Significance
Accurate incorporation of nonstandard amino acids (nsAAs) is central for genetic code expansion to increase the chemical diversity of proteins. However, aminoacyl-tRNA synthetases are polyspecific and facilitate incorporation of multiple nsAAs. We investigated and repurposed a natural protein degradation pathway, the N-end rule pathway, to devise an innovative system for rapid assessment of the accuracy of nsAA incorporation. Using this tool to monitor incorporation of the nsAA biphenylalanine allowed the identification of tyrosyl-tRNA synthetase (TyrRS) variants with improved amino acid specificity. The evolved TyrRS variants enhanced our ability to contain unwanted proliferation of genetically modified organisms. This posttranslational proofreading system will aid the evolution of orthogonal translation systems for specific incorporation of diverse nsAAs.
Keywords: genetic code expansion, protein degradation, N-end rule, nonstandard amino acids, synthetic biology
Abstract
Incorporation of nonstandard amino acids (nsAAs) leads to chemical diversification of proteins, which is an important tool for the investigation and engineering of biological processes. However, the aminoacyl-tRNA synthetases crucial for this process are polyspecific in regard to nsAAs and standard amino acids. Here, we develop a quality control system called “posttranslational proofreading” to more accurately and rapidly evaluate nsAA incorporation. We achieve this proofreading by hijacking a natural pathway of protein degradation known as the N-end rule, which regulates the lifespan of a protein based on its amino-terminal residue. We find that proteins containing certain desired N-terminal nsAAs have much longer half-lives compared with those proteins containing undesired amino acids. We use the posttranslational proofreading system to further evolve a Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (TyrRS) variant and a tRNATyr species for improved specificity of the nsAA biphenylalanine in vitro and in vivo. Our newly evolved biphenylalanine incorporation machinery enhances the biocontainment and growth of genetically engineered Escherichia coli strains that depend on biphenylalanine incorporation. Finally, we show that our posttranslational proofreading system can be designed for incorporation of other nsAAs by rational engineering of the ClpS protein, which mediates the N-end rule. Taken together, our posttranslational proofreading system for in vivo protein sequence verification presents an alternative paradigm for molecular recognition of amino acids and is a major advance in our ability to accurately expand the genetic code.
The ability to incorporate chemically diverse nonstandard amino acids (nsAAs) broadens the structural and functional diversity of proteins (1, 2). nsAAs with varied sidechains can serve as photo-crosslinking groups, spectroscopic and fluorescent probes, or reactive handles for conjugation (2). nsAA incorporation has also been applied to control proliferation of genetically modified organisms by introducing nsAA dependency in essential proteins (3, 4). These biocontainment approaches, known as “synthetic auxotrophy,” are important safeguards as we advance toward assembling genomes that contain large deviations from the standard genetic code (5), which can provide hosts with increased virus resistance (6, 7).
Site-specific incorporation of nsAAs requires a dedicated aminoacyl-tRNA synthetase (aaRS)-tRNA pair, also known as an orthogonal translation system (OTS), which must not cross-react with the host’s tRNAs and aaRSs. The substrate specificity of an aaRS is normally engineered through rounds of site-detected evolution to recognize a desired nsAA while discriminating any other AA in the cell (8). However, many engineered aaRSs fail to effectively discern between cognate nsAA substrates and standard AAs and other nsAAs. Thus, engineered aaRSs with low specificity can generate target proteins with different amino acids at the desired positions. Currently, most nsAAs are incorporated in response of a nonsense codon (e.g., UAG) within a gene encoding a target protein. Incorporation of nsAAs is then monitored by production of the full-length reporter protein as evaluated by standard gel electrophoresis or by fluorimetry. However, promiscuous aaRS-tRNA pairs can produce full-length target proteins even in the absence of the nsAA (9–13).
The aaRSs that facilitate nsAA incorporation may exhibit overlap of substrate specificities, which limits their simultaneous use for synthesis of proteins with different nsAAs (14–16). In contrast, the aaRS enzymes are highly specific for their natural cognate AA, and together with other pretranslational quality control processes allow a mistranslation rate of only 1 in 104 (8). Cross-talk of nsAA incorporation machinery with standard AAs may lower the effectiveness of previously demonstrated synthetic auxotrophy (3) as promiscuous activity of the biphenylalanine (BipA) incorporation machinery (17) can promote escape. Many other nsAA applications, such as protein double labeling, Förster resonance energy transfer (FRET), and antibody conjugation, require high fidelity incorporation to avoid heterogeneous protein production.
Currently, the identity of an incorporated amino acid is best determined by mass spectrometry of the desired recombinant protein. We sought to develop a new detection system with the following design criteria: (i) the ability to controllably mask and unmask misincorporation in vivo; (ii) compatibility with different reporter proteins; and (iii) customizability for most commonly used nsAAs. Here, we report how the N-end rule pathway of protein degradation, a natural protein regulatory and quality control pathway conserved across prokaryotes and eukaryotes (18–20), applies to commonly used nsAAs. The N-end rule states that the half-life of a protein is determined by its amino-terminal residue. Because components of the N-end rule pathway interact specifically with a subset of AAs, we hypothesized that nsAAs may be N-end stabilizing, whereas their standard AA analogs (Tyr/Phe/Trp/Leu/Lys/Arg) that are the most likely culprits for misincorporation are known to be N-end destabilizing residues in Escherichia coli, which result in protein half-lives on the timescale of minutes (19). We tested the effect of incorporation of commonly used nsAAs at the N terminus and used our findings to develop “posttranslational proofreading,” which enables high-accuracy discrimination of nsAA incorporation in vivo. Posttranslational proofreading is a remarkably modular, generalizable, and tunable system for specific protein recognition based on the identity of a single amino acid at the N terminus, which is a position increasingly targeted for applications in chemical biology (21). We demonstrated that the ability to optionally degrade proteins containing standard AA misincorporation events dramatically facilitates directed evolution for selective nsAA incorporation machinery.
