Significance
Translation termination is a crucial process during protein synthesis. Class I release factors (RFs) are in charge of recognizing stop codons and consequently hydrolyzing the peptidyl-tRNA at the ribosomal P site. High-resolution crystal- and cryo-EM structures of RFs bound to the ribosome revealed a network of potential interactions that is formed between the mRNA and RFs; however, it remained enigmatic which interactions are critical for accurate stop codon recognition and peptide release. By using chemically modified stop codon nucleotides, the importance and the contribution of single hydrogen bonds to stop codon recognition was investigated. This approach revealed a detailed picture of chemical groups defining a stop codon and contributing to the discrimination against sense codons during prokaryotic and eukaryotic translation termination.
Keywords: ribosome, translation, peptide release, release factor, mRNA modification
Abstract
Termination of protein synthesis is triggered by the recognition of a stop codon at the ribosomal A site and is mediated by class I release factors (RFs). Whereas in bacteria, RF1 and RF2 promote termination at UAA/UAG and UAA/UGA stop codons, respectively, eukaryotes only depend on one RF (eRF1) to initiate peptide release at all three stop codons. Based on several structural as well as biochemical studies, interactions between mRNA, tRNA, and rRNA have been proposed to be required for stop codon recognition. In this study, the influence of these interactions was investigated by using chemically modified stop codons. Single functional groups within stop codon nucleotides were substituted to weaken or completely eliminate specific interactions between the respective mRNA and RFs. Our findings provide detailed insight into the recognition mode of bacterial and eukaryotic RFs, thereby revealing the chemical groups of nucleotides that define the identity of stop codons and provide the means to discriminate against noncognate stop codons or UGG sense codons.
Protein synthesis is a crucial process in every living cell. The ribosome is a large ribonucleoprotein particle that is in charge of precisely orchestrating this immensely complex process. Whereas translation initiation and elongation depend on accurate mRNA/tRNA interactions (1, 2), translation termination depends on the specific and reliable recognition of stop codons through class I release factor proteins (RFs) (3, 4). Class I RFs bind to the ribosome upon the presence of UAA, UAG, or UGA codons at the ribosomal A site. Bacteria rely on two class I RFs—namely, RF1 and RF2—to terminate at all three stop codons. Whereas UAA is decoded by both RFs, UAG and UGA are exclusively recognized by RF1 and RF2, respectively (5). In archaea and eukaryotes, a single class I RF (aRF1 and eRF1, respectively) is sufficient for providing peptide release at all three stop codons (6). Upon binding of a class I RF to the ribosome, hydrolysis of the peptidyl-tRNA is triggered, and the nascent peptide is released (7–10). Subsequently, in bacteria a class II RF (RF3) binds to the posttermination complex and stimulates ribosome recycling (11–13). In contrast, eukaryotic eRF3 facilitates effective binding of eRF1 to the ribosome, and the ATP-binding cassette protein E1 triggers recycling (14–17).
Before the availability of high-resolution structures of termination complexes, it remained enigmatic how class I RFs interacted with stop codons at the ribosomal A site. Various biochemical and genetic studies in bacterial and eukaryotic translation systems were carried out to determine the protein domains and the corresponding amino acids that are crucial for stop codon recognition (reviewed in refs. 3 and 18).
In bacteria, the conserved motifs PxT and SPF of RF1 and RF2, respectively, were proposed to serve as “tripeptide anticodons” for the accurate decoding of stop codons (19). However, mutational studies also identified other amino acids that were involved in stop codon recognition, implicating that the tripeptide anticodon alone is not sufficient for providing translation termination (20). Remarkably, mutations altering the codon specificity of RFs were only identified outside of the potential codon recognition site (20–22).
In eukaryotes, eRF1 most likely evolved independently of the bacterial RFs, as suggested by the absence of sequence or structural similarities (23). However, one tripeptide motif—i.e., the glycine–glycine–glutamine (GGQ) motif—is universally conserved and might be a result of convergent evolution. The GGQ motif docks into the peptidyl-transferase center and thereby promotes peptide release (9, 10, 24). While this step during termination is identical in all organisms, stop codon recognition is achieved by specific interactions between RFs and mRNAs. Through mutational studies (25), cross-linking experiments (26, 27), and biochemical studies (28), highly conserved amino acid sequences, such as the TASNIKS, YxCxxxF, and GTS motifs, have been identified to be essential for stop codon recognition in eukaryotes. In accordance with bacterial RFs, residues outside of these motifs also significantly contributes to RF-mediated peptide release (27, 29–31). Together, these results indicated that stop codon recognition involves a network of interactions between the mRNA, rRNA, and RFs. However, the crucial interactions and, consequently, the structural prerequisites for stop codon recognition remained largely unresolved.
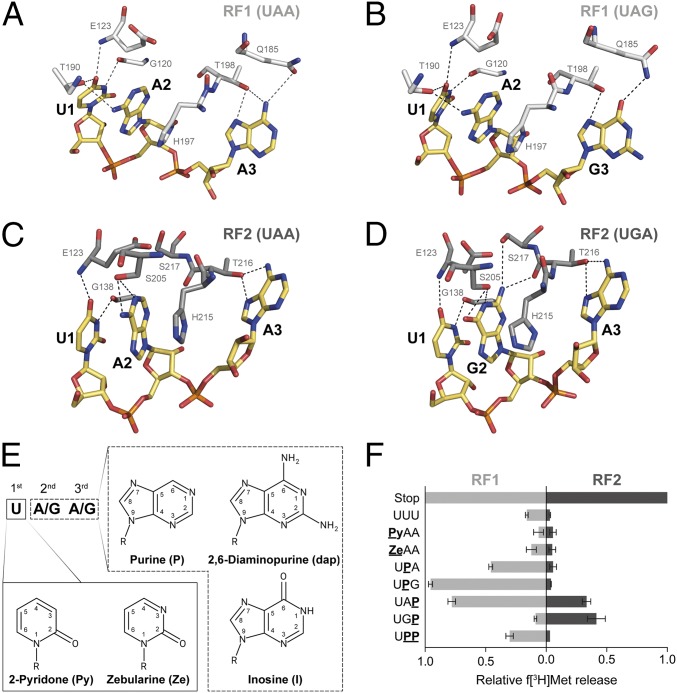
A significant step toward a better understanding of peptide release was achieved by the availability of high-resolution structures of bacterial and mammalian RFs bound to ribosomes (32–37). These structures provided intricate details of the potential hydrogen and stacking interactions that allow the efficient determination of a stop codon presented in the ribosomal A site by RFs (see Figs. 1 A–D and 4A). The combination of structural data with genetic and biochemical investigations enabled a detailed model of how stop codon recognition by RFs could be achieved. However, structural and standard mutational studies cannot always provide a complete picture of a highly complex process. High-resolution structures contribute invaluable insights into the molecular mechanisms of different biological processes; however, the importance of single interactions is mainly postulated based on the distances and positions of potential interaction partners and possibly provide only a snapshot of an intricate process.
