg, MD, USA), SaOS2 cells were cultured in McCoy's medium (Sigma, St. Louis, MO), and hFOB1.19 cells were cultured in Dulbecco's Modified Eagle's Medium: Nutrient Mixture F-12 (DMEM-F12) (Gibco, Gaithersburg, MD, USA). Both media were supplemented with 10% foetal bovine serum (10%FBS) (Gibco, Gaithersburg, MD, USA) and 100 U/ml penicillin/streptomycin (Invitrogen, Shanghai, China). The cells were incubated at 37°C in a humidified chamber supplanted with 5% CO2. Once adherent cells were more than 80% confluence, the cells were harvested with a 0.25% trypsin-0.02% EDTA solution (Gibco, Gaithersburg, MD, USA) and passaged.
Transfection
The MEG3 sequence was synthesized (based on the MEG3 sequence, GenBank™ NR_002766) and then subcloned into the pCDNA3.1 vector (GeneChem, Shanghai, China). Ectopic expression of MEG3 was achieved by transfection of pCDNA-MEG3, and the empty pCDNA vector was used as a control. MG63 cells were transfected with vectors using Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer's protocol. Then the cells were divided into three groups, which were the blank control group (normal) cultured without any handling, the negative control group (NC) transfected with an empty vector, and the lncRNA-MEG3 over-expression group (pCDNA-MEG3).
RNA extraction and RT-qPCR analysis
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The expression data of MEG3 were normalized to the expression of β-actin. The RT-qPCR kits were obtained from GenePharma Corporation (Suzhou, China) and the RT-qPCR reactions were performed according to the manufacturer's protocol. The reverse primers for PCR MEG3 and β-actin were provided by GenePharma Corporation (Suzhou, China), and the specific forward and reverse primers are shown in Table I. PCR was conducted at 95°C for 3 min, followed by 40 cycles of 95°C for 12 sec and 60°C for 40 sec. The specific products were detected and analysed in the Roche LightCycler 480 Detection System (Roche, Switzerland). The expression of MEG3 was calculated using the 2−ΔΔCt method. The experiment was repeated three times.
Table I.
Primer sequences for quantitative polymerase chain reaction.
| Gene | Primer sequences |
|---|---|
| MEG3 | F: 5′-GCTGGGTCGGCTGAAGAAC-3′ |
| R: 5′-CGTGGCTGTGGAGGGATTT-3′ | |
| β-actin | F: 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′ |
| R: 5′-CTGTCACCTTCACCGTTCCAGTTT-3′ |
MEG3, maternally expressed 3; F, forward; R, reverse.
Cell proliferation viability assay
The MTT assay was used to assess cell proliferation ability. MG63 cells were seeded in 96-well plates at 2×103 cells per well after the cells were successfully transfected. Six wells each were used for the NC and pCDNA-MEG3 groups. Cell proliferation was measured from 1 to 5 days. Then, 10 µl of a 5 mg/ml MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (KeyGEN Biotech, Nanjing, China) solution was added to each well, and the plates were incubated at 37°C for 4 h. To dissolve the formazan crystals, 150 µl of dimethylsulfoxide (DMSO) (KeyGEN Biotech, Nanjing, China) was added to each well. The absorbance of each well was measured at a spectral wavelength of 490 nm using a microplate reader (Spectra Thermo, Switzerland).
Flow cytometry analysis
MG63 cells were harvested and washed twice in ice-cold PBS 48 h after being transfected with pCDNA-MEG3 and empty vector. Cell apoptosis was determined by detecting phosphatidylserine exposure on the cellular plasma membranes using the fluorescent dye Annexin V-APC/7AAD Apoptosis Detection Kit (KeyGEN Biotech, Nanjing, China) according to the manufacturer's protocols. Then, approximately 5×105 cells were analysed in each of the samples and immediately analysed by bivariate flow cytometry using a FACScan-LSR equipped with CellQuest software.
Flow cytometry was also used for analysis of the cell cycle in the NC group and pCDNA-MEG3 group. Cells were harvested in the log phase of growth and washed twice with ice-cold PBS. After fixation in 70% ethanol for 24 h at 4°C, the cells were centrifuged and then stained with 50 µg/ml propidium iodide (PI) and 100 µg/ml RNase A (KeyGEN Biotech, Nanjing, China) for 30 min in the dark at 20°C. The samples were analysed with a BD FACSCalibur flow cytometer, and the data were analysed using CellQuest software.