Results
Evaluation of the BipA OTS Promiscuity.
Four primary OTS families have been developed for nsAA incorporation by suppression of UAG codons in targeted sequences (3, 4) (SI Appendix, Fig. S1A). We began by evaluating the promiscuity of the BipA OTS, which is comprised of the BipARS aminoacyl-tRNA synthetase and derived from the Methanocaldococcus jannaschii Tyr OTS (MjTyrRS and ) (17). We performed experiments where each standard AA was introduced individually at an elevated concentration in minimal media lacking BipA. These experiments suggested that Tyr and Leu were being misincorporated by the BipA OTS in the absence of BipA (SI Appendix, Fig. S1B). Similarly, we determined by mass spectrometry that target peptides produced upon expression of the BipA OTS but in the absence of BipA contained 90+% Tyr/Leu/Phe at the target site, with glutamine (Gln) also present due to expected near-cognate suppression (22, 23) (SI Appendix, Fig. S1C). This result confirmed that the BipA OTS was causing incorporation of standard AAs in the absence of an nsAA. As hypothesized, most of the standard AAs that we observed to be incorporated were expected to destabilize proteins if present at the N terminus according to the N-end rule pathway.
Application of the N-End Rule to Commonly Used nsAAs.
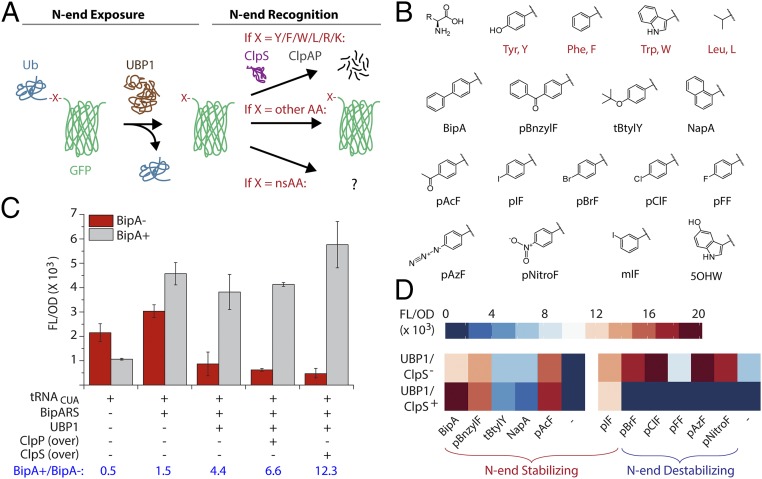
To investigate how the N-end rule applies to nsAAs, we constructed a reporter consisting of a cleavable ubiquitin domain (Ub) followed by one UAG codon, a conditionally strong N-degron (24, 25), and a superfolder green fluorescent protein (sfGFP) with a C-terminal His6x-tag (Fig. 1A). This reporter is designed such that nsAA incorporation is targeted at a site that is subsequently exposed as the N terminus. Depending on the identity of the incorporated AA in a given GFP protein, the protein will either be stabilized or destabilized. We genomically integrated this reporter into an E. coli strain that was genomically recoded to be devoid of UAG codons and their associated release factor (strain “C321.ΔA”) (6). The use of only one UAG codon increases assay sensitivity for promiscuity compared with the use of multi-UAG codon reporters (26), and genomic integration of the reporter increases reproducibility by eliminating plasmid copy number effects (27). We began experiments with a focus on BipA but eventually tested a panel of commonly used nsAAs (Fig. 1B). As a proof of concept, we tested coexpression of different components of the BipA OTS (“incorporation machinery”) with different components of the N-end rule pathway (“proofreading machinery”) in the presence and absence of BipA (BipA− or BipA+). Expression of the orthogonal alone was responsible for a moderate amount of GFP accumulation in cells based on normalized FL/OD signal (Fig. 1C). Expression of the BipARS together with resulted in nearly equivalent signal in BipA− or BipA+ cases. Expression of an N-terminally truncated yeast ubiquitin cleavase protein (UBP1) (28, 29) to expose the target residue as the N-terminal residue caused ∼fourfold reduction of the BipA− signal but no significant change in the BipA+ signal. The decrease in only the BipA− signal supported our hypothesis that BipA would be N-end stabilizing. BipA− signal decreased further upon overexpression of ClpS, the adaptor protein that directly binds to N-terminal destabilizing residues (Tyr/Phe/Trp/Leu) on protein substrates and delivers them to the ClpAP AAA+ protease complex for unfolding and degradation (30). ClpS overexpression may decrease substrate competition for proteins targeted for proofreading because ClpS is known to inhibit ClpAP-dependent degradation of other substrates such as SsrA-tagged proteins (31). Because ClpS overexpression resulted in lower growth rates in LB medium, we performed subsequent experiments in 2XYT medium, where we observed no differences in growth rates.