Fig. 1.
(A–D) Interactions between residues of bacterial RFs and the three stop codons at the decoding site. RF1 (white) and RF2 (gray) sense the stop codons (yellow) UAA (A) and UAG (B) or UAA (C) and UGA (D), respectively, by forming a set of hydrogen bonds (dashed lines; Escherichia coli numbering is used for RF residues; modified from refs. 32–35). (E) These interactions were modulated by the use of modified RNA bases within the stop codons. (F) The ability of RFs to recognize modified stop codons was determined by using a peptide release assay (error bars indicated SDs from the mean of three independent experiments).
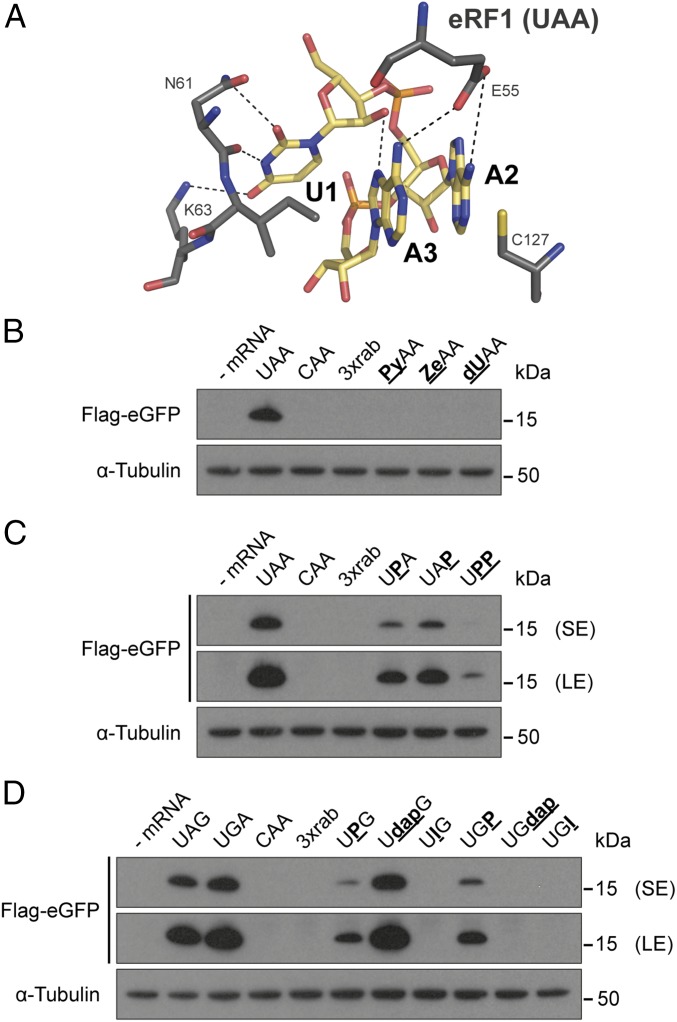
Fig. 4.
Eukaryotic termination at modified stop codons. (A) Potential hydrogen-bond interactions (dashed lines) between eRF1 (dark gray) and the UAA stop codon (yellow) in a U-turn-like confirmation (37). (B–D) Modified stop codons were incorporated in Flag-eGFP reporter mRNAs and were transfected into HEK293T cells. Twenty-four hours after transfection, the Flag-tagged translation products were detected by Western blotting. LE, long exposure; SE, short exposure.
Mutational studies are also indispensable for revealing crucial amino acids and protein domains for ligand binding or catalysis. Nevertheless, mutations might also disclose amino acids that are located in regions of proteins that only indirectly affect their function (20, 21, 31, 38). Furthermore, amino acid substitutions may not only disturb the direct interaction with a substrate, but also may potentially alter the overall structure of a protein domain, thereby indirectly influencing the association between interacting partners (39).
Here, we report a complementary approach that enables the direct chemical modification of stop codons and, thus, can contribute to a detailed understanding of stop codon recognition by RFs. By using atomic mutagenesis of stop codons, single interactions between the mRNA and the respective RF can be modified without significantly disturbing the overall geometry of the interaction (40). Various modified RNA derivatives (Fig. 1E) were introduced at all three positions of stop codons, and the ability of RFs to bind and provide termination activity was determined. This permitted us to single out crucial interactions strictly required for stop codon recognition.
Results
Minimal Requirements for Stop Codon Recognition in Bacteria.
To determine the termination activity at modified codons, the hydrolysis of radiolabeled f[3H]Met-tRNAfMet promoted by bacterial RFs was quantified (8, 41). The mRNAs used carried a Shine–Dalgarno sequence and an AUG start codon 5′ to the stop codon harboring the respective modification (41). After binding the mRNA and the f[3H]Met-tRNAfMet to the 70S ribosome, either RF1 or RF2 was added, and the amount of f[3H]Met released was determined by scintillation counting.
Initially, modifications at the first nucleotide were introduced by substituting U1 with pyridone (Py) or zebularine (Ze) (Fig. 1E). U1 has been reported to be the most strictly monitored nucleotide during translation termination and therefore was of special interest (42). Thereby, the hydrogen-bond network with the RFs sensing the first stop codon nucleotide—i.e., interactions via G120, E123, and T190 of RF1 and E123 and G138 of RF2—were disrupted (Fig. 1 A–D). Neither Py nor Ze within the stop codon was recognized by RF1 or RF2 (Fig. 1F).