Cell invasion
To detect the invasion ability of MG63 cells a transwell assay was used. In this assay, the number of cells that passed through a polycarbonate membrane (8-µm size) was calculated. The surface of the polycarbonate membrane of the chamber was covered with 20 ml Matrigel (Collaborative Research, Bedford, MA, USA). For the invasion assays, 2×105 cells plated in 200 ml of high-glucose DMEM without FBS were seeded in the upper chamber of the transwell invasion system (Corning Costar, Cambridge, MA, USA), and 500 µl of high-glucose DMEM containing 10% FBS was added to the lower chamber as a chemoattractant. After 24 h of incubation, cells that did not invade though the pores were wiped away with a cotton swab. Then, the cells on the lower surface of the chamber were fixed with 4% paraformaldehyde and stained with 1% crystal violet (KeyGEN Biotech, Nanjing, China). An inverted phase-contrast microscope (Olympus, Japan) was used to capture images and counts.
Western blot analysis
Cells from each group were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer. Then, the protein was collected by centrifugation and quantified. Equal amounts of protein were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Then, the protein blots were transferred to a PVDF membrane (Millipore, Bedford, MA, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline containing Tween (TBST) at room temperature for 2 h and incubated with different primary antibodies in TBST overnight at 4°C. After washing three times with TBST, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (1:2,000, Cell Signalling Technology) at room temperature for 120 min. The detected protein signals were visualized with an electrochemiluminescence (ECL) system Western blot kit (Thermo, Boston, MA, USA). Rabbit monoclonal antibodies against cyclin D1, caspase 3 and B cell lymphoma-2 (Bcl-2) were bought from Abcam (Cambridge, UK). A rabbit polyclonal antibody to matrix and matrix metalloproteinase 9 (MMP9) was bought from Proteintech (Chicago, IL, USA). Rabbit monoclonal antibodies against MDM2 and p53 were bought from Cell Signaling Technology (Danvers, MA, USA). A rabbit polyclonal antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Santa Cruz (Santa Cruz, CA, USA).
Statistical analysis
The experiments were repeated three times. The quantitative data were presented as the mean ± SEM. Statistical Package for the Social Sciences (SPSS) version 13.0 software (IBM, Armonk, NY, USA) was used for the data analysis. All analyses were performed with one-way analysis of variance (ANOVA). Pair-wise comparisons were performed by using t tests. A probability level of 0.05 was used to establish significance.
Results
MEG3 has low expression in an osteosarcoma cell line
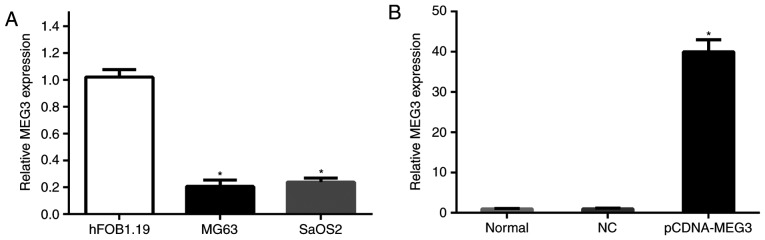
In order to test the expression of MEG3 in osteosarcoma cells, we detected the expression of MEG3 in the MG63, SaOS2 and hFOB1.19 cell lines via RT-qPCR. hFOB1.19 cells were used as a control group (Fig. 1A). The data showed that the expressions of MEG3 in MG63 cells and SaOS2 cells were significantly lower than that in normal hFOB1.19 cells (P<0.05). Therefore, the low expression of MEG3 was liable to have great importance in osteosarcoma cells.
Figure 1.
The relative expression (A) The expressions of MEG3 in MG63 cells and SaOS2 cells were significantly lower than that in normal hFOB1.19 cells (*P<0.05). (B) MG63 cells were transfected with pCDNA-MEG3 for 48 h. There is no significant difference between the NC group and normal group, while the level of MEG3 in the pCDNA-MEG3 group was increased by 40-fold compared with the NC group (*P<0.05). All of the data were presented as the mean ± SEM of three independent experiments.