Fig. 1.
Posttranslational proofreading proof of concept. (A) Scheme for proofreading consisting of N-end exposure and recognition steps applied to synthetic substrates. Ub is cleaved by ubiquitin cleavase UBP1 to expose the target site as N-terminal. ClpS is the native N-recognin in E. coli and ClpAP forms an AAA+ protease complex for degradation by the N-end rule pathway. (B) nsAAs used in this study (full chemical names in SI Appendix). (C) Incorporation assay showing fluorescence resulting from GFP expression normalized by optical density (FL/OD) in the absence/presence of BipA and expression of various OTS or N-end rule components. “Over” indicates overexpression of natively expressed components. Error bars represent SD, n = 3. (D) Heatmap of FL/OD signals obtained from an nsAA panel arranged roughly in descending size from left to right without proofreading occurring in Top row and with proofreading occurring in Bottom row. Left reflects activity of the Bipyridylalanine OTS and Right reflects activity of the p-acetyl-phenylalanine OTS. Heatmap values here and elsewhere are average of n = 3.
To examine how the N-end rule applies to a larger set of nsAAs, we used the Bipyridinylalanine OTS to screen 11 nsAAs because of its low nsAA− signal in our reporter assay (SI Appendix, Fig. S2). However, this OTS resulted in observable nsAA+ signal for only 5 of 11 tested phenyl-nsAAs, with preference for large hydrophobic side chains at the para position of phenylalanine (Fig. 1D). Notably, nsAA+ signal for these five nsAAs was unaffected by proofreading. The p-acetyl-phenylalanine OTS was used to test incorporation of the six remaining nsAAs and appeared to broadly increase signal for these six nsAAs with proofreading “off” (i.e., no expression of UBP1/ClpS). We observed marked differences in signal between proofreading off and “on” states based roughly on nsAA size. For p-iodo-phenylalanine and larger nsAAs, signal did not significantly change, and, therefore, p-iodo-phenylalanine and larger nsAAs appear N-end stabilizing. However, for p-bromo-phenylalanine and other smaller or polar phenyl-nsAAs such as p-azido-phenylalanine, signal was significantly diminished when proofreading was on relative to when it was off. The data suggest that smaller deviations from Tyr/Phe are tolerated by the ClpS binding pocket, making smaller and more polar nsAAs such as p-bromophenylalanine and p-azido-phenylalanine appear N-end destabilizing.
Engineering of the N-End Rule for Altered Recognition of nsAAs.
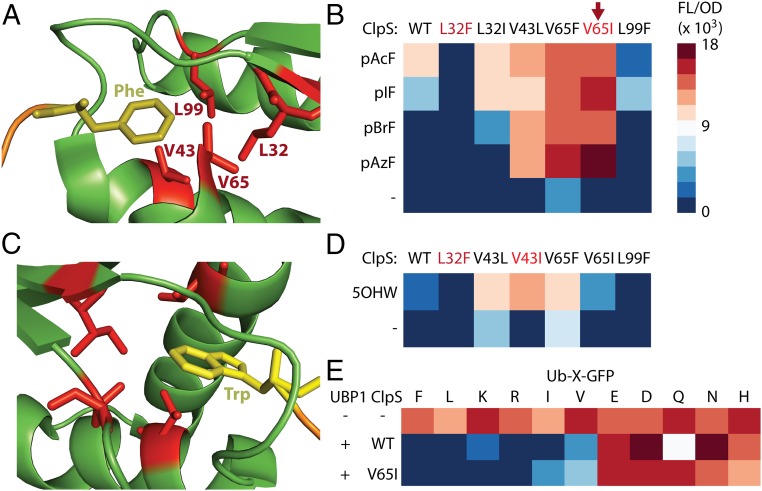
We hypothesized that we could engineer ClpS to alter N-end rule classification of these smaller nsAAs. We targeted four hydrophobic residues in the ClpS binding pocket for single-point mutagenesis covering Phe/Leu/Ile/Val using NTC-containing oligos (Fig. 2A). Sequence alignments of ClpS homologs across prokaryotes and eukaryotes showed conservation of these residues among related hydrophobic amino acids (SI Appendix, Fig. S3). By screening the resulting 12 single mutants in a ClpS-deficient version of our reporter strain with select nsAAs and the p-acetyl-phenylalanine OTS, we identified a variant (ClpSV65I) that resulted in stabilization of proteins containing all screened N-end phenyl nsAAs while destabilizing proteins containing standard AAs (Fig. 2B). In addition, we identified a variant (ClpSL32F) that resulted in complete destabilization of proteins containing all but the two largest screened N-end phenyl nsAAs (SI Appendix, Fig. S4). We also attempted to distinguish tryptophanyl analogs from Trp using the 5-hydroxytryptophan (5OHW) OTS (Fig. 2C). Although 5OHW appeared N-end destabilizing with wild-type ClpS, we observed that ClpSV43I and ClpSV65I improved discrimination of 5OHW from Trp in the ClpS-deficient strain (Fig. 2D). Given the desirable properties of ClpSV65I, we wanted to examine whether it alters N-end rule classification for standard AAs. We substituted the UAG codon in our GFP reporter for codons encoding a representative panel of standard AAs and found that ClpSV65I affects stability of these N-end standard AAs no differently than ClpS (Fig. 2E). Rational designs from our small library can precisely distinguish small modifications on a variety of chemical templates, such as nsAAs with phenyl as well as indole sidechains, showcasing the remarkably tunability of the proofreading strategy. Interestingly, overexpression of either ClpS or ClpSV65I leads to degradation of N-end Ile/Val, residues that are previously shown to be only weakly N-end destabilizing in vitro (32). Proteolysis of native proteins containing N-terminal Ile/Val may contribute to the toxicity observed from ClpS overexpression.