Subsequently, the second and third nucleotides of the stop codon were altered and investigated for their ability to promote termination. By introducing purine (P) into the stop codon, all exocyclic groups and their respective interactions were eliminated (Fig. 1E). With respect to RF1, this modification should eliminate hydrogen bonds between A2 and T190 or A3/G3 and Q185, depending on the position of P within the stop codon (43). However, the codons UPA or UPG still triggered release by RF1. Whereas the amount of f[3H]Met released was reduced to ∼50% at UPA, UPG almost reached wild-type (WT) levels (Fig. 1F). In addition, the loss of the exocyclic group at the third codon position of a UAP codon also allowed efficient release by RF1. Strikingly, the simultaneous presence of purines at the second and third codon positions (UPP) still provided detectable amounts of released f[3H]Met (Fig. 1F).
RF2 did not show this flexibility in terms of stop codon recognition. RF2 mainly interacted with the second and third nucleotides of UAA and UGA through S205 and T216 (Fig. 1 C and D). Eliminating the interaction of the carbonyl or amino group at C6 of A2/G2 with S205 strongly inhibited the release activity. However, UAP and UGP facilitated termination by RF2, but the levels of f[3H]Met released were reduced more than twofold (Fig. 1F).
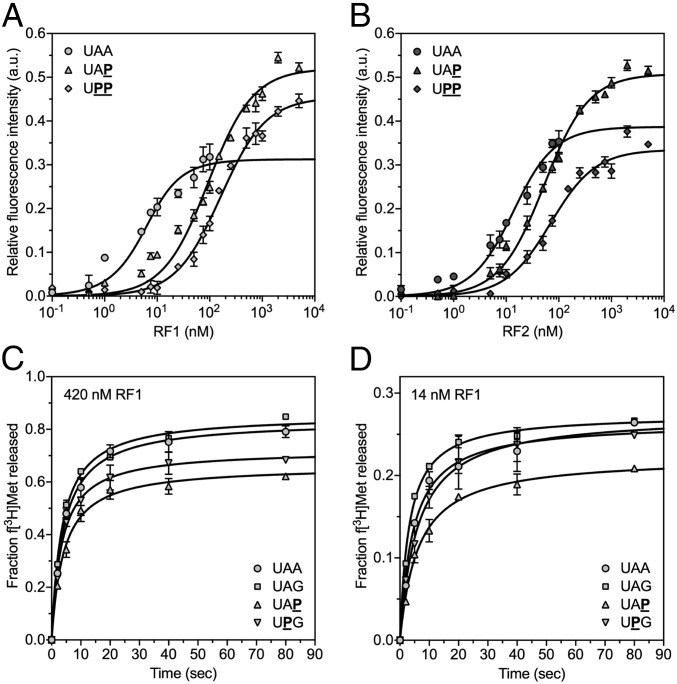
Previous studies using mutated versions of RF1 showed that substituting specific amino acids directly involved in the hydrogen-bond network significantly decreased the binding of RF1 to the ribosome but did not interfere with the rate of peptide release (41). Therefore, we investigated RF1 and RF2 for their ability to bind to the ribosome programmed with unmodified or modified stop codons in the A site using a fluorescence assay (Fig. 2 A and B) (41, 44). We estimated the KD of RF1 binding to the ribosome with an UAA stop codon in the A site to be <3 nM and the KD of RF2 under the above conditions to be 11.1 ± 2.1 nM. Binding UAP or UPP codons to the ribosome increased RF1 KD values compared with UAA >30- or 50-fold, respectively (UAP, KD = 91.3 ± 6.6 nM, and UPP, KD = 158.9 ± 13.9 nM; Fig. 2A and Table S1). RF2 showed a >4.5-fold increase in its KD for UAP and a sixfold increase for UPP (UAP, KD = 50.2 ± 3.0 nM, and UPP, KD = 67.5 ± 7.6 nM; Fig. 2B and Table S1). Thus, these findings indicated reduced binding of RF1 and RF2 to modified stop codon nucleotides. However, the activities of RF1 and RF2 in the peptide release assay did not reflect the KD differences between the RFs in the equilibrium binding experiments well.
Fig. 2.
The impact of modified stop codons in the 70S A site on binding and releases rates of RFs. (A) Increasing amounts of RF1 were added to ribosomes programmed with UAA (●), UAP (▲), or UPP (♦) stop codons in the A site. The relative changes in fluorescence activity were fit to the equilibrium KD equation (black line; error bars show the SDs from the mean in three independent experiments). (B) In analogy to RF1, RF2 binding kinetics to UAA (●), UAP (▲), or UPP (♦) were analyzed. The total amount of the supplemented RFs is indicated on the x axis. (C and D) Rates of RF1-mediated f[3H]Met release from 70S complexes formed with UAA (●), UAG (■), UAP (▲), or UPG (▼) were assayed under saturating (420 nM RF1) (C) and limiting (14 nM RF1) (D) amounts of RF1 (error bars depict the SDs from the mean in two independent experiments).
To analyze whether, besides binding, the catalysis by RFs is also affected, we determined the rate constants of RF-mediated peptide release for modified stop codons with endpoints similar to unmodified stop codons (Table S1). f[3H]Met release was triggered by the addition of saturating amounts of RF1 (Fig. 2C; 420 nM RF1). The concentration was approximately threefold higher than the highest observed KD—i.e., the KD of RF1 for UPP. Time courses were then repeated with limiting amounts of RF1 (Fig. 2D; 14 nM RF1), at a concentration approximately sevenfold below the lowest KD of RF1 for a modified stop codon—i.e., UAP. Under both conditions, RF1 rate constants were nearly identical for 70S complexes formed with UAA [420 nM RF1: kcat = (6.0 ± 0.1) × 10−2 s−1; 14 nM RF1: kcat = (1.9 ± 0.1) × 10−2 s−1] and UAG [420 nM RF1: kcat = (6.1 ± 0.1) × 10−2 s−1; 14 nM RF1: kcat = 2.0 × 10−2 s−1], which is in line with earlier reports (22, 44–46). The rates of release on UAP [420 nM RF1: kcat = (4.7 ± 0.1) × 10−2 s−1; 14 nM RF1: kcat = 1.6 × 10−2 s−1] and UPG codons [420 nM RF1: kcat = (5.2 ± 0.1) × 10−2 s−1; 14 nM RF1: kcat = (2.0 ± 0.1) × 10−2 s−1] were similar to UAA and UAG, respectively. These results suggest that modified stop codons do not compromise the catalytic activity of RFs, but interfere with binding of RFs to the ribosome.