MEG3 transfection and expression
We transfected MG63 cells with pCDNA-MEG3 to over-express MEG3 in the cells. In this way, we aimed to confirm the function of MEG3 in human osteosarcoma cells. After transfection for 48 h, we used RT-qPCR to detect the expression of MEG3 in the untreated normal group, the NC group transfected with empty vector and the pCDNA-MEG3 transfection group. The results revealed no significant difference between the NC group and normal group, while the level of MEG3 in the pCDNA-MEG3 group was increased by 40-fold compared with the NC group (P<0.05) (Fig. 1B). This result confirmed that the pCDNA-MEG3 vector was successfully transfected.
MEG3 promotes the apoptosis of MG63 cells
In order to detect the effect of MEG3 on apoptosis of MG63, we analysed cell apoptosis by flow cytometry, after 48 h of transfection of MG63 cells. The early apoptosis rate (Q4, showed in Fig. 2A) of the NC group and pCDNA-MEG3 group were 5.61±0.92% and 15.19±0.66%, respectively. The late apoptosis rate (Q2, showed in Fig. 2A) was 4.26±0.52% in the NC group and 11.64±0.79% in the pCDNA-MEG3 group. As shown in Fig. 2B, the total apoptosis rate of the pCDNA-MEG3 group was much higher compared with the NC group (P<0.05). These results indicated that the spontaneous apoptosis of MG63 cells with over-expressing MEG3 was significantly greater than that of the NC cells (P<0.05). Therefore, it was demonstrated that MEG3 could promote the apoptosis of MG63 cells.
Figure 2.
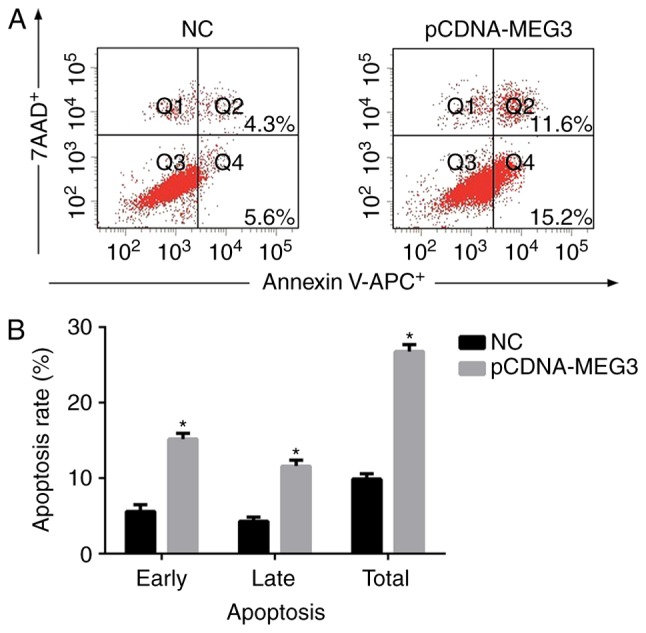
The effect of MEG3 on MG63 cells apoptosis. (A) After transfected MG63 cells for 48 h, cells were collected and stained with Annexin V-APC/7AAD and then examined by flow cytometry. The early apoptosis rate (Q4) of the NC group and pCDNA-MEG3 group were 5.61±0.92% and 15.19±0.66%, respectively. The late apoptosis rate (Q2) was 4.26±0.52% in the NC group and 11.64±0.79% in the pCDNA-MEG3 group. (B) The histogram showed the apoptosis rate obtained from NC and pCDNA-MEG3 group. The total apoptosis rate of the pCDNA-MEG3 group was higher compared with the NC group (*P<0.05).
MEG3 inhibits the proliferation of MG63 cells
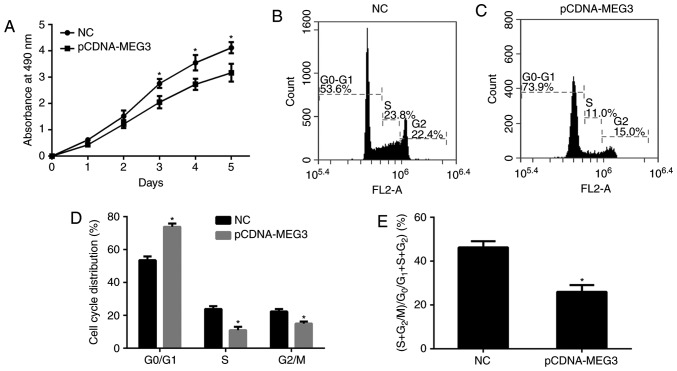
In order to explore the effects of MEG3, first, the MTT assay was conducted to compare the proliferation ability of the cells among all groups by drawing proliferation curves. We found that the proliferation ability of the pCDNA-MEG3 group appeared to be significantly reduced compared with that of the NC group (P<0.05) from the third day after transfection. The data showed that the greatest percentage reduction in proliferation ability was 0.95±0.03% (Fig. 3A). It was shown that MEG3 could inhibit the proliferation of MG63 cells.