Fig. 2.
Proofreading tunability achieved through rational ClpS engineering. (A) Cartoon generated from crystal structure of E. coli ClpS binding N-end Phe peptide (PDB ID code: 3O2B) showing four hydrophobic ClpS residues subjected to single-point mutations that sampled F/L/I/V. (B) Heatmap of FL/OD signals obtained using a ClpS− host expressing UBP1, the p-acetyl-phenylalanine OTS, and variants of ClpS in the presence of different nsAAs. (C) Cartoon generated from crystal structure of C. crescentus ClpS binding N-end Trp peptide (PDB ID code: 3GQ1). (D) FL/OD heatmap resulting from expression of UBP1, the 5-hydroxytryptophan OTS, and ClpS variants in the presence/absence of 5-hydroxytryptophan. Scale as in B. (E) FL/OD heatmap resulting from expression of UBP1/ClpS in strains with Ub-X-GFP reporter genes expressing standard AAs in place of X.
Application of Proofreading for Selective OTS Evolution.
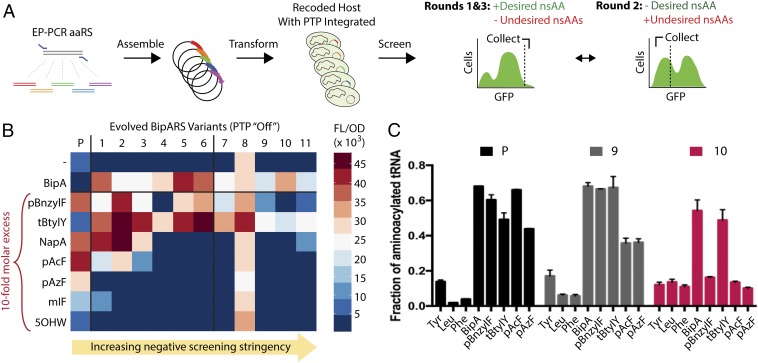
The ability of proofreading to discriminate incorporation of intended nsAAs from related standard AAs is useful for high-throughput screening of OTS libraries. To demonstrate this, we integrated the UBP1-clpSV65I expression cassette into our ClpS-deficient reporter strain and used this strain (“C321.Nend”) to improve the selectivity of the parental BipA OTS. Previous efforts to engineer MjTyrRS variants like BipARS focused on site-directed mutagenesis on positions near the amino acid binding pocket (10, 33). To generate a novel BipARS library, we used error-prone PCR to introduce two to four mutations throughout the bipARS gene. These libraries were transformed into C321.Nend and screened with three rounds of fluorescence-activated cell sorting (FACS): (i) positive sort for GFP+ cells in BipA+, (ii) negative sort for GFP− cells in BipA− to expose the tendency for misincorporation, and (iii) final positive sort for GFP+ cells in BipA+ (Fig. 3A). To obtain variants exhibiting decreased promiscuity against other nsAAs, we altered the negative screening stringency by varying addition of undesired nsAAs, which changed the profile of isolated variants (SI Appendix, Fig. S5). Purification and retransformation of the 11 most enriched variants into strain C321.Ub-UAG-sfGFP (which lacks proofreading machinery) showed that most of our variants increased BipA+ signal and decreased BipA− signal compared with the parental OTS (Fig. 3B and SI Appendix, Table S1, variants 1–6). Supplementation with undesired nsAAs enriched for mutants with even greater selectivity against standard AAs and undesired nsAAs (variants 4, 9–11) but also gave rise to an extremely promiscuous variant (variant 8), suggesting that these conditions may be nearly too harsh and facilitate emergence of cheaters. One mutant that was only isolated in higher stringencies (variant 10) exhibited high activity on BipA and no observable activity on any other nsAAs except tert-Butyl-tyrosine (tBtylY), whose structure is very similar to BipA and contains the inert tert-Butyl protecting group typically removed for further modification (Fig. 3B). SDS/PAGE of Ub-X-GFP, resulting from the variant 10 OTS after expression and affinity purification, showed no observable BipA− protein production in contrast to the parental BipA OTS, which shows a distinct BipA− band (SI Appendix, Fig. S6A). Furthermore, mass spectrometry confirmed site-specific BipA incorporation in the BipA+ condition (SI Appendix, Fig. S6 B–D).
Fig. 3.