Bacterial RF1 and RF2 Recognize Distinct Sets of Modified Stop Codons.
To further analyze the mechanism of stop codon recognition, we moved the experimental setup into a more biologically authentic setting. Instead of using a minimal P-site substrate to investigate the termination efficiency, an in vitro translation (IVT) assay, dependent on a bona fide mRNA, was used. This translation assay was based on a recombinant bacterial translation system, in which RFs can be added separately (47).
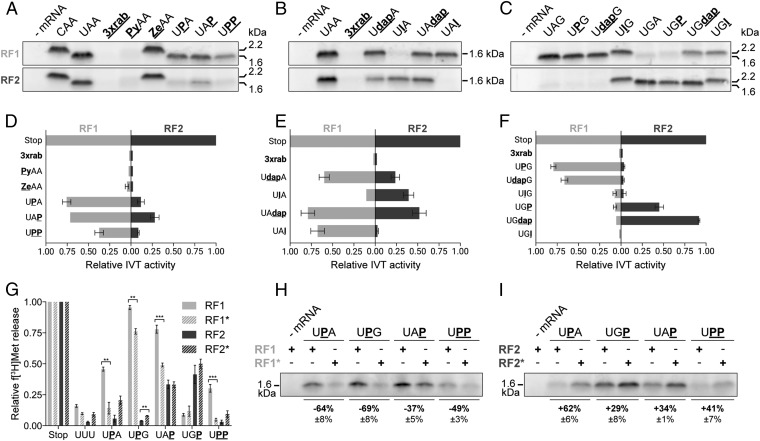
To introduce modified stop codons into an mRNA, a suitable reporter ErmCL mRNA was designed (48, 49). This mRNA harbored two stop codons: a chemically modified stop codon at codon 15 and an unmodified termination signal (UAA) at codon 20. The modification of interest was introduced through splinted ligation of a chemically synthesized oligonucleotide, carrying the modification to the unmodified 5′ fragment of the mRNA (48). A successful termination event at the modified stop codon at codon 15 resulted in a ∼1.6-kDa-sized peptide, whereas a read-through event generated a peptide with a molecular mass of ∼2.2 kDa. Therefore, translation products could be resolved by standard Tricine-SDS/PAGE (Fig. 3) (50). When the modified stop codon was neither recognized by RFs nor tRNAs, only barely detectable amounts of peptides were formed due to the stable stalling of ribosomes at the modified codon [a triple ribose abasic codon (3xrab) was used as a control to assess the background resulting from ribosome stalling; Fig. 3] (51).
Fig. 3.
Termination activities of bacterial RFs at modified stop codons using IVT assays. (A–C) The efficiency of termination at the modified stop codons was deduced from the yield of a 1.6-kDa-sized ErmCL peptide determined by Tricine-SDS/PAGE. Read-through of the modified stop codon resulted in termination at a second unmodified UAA stop codon reflected by a 2.2-kDa translation product (A and C). (D–F) Translation products shown in A–C were quantified (error bars show SDs from the mean of three independent experiments). (G–I) The release activities of RF mutants—i.e., RF1* and RF2*—were tested in the peptide release assay (G) and in the IVT-based assay (H and I). In G, significant differences between RF1, RF2 and RF1*, RF2*, respectively, were determined by using the two-tailed unpaired Student’s t test. ***P < 0.001; **P < 0.01.
In accordance with the minimal peptide release assay, neither the substitution of Py nor Ze for nucleotide U1 provided termination activity through RF1 or RF2, respectively. Whereas Py led to stalling at the modified codon, Ze caused read-through and resulted in a 2.2-kDa peptide product (Fig. 3 A and D). At the second and third positions, RF1 triggered peptidyl-tRNA hydrolysis in the absence of any exocyclic group, independent of the type of stop codon. As also observed in the minimal termination assay, UPP was suitable to be recognized by RF1, providing ∼40% termination activity compared with an unmodified UAA codon (Fig. 3 A and D). In contrast, the presence of P was only tolerated by RF2 at the third codon position, but it reduced the peptide yield more than threefold, thus confirming the importance of hydrogen bonding between RF2 and the exocyclic groups of the second and third nucleotides (Fig. 3 A and D).
Previously, RF variants that exhibited altered stop codon recognition fidelities have been reported. In the case of RF1, a deletion in domain 4 close to the switch loop region (Δ302–304) caused higher accuracy at near-stop codons (RF1ha, designated as RF1* throughout this work) (22). However, the deleted amino acids were not directly interacting with the mRNA and therefore only indirectly influenced stop codon recognition. A set of purine-modified stop codons was analyzed by using RF1*, revealing a higher sensitivity toward base derivatives (Fig. 3 G and H and Fig. S1 A and B). It should be noted that this increased sensitivity was not reflected by the decreased binding of RF1* to modified stop codons (UAA, KD = 11.4 ± 2.1 nM; UAP, KD = 47.7 ± 4.1 nM; and UPP, KD = 68.9 ± 7.5 nM; Fig. S2 and Table S1). In accordance with the determined KD values of the WT RFs, these results point to an additional impact of the modifications on the release activity—e.g., by impeding the closed to open conformational change of the RFs (41).