Figure 3.
The effect of MEG3 on MG63 cell proliferation. (A) The cell proliferation viability was determined by MTT assay. 490 nm absorption was assayed after culture from 1 to 5 days. The proliferation ability of the pCDNA-MEG3 group appeared to be significantly reduced from the third day after transfection. The greatest percentage reduction in proliferation ability was 0.95±0.03%. The statistical tests were based on the NC group (*P<0.05). (B and C) Cell cycle distribution of different group cells was measured using flow cytometry analysis. The G0/G1 phase constituted 73.92±1.90% of the cells in the pCDNA-MEG3 group and 53.61±2.30% in the NC group. S phase cells accounted for 11.01±2.10% of the cells in the pCDNA-MEG3 group and 23.93±1.70% in the NC group. G2 phase cells accounted for 15.01±1.30% of the cells in the pCDNA-MEG3 group and 22.41±1.50% in the NC group. (D) The histogram showed the cells of G0/G1 phase more frequent in the pCDNA-MEG3 group were than in the NC group, while the cells of the S phase were relatively less frequent (*P<0.05). (E) The histogram showed the proliferation index in different groups. The cell proliferation index ((S+G2)/(G0/G1+S+G2)) had decreased by 10.26±3.12% in the pCDNA-MEG3 group. The statistical tests were based on the NC group (*P<0.05). All of the data were presented as the mean ± SEM of three independent experiments.
In order to study the changes to cells, we analysed the cell cycle distribution by means of flow cytometry. The results showed that the G0/G1 phase constituted 73.92±1.90% of the cells in the pCDNA-MEG3 group and 53.61±2.30% in the NC group. S phase cells accounted for 11.01±2.10% of the cells in the pCDNA-MEG3 group and 23.93±1.70% in the NC group. G2 phase cells accounted for 15.01±1.30% of the cells in the pCDNA-MEG3 group and 22.41±1.50% in the NC group (Fig. 3B, C). By analysing the data, we found that the cells of G0/G1 phase more frequent in the pCDNA-MEG3 group were than in the NC group, while the cells of the S phase were relatively less frequent (P<0.05; Fig. 3D). These results also showed that the cell proliferation index ((S+G2)/(G0/G1+S+G2)) had decreased by 10.26±3.12% in the pCDNA-MEG3 group compared with the NC group (Fig. 3E), indicating that the up-regulation of MEG3 could inhibit the proliferation ability of MG63 cells.
MEG3 inhibits the invasion of MG63 cells
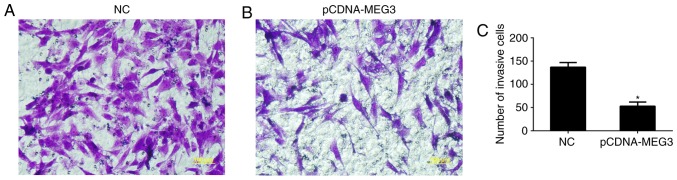
The transwell invasion assay was used to detect the differences in cell invasion ability between the NC group and pCDNA-MEG3 group. By transferring Matrigel from the upper chamber to the lower chamber, the number of cells in the NC group and pCDNA-MEG3 group were 137±10.13 and 53±8.94, respectively (Fig. 4A, B). In this experiment, the number of MG63 cells in the lower chamber was significantly smaller in the pCDNA-MEG3 group than in the NC group (P<0.05) (Fig. 4C). This procedure indicated that the over-expression of MEG3 could inhibit the invasion ability of MG63.
Figure 4.
The effect of MEG3 on MG63 cell invasion. (A and B) Cells that passed through the Matrigelcoated polycarbonate membrane were stained with crystal violet and observed under an inverted phase contrast microscope. The number of cells in the NC group and pCDNA-MEG3 group were 137±10.13 and 53±8.94. (C) The histogram showed the number of MG63 cells passed through Transwell invasion chambers was significantly smaller in the pCDNA-MEG3 group. All statistical tests were based on the NC group (*P<0.05). All of the data were presented as the mean ± SEM of three independent experiments.