Selective BipA OTS evolution using proofreading. (A) FACS evolution scheme with error-prone PCR aminoacyl-tRNA synthetase libraries transformed into hosts with posttranslational proofreading (“PTP”, using ClpSV65I) genomically integrated, followed by three sorting rounds. (B) Evaluation of enriched evolved BipARS variants in clean backgrounds on a panel of nsAAs ([BipA] = 100 μM, [rest] = 1 mM, which are their standard concentrations). The parental variant is noted as “P.” (C) In vitro amino acid substrate specificity profile of BipA OTS variants. Error bars = SD, n = 3.
We sequenced the aminoacyl-tRNA synthetase and tRNA regions of all enriched OTS variants and discovered spontaneous tRNA mutations in our most selective variants, such as 4, 9, and 10, perhaps arising because of our use of a MutS-deficient host (SI Appendix, Fig. S7A and Table S1). None of the observed tRNA mutations occurred at the anticodon region, indicating that these tRNAs still lead to incorporation at the UAG site and do not cause off-target incorporation. Because OTS variants 4, 9, and 10 caused limited fluorescent protein production in the absence of BipA, we can also confidently state that these tRNAs are not being acylated by endogenous synthetases. When we reverted these tRNA mutations, each corresponding BipA OTS became more promiscuous (SI Appendix, Fig. S7B), suggesting that observed tRNA mutations increase selectivity. The G51 position (G50 in E. coli nomenclature) mutated in tRNA variant 10 is the most significant base pair in determining acylated tRNA binding affinity to elongation factor Tu (EF-Tu), which influences ribosomal incorporation selectivity downstream of the aminoacyl-tRNA synthetase (34, 35). To more rigorously assess OTS selectivity, we purified BipARS and tRNA for the parental, variant 9, and variant 10 OTSs. The observed in vitro substrate specificity as determined by tRNA aminoacylation is in excellent agreement with our in vivo assays (Fig. 3C), and the data suggests that BipARS and tRNA variants each contribute to selectivity improvements (SI Appendix, Fig. S7 C and D). The variant 10 OTS exhibited the highest selectivity for BipA and was chosen for subsequent applications.
Demonstration of Enhanced Biocontainment Using the More Selective OTS.
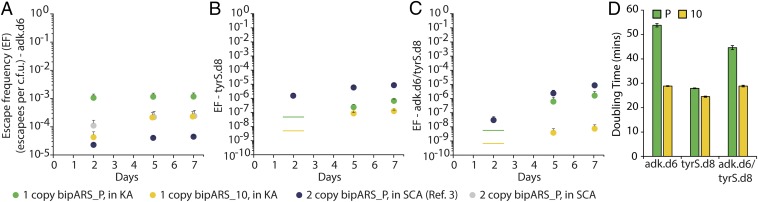
To demonstrate the utility of a more selective OTS for biocontainment based on synthetic auxotrophy, we substituted the parental BipA OTS construct previously used in three biocontained strains that exhibit observable escape frequencies with plasmids containing either parental or variant 10 OTS. These three biocontained strains (adk.d6, tyrS.d8, and adk.d6/tyrS.d8) harbor computational redesigns of two essential genes (adk and tyrS) to make their stability dependent on BipA (3). The effectiveness of synthetic biocontainment is evaluated by growing cells in permissive media that contains nsAA and, subsequently, plating on nonpermissive media that does not contain nsAA. In this manner, the fraction of cells that gain the undesired ability to grow without nsAA can be measured and this is the escape frequency.
We monitored escape frequencies on nonpermissive media for 7 d and observed lower escape frequencies for strains containing the variant 10 OTS at all measured time-points (Fig. 4 A–C and SI Appendix, Fig. S8). The difference in escape frequency was most apparent for the adk.d6/tyrS.d8 strain, which exhibited a 7-d escape frequency of 7.4 × 10−9, a value more than two orders of magnitude lower than observed for any recoded E. coli strain containing only two genes engineered to depend on an nsAA. Furthermore, the fitness of all three strains improved with the variant 10 OTS, with doubling time decreasing by nearly twofold (Fig. 4D). The improved fitness is likely due to decreased formation of destabilized Adk.d6 and TyrS.d8 proteins containing misincorporation events, which should reduce burden on degradation machinery. Finally, variant 10 also delayed onset of growth of adk.d6/tyrS.d8 on noncognate nsAAs (SI Appendix, Table S2). We expect these benefits to carry over to all strains which employ variant 10 over the parental OTS. The increase in containment efficacy and growth rate is significant for potential industrial uses of the biocontained strain because it translates into the ability to safely grow 100-fold more cells in a reactor volume without concern for an escapee that could propagate upon accidental environmental release. Furthermore, these biocontained cells will grow more rapidly than first generation biocontained cells, which can accelerate the rate of industrial production of metabolites or proteins.
Fig. 4.
Effect of evolved BipA OTS on biocontainment strain escape and fitness. (A) Escape frequencies over time for adk.d6 strains transformed with constructs indicated in legend below plots. Green and yellow circles compare escape frequencies of parent and evolved variants and are most relevant for this study. Navy circles represent previously published data (from ref. 3). Gray circles for adk.d6 represent our repeat of previously published data. KA, kanamycin+arabinose; SCA, SDS+chloramphenicol+arabinose. Error bars in A–C represent SEM, n = 3. (B) Escape frequencies over time for tyrS.d8 strains. Lines represent assay detection limit in cases where no colonies were observed. (C) Escape frequencies over time for adk.d6/tyrS.d8 strains. (D) Doubling time for biocontained strains with parental (P) or variant 10 OTS. Error bars = SD, n = 3.