A mutated version of RF2 has been reported (E167K, designated as RF2* throughout this work) that lost its specificity and provided release activity at all three canonical stop codons in the f[3H]Met release assay (Fig. S1A) (20). In accordance with this observation, RF2* also showed an increased tolerance toward P within stop codons (Fig. 3 G and I). However, in the IVT assay, the release activity of RF2* at UAG stop codons could not be reproduced (Fig. S1C).
Discrimination of RF1 and RF2 Against Noncognate Stop Codons and Sense Codons.
To further characterize stop codon recognition, we investigated how RF1 and RF2 would discriminate either against UGA and UAG, respectively, or against UGG (tryptophan; Trp) sense codons (42, 44). Earlier works proposed that RFs use the 2-amino group of a G nucleotide to sense the presence of a G at this position (19, 52). This conclusion was based on a study using the peptide release assay at stop codons carrying inosine (I) at the second and third positions (Fig. 1E) (19). Ito et al. observed that RF1 efficiently catalyzed termination at UIA codons, suggesting that the presence of a carbonyl oxygen at C6 does not signal the presence of G (19). Consequently, the 2-amino group was postulated as the main discriminator. However, computational or structural studies did not support this model (43, 53). To further clarify the role of the 2-amino group, we introduced this group through the incorporation of 2,6-diaminopurine (dap) at the UAA codon (Fig. 1E). Indeed, the presence of this amino group at the second nucleotide strongly reduced f[3H]Met release by RF1 (Fig. S3A).
Strikingly, RF2 was strongly inhibited by the presence of a 2-amino group at the second codon nucleotide (Fig. S3A). However, at the third nucleotide, the 2-amino group did not interfere with RF2-mediated termination, contradicting a general discriminatory role. It should be noted that UAI did not result in detectable quantities of released f[3H]Met (Fig. S3A) (19).
Due to these conflicting results, I- and dap-harboring stop codons were also tested in the IVT-based assay. In contrast to the minimal release assay, RF1 showed termination activity even in the presence of the 2-amino group using dap at the second codon position, whereas inosine did not provide termination activity (Fig. 3 B and E). Strikingly, an inhibitory effect of the 2-amino group was only observed in absence of the amino group at position 6 (Fig. S3B). At the third codon position, both exocyclic groups did not interfere with termination (Fig. 3 B and E).
RF2 only poorly recognized UdapA and UIA codons, consistent with the finding that RF2 was generally more sensitive to modulations of the stop codon (Fig. 3 B and E). At the UAI codon, peptide release could not be detected, whereas UAdap stimulated RF2-mediated release, indicating a strong discriminatory role of the carbonyl group for RF2 (Fig. 3 B and E).
In addition to characterizing recognition of UAG and UGA by RF1 and RF2, respectively, the discrimination against the UGG sense codon was investigated. Therefore, the UGG codon was modified to harbor P, dap, or I at the second or third codon positions and was introduced into the ErmCL mRNA context. In the IVT assay, all possible competing ternary complexes were present, potentially interfering with the termination reaction. By substituting the G at the second position with a P or dap, RF1 still provided termination activity, whereas UIG led to read-through, causing the incorporation of Trp as confirmed by mass spectrometry (MS) (Fig. 3 C and F and Fig. S4A). Strikingly, read-through was also observed at UGdap and UGP, which resulted in the incorporation of Trp at position 15 (Fig. S4B). By increasing the concentration of tRNAs, thereby leading to a stronger competition with RFs, we were not able to stimulate read-through of canonical or modified stop codons (Fig. S5).
Eukaryotic Release Factor Activity Depends on an Elaborate Hydrogen-Bond Network.
Due to the independent evolution of prokaryotic and eukaryotic RFs and an altered stop codon recognition motif, eRF1 was also investigated. In 2015, high-resolution structures were published, suggesting specific interactions between eRF1 and the stop codon (Fig. 4A) (36, 37). These studies revealed a U-turn-like confirmation of the stop codon and several hydrogen bonds between mRNA and eRF1 to be crucial for stop codon recognition. Based on this structural information, we incorporated modified nucleotides into stop codons to eliminate several of the proposed interactions. Therefore, we adapted a truncated Flag-eGFP mRNA by ligating a synthetic RNA oligonucleotide encoding a modified stop codon to its 3′ end (Fig. S6).
The modified mRNAs were transfected into HEK293T cells, and the translation products were subsequently analyzed by Western blotting. In the case of termination at a modified stop codon, a 9-kDa peptide product was expected. Stalling at the stop codon and read-through events led to the absence of a detectable product due to subsequent degradation (Fig. 4 B–D) (54–56).
As in the prokaryotic termination system, Ze and Py at the first stop codon nucleotide, which disrupts the interactions with N61 and K63, inhibited termination by eRF1 (Fig. 4B). Due to the possible involvement of the 2′-OH to stabilize the U-turn motif, a deoxy-UAA–containing mRNA (dUAA) was investigated (36). Indeed, dUAA was not able to induce termination, underlining the importance of the interaction of the 2′-OH of U1 with N7 of A3 (Fig. 4B).
In contrast, at the second and third nucleotides, purine was recognized by eRF1. Additionally, UPP was able to trigger termination to a reduced extent (Fig. 4C), although UPP cannot form the interactions via the 2-amino groups of A2 and A3 with E55 (Fig. 4A). Inosine, in the context of UIG or UGI codons, did not result in a defined peptide product, most likely causing stop codon read-through leading to the incorporation of Trp at the modified codon (Fig. 4D). At the UdapG codon, eRF1 triggered termination, whereas at a UGdap codon, no termination activity was observed. By using an IVT assay based on HeLa extracts, we were able to differentiate between stalling and read-through events (Fig. S7). Thus, read-through was observed at ZeAA, UIG, UGI, and, strikingly, also at UGdap codons (Fig. S7).
Discussion
Efficient and accurate stop codon recognition by RFs involves an elaborate network of interactions between stop codon nucleotides and specific residues within RFs. By genetic and structural studies, a number of residues have been identified that are crucial for the binding of RFs to stop codons and subsequently for triggering peptidyl-tRNA hydrolysis (7, 19, 20, 32–35, 57).