MEG3 activates p53 and reduces the expression of MDM2
To further study the mechanism of MEG3 in MG63 cells, we detected the expression of p53 protein in MEG3-transfected MG63 cells via Western blotting analysis, and the expression of p53 in the pCDNA-MEG3 group was higher than in the NC group (P<0.05). This result indicated that over-expression of MEG3 could activate the transcriptional activity of p53. Western blotting analysis also showed that the expression of the oncogene MDM2 was reduced upon the up-regulation of MEG3 (P<0.05) (Fig. 5A, B). These data suggested that p53 and MDM2 may be involved in the regulation of malignancy by MEG3.
Figure 5.
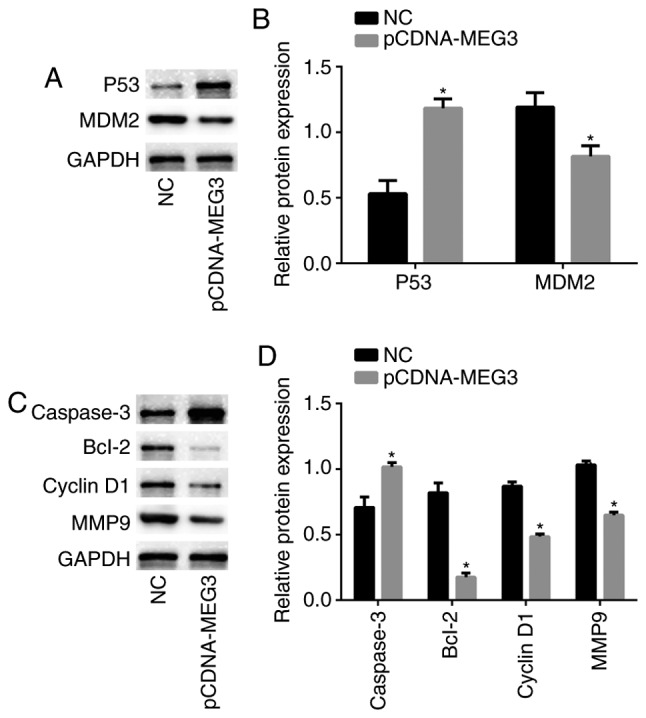
Related proteins expression in MG63 cells. (A and C) Western blotting was performed to detect the expression of p53, MDM2, cyclin D1, caspase 3, Bcl-2 and MMP9 in MG63 cells in different groups, GAPDH was used as a loading control to ensure uniform protein loading in all lanes. (B and D) The histogram showed the relative protein expression obtained for each group. All statistical tests were based on the NC group (*P<0.05). All of the data were presented as the mean ± SEM of three independent experiments.
MEG3 regulates the expression of caspase 3, Bcl-2, cyclin D1, and MMP9 in MG63 cells
As the above experiments had shown, the over-expression of MEG3 could inhibit the proliferation and invasion and induce the apoptosis of MG63 cells. In order to explore the biological mechanism of MEG3 at the molecular level, we used western blotting analysis to detect the expression of proteins in downstream pathways related to proliferation, invasion and apoptosis in MG63 cells. Among the proteins related to apoptosis, the expression of caspase 3 in the pCDNA-MEG3 group was significantly up-regulated, while the expression of Bcl-2 in the pCDNA-MEG3 group was significantly down-regulated. Cyclin D1, a protein that is related to cell cycles, had reduced expression in the pCDNA-MEG3 group; MMP9, a protein related to invasion, also showed lower expression in the pCDNA-MEG3 group (Fig. 5C, D). The above data were compared with the NC group as a control, and it was shown that the caspase 3, Bcl-2, cyclin D1, and MMP9 proteins were involved in the regulation of proliferation, invasion and apoptosis of MG63 cells. And we considered that changes in expression levels of these proteins are regulated by MEG3 through p53 and MDM2.
Discussion
lncRNA was once considered to be a type of nucleic acid without any function. However, increasing numbers of studies related to lncRNA gradually revealed that it plays important roles in biological process, and recently, especially in the occurrence, development and the degree of malignancy of various tumours (27). Some studies revealed that the lncRNA MEG3 has low expression in various tumour cells, and in the above experiments, we discovered that the expression of MEG3 in the MG63 cell line and SaOS2 cell line were lower compared with a normal human foetal osteoblast cell line. This result allowed us to infer that MEG3 might be involved in the progression of osteosarcoma cells and encouraged us to focus on the role of MEG3 in biological processes.