Discussion
We have demonstrated how the N-end rule pathway of protein degradation applies to commonly used nsAAs and how the pathway can be engineered for altered N-end rule classification of these molecules. We harnessed these findings to develop our posttranslational proofreading method, which eliminates most false-positive protein expression (at the N terminus) and, therefore, improves the ability to determine and increase the selectivity of OTSs used for nsAA incorporation. Furthermore, the capability of proofreading to distinguish among nsAAs will facilitate future efforts to simultaneously harness more than 21 amino acids. We validated proofreading during evolution of the BipA OTS, which resulted not only in greater selectivity in vivo and in vitro, but also in enhanced biocontainment efficacy and greater strain fitness in all tested biocontained strains. Compared with strategies that feature toxin-antitoxin systems or metabolic auxotrophs (36–40), the strategy of synthetic auxotrophy has been the most effective biocontainment strategy reported in the literature in terms of limiting cell growth to conditions that are not naturally available and resulting in escape rates below a detection limit of 10−12 (3). Our work shows how OTS selectivity influences the effectiveness of synthetic auxotrophy and generates the most selective OTS for the most effective biocontainment strategy available.
In addition to providing an innovative paradigm for OTS evaluation and evolution, posttranslational proofreading can be transformative for applications in which the identity of a single amino acid is critical, such as screening of natural synthetases for nsAA acceptance (41), sense codon reassignment (42, 43), posttranslational modifications (44), and for industrial uses where purity is extremely important, such as nsAA-containing biologics (45). The ability to discriminate between nsAAs and standard AAs may prove especially useful for reassigning sense codons, where novel screening and analytical methods are required because UAG readthrough is independent of the nature of the AA. For industrial production of proteins containing nsAAs, significant cost savings may be obtained through biosynthesis of nsAAs rather than supplementation (46), and the use of nsAA biosynthetic pathways adds additional motivation to the need for selective OTSs that do not recognize structurally similar precursors to the desired nsAA. Proofreading may also find use in translational regulation and as an orthogonal biocontainment strategy.
It may be possible to expand the applicability of proofreading to all 20 standard AAs given that they are all known to be N-end destabilizing under certain contexts (47). The feasibility of increasing the set of N-end destabilizing AAs by engineering or importing conditionally expressed N-recognins across organisms has only begun to be explored. An engineered methionine aminopeptidase with broader substrate specificity has been expressed successfully in E. coli, presumably increasing the number of native substrates of N-end rule degradation (48). In addition, aspartate and glutamate have previously been converted to N-end destabilizing residues in E. coli using the bacterial aminoacyl-transferase Bpt from Vibrio vulnificus (49). These results suggest that E. coli can tolerate some increases in the number of native proteins that are likely subject to N-end degradation despite the impact that potential degradation of essential proteins may have on cell viability. Future attempts to engineer the N-end rule may shed additional light on how pathway components evolved. Because small changes in N-recognin binding pocket size can strongly influence recognition of unnatural analogs, our work suggests that natural N-recognin homologs may vary considerably in their nsAA recognition profiles. Characterization of natural diversity may increase the number of useful modules for future proofreading efforts.
Materials and Methods
Orthogonal Translation System Plasmid Construction.
Two copies of orthogonal MjTyrRS-derived aaRSs and opt were kindly provided in pEVOL plasmids by Peter Schultz (Scripps Institute) (10). Engineered aaRSs used in this study were the following: BipARS (17), BipyARS (17), pAcFRS (50), pAzFRS (51), and NapARS (52). The pEVOL plasmids were maintained using chloramphenicol. Original plasmids harboring two aaRS copies were used for synthetase promiscuity comparison experiments (Figs. 2 and 3). For generation and characterization of synthetase variants, plasmids harboring only one aaRS copy under inducible expression were constructed using Gibson assembly (53). The ScWRS-R3-13 AARS was synthesized as codon-optimized for expression in E. coli and cloned into the pEVOL plasmid along with its associated tRNA (54, 55). In all cases, tRNA is constitutively expressed and aaRS expression is either arabinose inducible or constitutive.
Posttranslational Proofreading Plasmid Construction.
An N-terminally truncated form of the UBP1 gene from Saccharomyces cerevisiae (28, 29) (ScUBP1trunc or simply UBP1) was synthesized as codon-optimized for expression in E. coli and cloned into the pZE21 vector (Kanamycin resistance, ColE1 origin, TET promoter) (Expressys). The E. coli genes clpS and clpP were PCR amplified from E. coli MG1655 and cloned into artificial operons downstream of the UBP1 gene in the pZE21 vector using Gibson assembly. Artificial operons were created by inserting the following RBS sequence between the UBP1 and clp genes: TAATAAAAGGAGATATACC. This RBS was originally designed using the RBS calculator (56) and previously validated in the context of another artificial operon (57). Rational engineering of ClpS variants was performed by dividing the clpS gene into two amplicons where the second amplicon contained a degenerate NTC or NTT sequence in the oligo corresponding to each codon of interest. The four initial positions of interest in the clpS gene correspond to amino acids 32, 43, 65, and 99. In each case, Gibson assembly was used to ligate both amplicons and the backbone plasmid. The pZE/UBP1/ClpS and pZE/UBP1/ClpS_V65I plasmids are available from Addgene (catalog nos. 98566 and 98567).