In this study, rather than using mutant versions of RFs, we aimed to characterize the relevance of the interactions between RF residues and various stop codons by atomic mutagenesis of the mRNA. The precise site- and position-specific substitution of chemical groups thereby can reduce or weaken specific proposed interactions (40, 58–60).
Using this approach, we were able to investigate and identify the contribution of single interactions between bacterial and eukaryotic RFs and stop codons to enable translation termination (Fig. 5). As has been proposed, the U1 position of the stop codon was stringently recognized by hydrogen bonding with conserved RF1/RF2 residues (44, 53). Indeed, all modifications that were introduced at the U1 position strongly inhibited translation termination, consistent with the importance of these interactions. However, at the second and third codon nucleotides, RF1 and RF2 showed distinct differences in stop codon recognition. Therefore, RF1 does not strictly depend on the presence of exocyclic groups at the second or third codon positions (Fig. 5). Even the combination of two purines (UPP) still provided detectable RF1-mediated termination activity (Figs. 1 and 3).
Fig. 5.
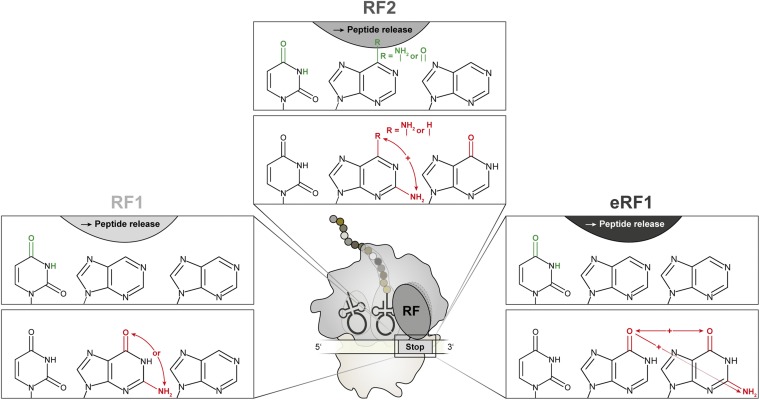
Summary of the chemical requirements of stop codon nucleotides for recognition by prokaryotic RF1 (Left), RF2 (Center), and eukaryotic eRF1 (Right). The pyrimidine–purine–purine matrix of all stop codons is schematically depicted (black). Exocyclic groups that were identified to be crucial for stop codon recognition and peptide release by RFs are illustrated in green (Upper). Chemical groups that were discriminated by the respective RF are shown in red (Lower). + indicates inhibitory effects mediated by a combination of distinct exocyclic groups of the stop codon.
From structural studies, T190 was reported to be within hydrogen-bonding distance of the amino group of A2, and it was postulated that the source of discrimination against G2 was the inability to form an interaction with the carbonyl oxygen of G (35). In light of our results, this seems less likely, as the absence of this interaction still provided almost WT-like RF1 activity (Fig. 3 A and D). Thus, it is possible that the presence of a carbonyl oxygen at G2 leads to a repulsion with T190, thereby providing the means for discrimination, as postulated by MD simulations (Fig. 5) (53). At the third codon position, the hydrogen bond between the N7 of purine and T198 seemed sufficient to position the base in a productive conformation.
In contrast to the rather flexible codon recognition of RF1, RF2 strongly depended on the presence of either an amino or carbonyl group at the second codon nucleotide (Fig. 5). The interaction with S205, which is part of the SPF tripeptide anticodon motif, still can form a hydrogen bond with the N1 of the purine at position 2, but this seemed insufficient to provide termination (Figs. 1 and 3). At the third codon position, the absence of the 6-amino group strongly reduced the ability for peptidyl-tRNA hydrolysis, but residual activity could still be detected (Fig. 3A), suggesting that the remains of the bifurcated interactions with T215 can partly compensate for the loss of the exocyclic amino group.
As a consequence, the ability to discriminate against noncognate stop codons or sense codons does not seem to only rely on the presence of defined chemical interactions, but might rather depend on the exclusion of unsuitable residues. Whereas purine at the second codon nucleotide provided RF1-mediated release, the presence of a carbonyl oxygen at the second nucleotide (UIA) abolished termination (Fig. 3 B and E). This contradicted earlier observations reporting termination activity at UIA and the consequential postulation that the 2-amino group is pivotal for perceiving G’s (19). Whereas the minimal peptide release assay indeed pointed toward an exclusive discriminatory role of the amino group in the case of RF1 (Fig. S3A), the more authentic IVT-based assay excluded this possibility (Fig. 3 C and F). Dap did not inhibit termination at the second or third codon position (Fig. 3 E and F). It is noteworthy that the potential inhibitory role of the 2-amino group was overruled by the stimulatory role of the 6-amino group and could only be observed in the case of 2-aminopurine, and not in the case of 2,6-daminopurine (Fig. S3B).
The impact of the 2-amino group present at the second or third position of the stop codon on RF2-mediated release was found to be less ambiguous. Even at the second codon position, this group was inhibitory to recognition by RFs, although RF2 terminates at UGA, which is consistent with the general low tolerance of RF2 toward modifications. At the third codon position, the 2-amino group promoted termination, supporting the hypothesis that the 2-amino group does not indicate the presence of a G. Inosine, in contrast to earlier observations (19), did not promote termination through RF2 when positioned at the third nucleotide (Fig. 3). As proposed by MD simulations, an unfavorable hydrophobic contact of the carbonyl oxygen with V190 potentially contributes to this discrimination (53).
In the case of discriminating against a Trp sense codon (UGG), the carbonyl oxygen also has an important role. At UIG and UGI codons, only read-through events were observed (Fig. 3 C and F). In contrast, UdapG and UGdap were efficiently recognized by RF1 and RF2, respectively. Strikingly, RF2 terminated at UGdap, whereas in the presence of only RF1, read-through was observed. Consequently, the absence of RF2 caused UGdap to be interpreted as a sense codon by the translation machinery.