Based on studies of the development of malignant tumour cells, cell proliferation, apoptosis, migration and invasion are mainly involved. Luo et al claimed that MEG3 can inhibit the proliferation of cancer prostate cells and induce its apoptosis (15). Peng et al maintained that the ectopic expression of MEG3 inhibits proliferation, migration and invasion, and promotes cell apoptosis in gastric cancer (16). These studies show that MEG3 has played a role in the regulation of the proliferation, apoptosis, migration and invasion of cancer cells. Thus, we speculate that in osteosarcoma cells, MEG3 can also promote apoptosis and inhibit proliferation, migration and invasion. In order to confirm our speculation, we transfected MG63 cells with vectors and increased the expression level of MEG3 through human intervention. The results showed that the over-expression of MEG3 in the MG63 cell line had the same effect as our expectations.
Studies have indicated that MEG3 usually plays the role of tumour suppressor by activating p53 (17,18). p53 is a well-known tumour suppressor that regulates the expression of many target genes. Kai-hua et al claimed that MEG3 inhibits NSCLC cell proliferation and induces apoptosis by affecting p53 expression (19). Zhu et al indicated that MEG3 interacts with p53 protein and regulates p53 target genes in hepatoma cells (20). We detected the expression of p53 in MG63 cells before and after transfection, and discovered that the expression level of p53 also increased after MEG3 up-regulation.
As we know, MDM2 is a major suppressor of p53 expression (22). The mechanism by which MDM2 suppresses p53 has classically been thought to involve two distinct processes: binding of MDM2 to the N-terminal domain of p53 and thus masking the access of p53 to the transcriptional machinery, and ubiquitination of p53 via MDM2 and thereby targeting p53 for proteasomal degradation (28–30). Our experiments also concluded that the expression of MDM2 in the pCDNA-MEG3 group was significantly lower than that in the control group. We speculate that the activation of p53 by MEG3 might be caused by MEG3-mediated inhibition of MDM2, but further study on the specific mechanism by which MEG3 modulates MDM2 is still needed.
After obtaining the results of the expression of p53 and MDM2, we detected the expression of related proteins in downstream pathways to study the molecular mechanism of tumour inhibition. Due to the regulation of cell apoptosis and proliferation by MEG3, first, we detected the protein expression level of caspase 3, Bcl-2 and cyclin D1 in cells to determine whether they were involved in the process of MEG3-mediated regulation. Caspase 3 is a kind of terminal caspase that is an essential element of the apoptotic pathway. The specific inhibition of the activity of caspase 3 can inhibit apoptosis (31). Bcl-2 is generally considered as an anti-apoptotic gene; it has the effect of inhibiting cell loss and preventing cell apoptosis by blocking the release of cytochrome C and inhibiting caspase 3 activity (32). Cyclin D1 in complex with cdk4 is essential for G1/S phase transition and is a major positive regulator of the critical G1 restriction point in the cell cycle (33). In the experiment shown in Fig. 5, we discovered that up-regulation of MEG3 increased the expression of p53 and decreased the expression of MDM2 while the expression of caspase 3 increased and the expression of Bcl-2 and cyclin D1 decreased. We hypothesized that MEG3 might induce MG63 cell apoptosis by up-regulating caspase 3 and down-regulating Bcl-2 and might inhibit cell proliferation by down-regulating cyclin D1 through p53-dependent pathways. Furthermore, matrix metalloproteinases (MMPs) are a family of highly homologous zinc-dependent endopeptidases that play a key role in tumour invasion and metastasis. MMPs can degrade all kinds of proteins in the extracellular matrix (ECM) and destroy the tissue barriers during tumour cell invasion (34). Chen et al found that MDM2 over-expression induced MMP9 expression in a dose-dependent manner (35). Our experimental data showed that the up-regulation of MEG3 enabled low expression of MMP9, which may be affected by the down-regulation of MDM2.
In conclusion, MEG3 has low expression in osteosarcoma cells, while the up-regulation of MEG3 can induce the apoptosis of MG63 cells and inhibit cell proliferation, invasion and migration. Furthermore, MDM2 and p53 play an important role in this process.