Reporter Plasmid Construction.
Three reporter constructs were initially cloned into pZE21 vectors before use as templates for PCR amplification and genomic integration. The first of these consists of a Ubiquitin-*-LFVQEL-sfGFP-His6x fusion (“Ub-UAG-sfGFP”) downstream of the TET promoter. The second has an additional UAG codon internal to the sfGFP at position Y151* (“Ub-UAG-sfGFP_151UAG”). The third has an ATG codon (encoding Met) in place of the first UAG (“Ub-M-sfGFP_151UAG”).
More materials and methods are included in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Daniel J. Mandell (Harvard), Dr. Ethan Garner (Harvard), Dr. Karl Schmitz [Massachusetts Institute of Technology (MIT)], Georgia Squyres (Harvard), Alex Bisson (Harvard), Bernardo Cervantes (MIT), Dr. Yekaterina Tarasova (MIT), Dr. Abubakar Jalloh, Juhee Park (MIT), and Dr. Irene M. B. Reizman (Rose-Hulman) for insightful discussions or manuscript comments; Chad Araneo for FACS assistance; and Dr. Bogdan Budnik for mass spectrometry assistance. This project was graciously funded by US Department of Energy Grant DE-FG02-02ER63445 (to G.M.C.) and National Institutes of Health Grants R01GM22854 and R35GM122560 (to D.S.).
Footnotes
Conflict of interest statement: G.M.C. has related financial interests in ReadCoor, EnEvolv, and GRO Biosciences. A.M.K. and G.M.C. have filed a provisional patent on posttranslational proofreading, and A.M.K., D.S., E.K., and G.M.C. have filed a provisional patent on evolved BipA OTS variants. For a complete list of G.M.C.’s financial interests, please visit arep.med.harvard.edu/gmc/tech.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715137115/-/DCSupplemental.
References
- 1.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 2.Dumas A, Lercher L, Spicer CD, Davis BG. Designing logical codon reassignment–Expanding the chemistry in biology. Chem Sci (Camb) 2015;6:50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandell DJ, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rovner AJ, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 6.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma NJ, Isaacs FJ. Genomic recoding broadly obstructs the propagation of horizontally transferred genetic elements. Cell Syst. 2016;3:199–207. doi: 10.1016/j.cels.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibba M, Söll D. Aminoacyl-tRNAs: Setting the limits of the genetic code. Genes Dev. 2004;18:731–738. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 9.Oki K, Sakamoto K, Kobayashi T, Sasaki HM, Yokoyama S. Transplantation of a tyrosine editing domain into a tyrosyl-tRNA synthetase variant enhances its specificity for a tyrosine analog. Proc Natl Acad Sci USA. 2008;105:13298–13303. doi: 10.1073/pnas.0803531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Antonczak AK, et al. Importance of single molecular determinants in the fidelity of expanded genetic codes. Proc Natl Acad Sci USA. 2011;108:1320–1325. doi: 10.1073/pnas.1012276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nehring S, Budisa N, Wiltschi B. Performance analysis of orthogonal pairs designed for an expanded eukaryotic genetic code. PLoS One. 2012;7:e31992. doi: 10.1371/journal.pone.0031992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monk JW, et al. Rapid and inexpensive evaluation of nonstandard amino acid incorporation in Escherichia coli. ACS Synth Biol. 2017;6:45–54. doi: 10.1021/acssynbio.6b00192. [DOI] [PubMed] [Google Scholar]
- 14.Fan C, Ho JML, Chirathivat N, Söll D, Wang Y-S. Exploring the substrate range of wild-type aminoacyl-tRNA synthetases. ChemBioChem. 2014;15:1805–1809. doi: 10.1002/cbic.201402083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo L-T, et al. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc Natl Acad Sci USA. 2014;111:16724–16729. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y-S, et al. The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Mol Biosyst. 2011;7:714–717. doi: 10.1039/c0mb00217h. [DOI] [PubMed] [Google Scholar]
- 17.Xie J, Liu W, Schultz PG. A genetically encoded bidentate, metal-binding amino acid. Angew Chem Int Ed Engl. 2007;46:9239–9242. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]
- 18.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 19.Tobias JW, Shrader TE, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 20.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen CB, Francis MB. Targeting the N terminus for site-selective protein modification. Nat Chem Biol. 2017;13:697–705. doi: 10.1038/nchembio.2416. [DOI] [PubMed] [Google Scholar]
- 22.Johnson DBF, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donoghue P, et al. Near-cognate suppression of amber, opal and quadruplet codons competes with aminoacyl-tRNAPyl for genetic code expansion. FEBS Lett. 2012;586:3931–3937. doi: 10.1016/j.febslet.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KH, Sauer RT, Baker TA. ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev. 2007;21:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang KH, Oakes ESC, Sauer RT, Baker TA. Tuning the strength of a bacterial N-end rule degradation signal. J Biol Chem. 2008;283:24600–24607. doi: 10.1074/jbc.M802213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amiram M, et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat Biotechnol. 2015;33:1272–1279. doi: 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Million-Weaver S, Alexander DL, Allen JM, Camps M. Quantifying plasmid copy number to investigate plasmid dosage effects associated with directed protein evolution. Methods Mol Biol. 2012;834:33–48. doi: 10.1007/978-1-61779-483-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobias JW, Varshavsky A. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12021–12028. [PubMed] [Google Scholar]