Because of the remarkably different requirements of bacterial RF1 and RF2, it was especially appealing to investigate eRF1 due to its separate evolution and highly different structure (23). One major difference to bacteria is the geometry of the stop codon during eRF1-mediated termination. In eukaryotes, the stop codon forms a U-turn motif in contrast to an almost linear arrangement during bacterial RF binding (36, 37). This U-turn motif is stabilized by hydrogen bonds formed between the 2′-OH of U1 and the N7 of A3 (Fig. 4A). Indeed, the incorporation of dU at the stop codon inhibited stop codon recognition by eRF1 (Fig. 4B). In a bacterial system, the introduction of deoxy-nucleotides into stop codons did not inhibit termination, reflecting the different codon geometry (10). Also, additional modifications at the U1 position led to either inhibition of termination (PyAA) or read-through events (ZeAA). This is consistent with a model in which the interaction of U1 with the NIKS motif is crucial for the specific selection of stop codons.
For the recognition of purines at stop codons, eRF1 showed, similarly to bacterial RF1, a rather high flexibility toward the absence of single hydrogen bonds at the second or third position (Fig. 5). The presence of P in stop codons still provided eRF1-mediated termination, even at UPP (Fig. 4 C and D). Additionally, in the case of eRF1, it was the presence of a carbonyl oxygen rather than the absence of hydrogen bonds that set the basis for recognizing stop codons and discriminating against sense codons (Fig. 5) (37).
Even though we could identify the chemical groups of the stop codons that allowed discrimination, it still remains enigmatic which RF domains are responsible for the specificity, even more so since some eukaryotic organisms reprogrammed their genetic code. In these eukaryotes, stop codons became sense codons, and, consequently, the specificity of RFs had to be adjusted as well (61–63). It is still unclear how such a reprogramming is executed because no distinct amino acid exchanges or motifs have been described that would cause altered stop codon specificity (63). Strikingly, amino acids that form direct interactions with the stop codons were not yet identified to contribute to the specificity of class I RFs. A study selecting for omnipotent bacterial RFs revealed E167K of RF2 to generate this extended function (28). Interestingly, this residue also does not directly contact the stop codon and only indirectly causes this effect. Additionally, purine modifications within stop codons were more efficiently recognized and bound by RF2* (Fig. 3 G and I). On the other hand, RF1*, carrying a deletion at domain 4 and again not in direct contact with the stop codon, exhibited a higher accuracy toward the discrimination of modifications (22). This indicated that residues of RFs that form direct contacts with stop codons are only the first crucial instance of a complicated stop codon recognition process orchestrated by a network of interactions, including various amino acid residues and protein domains.
By using modified stop codon nucleotides in various translation systems, we have identified the minimal crucial hydrogen-bond network between stop codons and RFs of prokaryotes and eukaryotes. Whereas various interactions are not essential for stop codon recognition and their absence did not reduce termination activity, the presence of carbonyl oxygens is vital for discriminating stop codons from sense codons. Thus, this study enables a better understanding of the tightly regulated and complex mechanisms of stop codon recognition (Fig. 5).
Methods
Detailed methods are provided in SI Methods. Peptide release assays (8), RF KD titrations (41, 44, 64), mRNA ligations (48), and IVTs (48) were performed as described.
Note.
During the revision of this manuscript Lind et al. (65) published MD free energy calculations for eRF1 that confirmed the importance of U1 for stop codon recognition. The authors postulated that the high preference for U over C is mainly caused by a repulsive interaction between the amino group of C and Lys63 (65). Whereas this repulsion might be conducive to the discrimination, the hydrogen bonds of U1 with Lsy63 and Asp61 also appear to be fundamental to the recognition process of eRF1 (Fig. 4 A and B).
Supplementary Material
Acknowledgments
We thank Norbert Polacek for valuable comments on the manuscript, suggestions, and help; Martina Hölzl for technical assistance; Andrei Korostelev for providing RF1*; the Biomolecular & Proteomics Mass Spectrometry Facility at the University of California at San Diego, especially Majid Ghassemian and Orazio Barna, for validation of the RF2 Gln252 methylation; Simon Spiegl and Georg Altenbacher for their help with mammalian cell culture and for providing antibodies; and the whole S.J. laboratory for helpful discussions and for providing a stimulating environment. This work was supported by Austrian Science Fund Grants P 22658-B12 and P 28494-BBL (to M.D.E.) and SFB F4411 (to A.H.); and European Molecular Biology Organization Short-Term Fellowship ASTF 553-2016 (to T.P.H.). J.W. was supported by the Swiss National Science Foundation Grant 31003A_166527.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714554115/-/DCSupplemental.
References
- 1.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 2.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura Y, Ito K. tRNA mimicry in translation termination and beyond. Wiley Interdiscip Rev RNA. 2011;2:647–668. doi: 10.1002/wrna.81. [DOI] [PubMed] [Google Scholar]
- 4.Youngman EM, McDonald ME, Green R. Peptide release on the ribosome: Mechanism and implications for translational control. Annu Rev Microbiol. 2008;62:353–373. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 5.Klaholz BP. Molecular recognition and catalysis in translation termination complexes. Trends Biochem Sci. 2011;36:282–292. doi: 10.1016/j.tibs.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci USA. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amort M, et al. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 2007;35:5130–5140. doi: 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos N, Zhu J, Donohue JP, Korostelev AA, Noller HF. Crystal structure of the 70S ribosome bound with the Q253P mutant form of release factor RF2. Structure. 2013;21:1258–1263. doi: 10.1016/j.str.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw JJ, Trobro S, He SL, Åqvist J, Green R. A role for the 2′ OH of peptidyl-tRNA substrate in peptide release on the ribosome revealed through RF-mediated rescue. Chem Biol. 2012;19:983–993. doi: 10.1016/j.chembiol.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koutmou KS, McDonald ME, Brunelle JL, Green R. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA. 2014;20:609–620. doi: 10.1261/rna.042523.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franckenberg S, Becker T, Beckmann R. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr Opin Struct Biol. 2012;22:786–796. doi: 10.1016/j.sbi.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Shi X, Joseph S. Mechanism of translation termination: RF1 dissociation follows dissociation of RF3 from the ribosome. Biochemistry. 2016;55:6344–6354. doi: 10.1021/acs.biochem.6b00921. [DOI] [PubMed] [Google Scholar]
- 14.Barthelme D, et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci USA. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyler DE, Wehner KA, Green R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem. 2013;288:29530–29538. doi: 10.1074/jbc.M113.487090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura Y, Ito K. How protein reads the stop codon and terminates translation. Genes Cells. 1998;3:265–278. doi: 10.1046/j.1365-2443.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- 19.Ito K, Uno M, Nakamura Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 20.Ito K, Uno M, Nakamura Y. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc Natl Acad Sci USA. 1998;95:8165–8169. doi: 10.1073/pnas.95.14.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young DJ, Edgar CD, Poole ES, Tate WP. The codon specificity of eubacterial release factors is determined by the sequence and size of the recognition loop. RNA. 2010;16:1623–1633. doi: 10.1261/rna.2117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svidritskiy E, Korostelev AA. 2017. Conformational control of translation termination on the 70S ribosome. bioRxiv:10.1101/226837.