References
- 1.Endo-Munoz L, Cumming A, Sommerville S, Dickinson I, Saunders NA. Osteosarcoma is characterised by reduced expression of markers of osteoclastogenesis and antigen presentation compared with normal bone. Br J Cancer. 2010;103:73–81. doi: 10.1038/sj.bjc.6605723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielack SS, Marina N, Ferrari S, Helman LJ, Smeland S, Whelan JS, Reaman GH. Osteosarcoma: The same old drugs or more? J Clin Oncol. 2008;26:3102–3103. doi: 10.1200/JCO.2008.17.1108. [DOI] [PubMed] [Google Scholar]
- 3.Gill J, Ahluwalia MK, Geller D, Gorlick R. New targets and approaches in osteosarcoma. Pharmacol Ther. 2013;137:89–99. doi: 10.1016/j.pharmthera.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Allison DC, Carney SC, Ahlmann ER, Hendifar A, Chawla S, Fedenko A, Angeles C, Menendez LR. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012:704872. doi: 10.1155/2012/704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perkel JM. Visiting ‘noncodarnia’. Biotechniques. 2013;54:303–304. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 10.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 11.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One. 2012;7:e49462. doi: 10.1371/journal.pone.0049462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribarska T, Goering W, Droop J, Bastian KM, Ingenwerth M, Schulz WA. Deregulation of an imprinted gene network in prostate cancer. Epigenetics. 2014;9:704–717. doi: 10.4161/epi.28006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greife A, Knievel J, Ribarska T, Niegisch G, Schulz WA. Concomitant downregulation of the imprinted genes DLK1 and MEG3 at 14q32.2 by epigenetic mechanisms in urothelial carcinoma. Clin Epigenetics. 2014;6:29. doi: 10.1186/1868-7083-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo G, Wang M, Wu X, Tao D, Xiao X, Wang L, Min F, Zeng F, Jiang G. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell Physiol Biochem. 2015;37:2209–2220. doi: 10.1159/000438577. [DOI] [PubMed] [Google Scholar]
- 16.Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 19.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, Wei L, Jin Y, Fu H, Wu Y, Zheng X. Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PloS One. 2015;10:e0139790. doi: 10.1371/journal.pone.0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 22.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/S0959-437X(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 23.Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p53-dependent gene expression. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu LX, Deng W, Zhou XT, Chen RP, Xiang MQ, Guo YT, Pu ZJ, Li R, Wang GF, Wu LF. The mechanism of adenosine-mediated activation of lncRNA MEG3 and its antitumor effects in human hepatoma cells. Int J Oncol. 2016;48:421–429. doi: 10.3892/ijo.2015.3248. [DOI] [PubMed] [Google Scholar]
- 25.Lv D, Sun R, Yu Q, Zhang X. The long non-coding RNA maternally expressed gene 3 activates p53 and is downregulated in esophageal squamous cell cancer. Tumour Biol. 2016 doi: 10.1007/s13277-016-5426-y. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P, Lu B, Liu G, Wang Z. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PloS One. 2015;10:e0114586. doi: 10.1371/journal.pone.0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst C, Morton CC. Identification and function of long non-coding RNA. Front Cell Neurosci. 2013;7:168. doi: 10.3389/fncel.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 30.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 31.Marchetti P, Castedo M, Susin SA, Zamzami N, Hirsch T, Macho A, Haeffner A, Hirsch F, Geuskens M, Kroemer G. Mitochondrial permeability transition is a central coordinating event of apoptosis. J Exp Med. 1996;184:1155–1160. doi: 10.1084/jem.184.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HJ, Lee EK, Seo YE, Shin YH, Kim HS, Chun YH, Yoon JS, Kim HH, Han MY, Kim CK, et al. Roles of Bcl-2 and caspase-9 and −3 in CD30-induced human eosinophil apoptosis. J Microbiol Immunol Infect. 2017;50:145–152. doi: 10.1016/j.jmii.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 34.Swarnakar S, Paul S, Singh LP, Reiter RJ. Matrix metalloproteinases in health and disease: Regulation by melatonin. J Pineal Res. 2011;50:8–20. doi: 10.1111/j.1600-079X.2010.00812.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Qiu J, Yang D, Lu J, Yan C, Zha X, Yin Y. MDM2 promotes invasion and metastasis in invasive ductal breast carcinoma by inducing matrix metalloproteinase-9. PloS One. 2013;8:e78794. doi: 10.1371/journal.pone.0078794. [DOI] [PMC free article] [PubMed] [Google Scholar]