- 29.Wojtowicz A, et al. Expression of yeast deubiquitination enzyme UBP1 analogues in E. coli. Microb Cell Fact. 2005;4:17. doi: 10.1186/1475-2859-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Román-Hernández G, Hou JY, Grant RA, Sauer RT, Baker TA. The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol Cell. 2011;43:217–228. doi: 10.1016/j.molcel.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 32.Wang KH, Roman-Hernandez G, Grant RA, Sauer RT, Baker TA. The molecular basis of N-end rule recognition. Mol Cell. 2008;32:406–414. doi: 10.1016/j.molcel.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 34.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 35.Schrader JM, Chapman SJ, Uhlenbeck OC. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol. 2009;386:1255–1264. doi: 10.1016/j.jmb.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronchel MC, Ramos JL. Dual system to reinforce biological containment of recombinant bacteria designed for rhizoremediation. Appl Environ Microbiol. 2001;67:2649–2656. doi: 10.1128/AEM.67.6.2649-2656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, Wu Y-J. A fluorescent, genetically engineered microorganism that degrades organophosphates and commits suicide when required. Appl Microbiol Biotechnol. 2009;82:749–756. doi: 10.1007/s00253-009-1857-3. [DOI] [PubMed] [Google Scholar]
- 38.Pasotti L, Zucca S, Lupotto M, Cusella De Angelis MG, Magni P. Characterization of a synthetic bacterial self-destruction device for programmed cell death and for recombinant proteins release. J Biol Eng. 2011;5:8. doi: 10.1186/1754-1611-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright O, Delmans M, Stan G-B, Ellis T. GeneGuard: A modular plasmid system designed for biosafety. ACS Synth Biol. 2015;4:307–316. doi: 10.1021/sb500234s. [DOI] [PubMed] [Google Scholar]
- 40.Chan CTY, Lee JW, Cameron DE, Bashor CJ, Collins JJ. ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat Chem Biol. 2016;12:82–86. doi: 10.1038/nchembio.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanrikulu IC, Schmitt E, Mechulam Y, Goddard WA, 3rd, Tirrell DA. Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc Natl Acad Sci USA. 2009;106:15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnakumar R, Ling J. Experimental challenges of sense codon reassignment: An innovative approach to genetic code expansion. FEBS Lett. 2014;588:383–388. doi: 10.1016/j.febslet.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 43.Biddle W, Schmitt MA, Fisk JD. Evaluating sense codon reassignment with a simple fluorescence screen. Biochemistry. 2015;54:7355–7364. doi: 10.1021/acs.biochem.5b00870. [DOI] [PubMed] [Google Scholar]
- 44.Losfeld M-E, Soncin F, Ng BG, Singec I, Freeze HH. A sensitive green fluorescent protein biomarker of N-glycosylation site occupancy. FASEB J. 2012;26:4210–4217. doi: 10.1096/fj.12-211656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmerman ES, et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug Chem. 2014;25:351–361. doi: 10.1021/bc400490z. [DOI] [PubMed] [Google Scholar]
- 46.Völler J-S, Budisa N. Coupling genetic code expansion and metabolic engineering for synthetic cells. Curr Opin Biotechnol. 2017;48:1–7. doi: 10.1016/j.copbio.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Chen S-J, Wu X, Wadas B, Oh J-H, Varshavsky A. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science. 2017;355:eaal3655. doi: 10.1126/science.aal3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y-D, Jeng J-C, Wang C-F, Wang S-C, Chang S-T. Removal of N-terminal methionine from recombinant proteins by engineered E. coli methionine aminopeptidase. Protein Sci. 2004;13:1802–1810. doi: 10.1110/ps.04679104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graciet E, et al. Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen. Proc Natl Acad Sci USA. 2006;103:3078–3083. doi: 10.1073/pnas.0511224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin JW, et al. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Brock A, Schultz PG. Adding L-3-(2-Naphthyl)alanine to the genetic code of E. coli. J Am Chem Soc. 2002;124:1836–1837. doi: 10.1021/ja012307j. [DOI] [PubMed] [Google Scholar]
- 53.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 54.Hughes RA, Ellington AD. Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucleic Acids Res. 2010;38:6813–6830. doi: 10.1093/nar/gkq521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellefson JW, et al. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat Biotechnol. 2014;32:97–101. doi: 10.1038/nbt.2714. [DOI] [PubMed] [Google Scholar]
- 56.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kunjapur AM, Tarasova Y, Prather KLJ. Synthesis and accumulation of aromatic aldehydes in an engineered strain of Escherichia coli. J Am Chem Soc. 2014;136:11644–11654. doi: 10.1021/ja506664a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.