- 23.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mora L, et al. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol. 2003;47:267–275. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- 25.Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulygin KN, et al. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA. 2010;16:1902–1914. doi: 10.1261/rna.2066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito K, et al. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc Natl Acad Sci USA. 2002;99:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conard SE, et al. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA. 2012;18:1210–1221. doi: 10.1261/rna.031997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolosov P, et al. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchet S, et al. New insights into stop codon recognition by eRF1. Nucleic Acids Res. 2015;43:3298–3308. doi: 10.1093/nar/gkv154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 33.Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weixlbaumer A, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matheisl S, Berninghausen O, Becker T, Beckmann R. Structure of a human translation termination complex. Nucleic Acids Res. 2015;43:8615–8626. doi: 10.1093/nar/gkv909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oparina NJ, Kalinina OV, Gelfand MS, Kisselev LL. Common and specific amino acid residues in the prokaryotic polypeptide release factors RF1 and RF2: Possible functional implications. Nucleic Acids Res. 2005;33:5226–5234. doi: 10.1093/nar/gki841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studer RA, Dessailly BH, Orengo CA. Residue mutations and their impact on protein structure and function: Detecting beneficial and pathogenic changes. Biochem J. 2013;449:581–594. doi: 10.1042/BJ20121221. [DOI] [PubMed] [Google Scholar]
- 40.Erlacher MD, et al. Chemical engineering of the peptidyl transferase center reveals an important role of the 2′-hydroxyl group of A2451. Nucleic Acids Res. 2005;33:1618–1627. doi: 10.1093/nar/gki308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trappl K, Mathew MA, Joseph S. Thermodynamic and kinetic insights into stop codon recognition by release factor 1. PLoS One. 2014;9:e94058. doi: 10.1371/journal.pone.0094058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hetrick B, Lee K, Joseph S. Kinetics of stop codon recognition by release factor 1. Biochemistry. 2009;48:11178–11184. doi: 10.1021/bi901577d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Svidritskiy E, Madireddy R, Korostelev AA. Structural basis for translation termination on a pseudouridylated stop codon. J Mol Biol. 2016;428:2228–2236. doi: 10.1016/j.jmb.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 48.Hoernes TP, et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016;44:852–862. doi: 10.1093/nar/gkv1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 50.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 51.Ueda T, Kanamori T, Ohashi H. Ribosome display with the PURE technology. Methods Mol Biol. 2010;607:219–225. doi: 10.1007/978-1-60327-331-2_18. [DOI] [PubMed] [Google Scholar]
- 52.Nakamura Y, Ito K, Ehrenberg M. Mimicry grasps reality in translation termination. Cell. 2000;101:349–352. doi: 10.1016/s0092-8674(00)80845-4. [DOI] [PubMed] [Google Scholar]
- 53.Sund J, Andér M, Åqvist J. Principles of stop-codon reading on the ribosome. Nature. 2010;465:947–950. doi: 10.1038/nature09082. [DOI] [PubMed] [Google Scholar]
- 54.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 55.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 56.Guydosh NR, Green R. Translation of poly(A) tails leads to precise mRNA cleavage. RNA. 2017;23:749–761. doi: 10.1261/rna.060418.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tate W, Greuer B, Brimacombe R. Codon recognition in polypeptide chain termination: Site directed crosslinking of termination codon to Escherichia coli release factor 2. Nucleic Acids Res. 1990;18:6537–6544. doi: 10.1093/nar/18.22.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clementi N, Chirkova A, Puffer B, Micura R, Polacek N. Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation. Nat Chem Biol. 2010;6:344–351. doi: 10.1038/nchembio.341. [DOI] [PubMed] [Google Scholar]
- 59.Koch M, et al. The integrity of the G2421-C2395 base pair in the ribosomal E-site is crucial for protein synthesis. RNA Biol. 2015;12:70–81. doi: 10.1080/15476286.2015.1017218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koch M, Willi J, Pradère U, Hall J, Polacek N. Critical 23S rRNA interactions for macrolide-dependent ribosome stalling on the ErmCL nascent peptide chain. Nucleic Acids Res. 2017;45:6717–6728. doi: 10.1093/nar/gkx195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alkalaeva E, Mikhailova T. Reassigning stop codons via translation termination: How a few eukaryotes broke the dogma. BioEssays. 2017;39:1600213. doi: 10.1002/bies.201600213. [DOI] [PubMed] [Google Scholar]
- 62.Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, Baranov PV. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in Condylostoma magnum. Mol Biol Evol. 2016;33:2885–2889. doi: 10.1093/molbev/msw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swart EC, Serra V, Petroni G, Nowacki M. Genetic codes with no dedicated stop codon: Context-dependent translation termination. Cell. 2016;166:691–702. doi: 10.1016/j.cell.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Field A, Hetrick B, Mathew M, Joseph S. Histidine 197 in release factor 1 is essential for a site binding and peptide release. Biochemistry. 2010;49:9385–9390. doi: 10.1021/bi1012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lind C, Oliveira A, Åqvist J. Origin of the omnipotence of eukaryotic release factor 1. Nat Commun. 2017;8:1425. doi: 10.1038/s41467-017-01757-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.