Significance
In flowering plants, the female germ line begins as a single cell known as the megaspore mother cell (MMC) in each ovule. The mechanisms that restrict MMC fate to a single cell remain largely unknown. We show that the Arabidopsis cytochrome P450 gene KLU acts through the chromatin remodeling complex SWR1 to promote WRKY28 expression in ovule primordia. We show that WRKY28 is expressed in a few somatic cells surrounding the MMC and is required to inhibit these cells from acquiring the MMC-like cell fate. Consistent with non–cell-autonomous KLU activity, KLU-expressing cells and WRKY28-expressing cells are neither identical nor adjacently positioned. Our study demonstrates that cell–cell interactions involving only somatic cells in ovule primordia ensure the specification of a single MMC.
Keywords: megaspore mother cell, cytochrome P450 KLU, SWR1, WRKY28
Abstract
Germ-line specification is essential for sexual reproduction. In the ovules of most flowering plants, only a single hypodermal cell enlarges and differentiates into a megaspore mother cell (MMC), the founder cell of the female germ-line lineage. The molecular mechanisms restricting MMC specification to a single cell remain elusive. We show that the Arabidopsis transcription factor WRKY28 is exclusively expressed in hypodermal somatic cells surrounding the MMC and is required to repress these cells from acquiring MMC-like cell identity. In this process, the SWR1 chromatin remodeling complex mediates the incorporation of the histone variant H2A.Z at the WRKY28 locus. Moreover, the cytochrome P450 gene KLU, expressed in inner integument primordia, non–cell-autonomously promotes WRKY28 expression through H2A.Z deposition at WRKY28. Taken together, our findings show how somatic cells in ovule primordia cooperatively use chromatin remodeling to restrict germ-line cell specification to a single cell.
Germ-line cell specification is a critical process in sexually reproducing organisms. Unlike animals, in which germ-line cells are set aside early during embryogenesis, flowering plants specify germ-line cells from somatic cells in the adult stage (1, 2). Only one distal somatic cell of the nucellus in ovules of flowering plants differentiates into a megasporocyte [also termed megaspore mother cell (MMC)] to initiate the female germ-line lineage (3).
A lateral inhibition mechanism mediated by a ligand-receptor system in the MMC and adjacent somatic cells prevents the differentiation of multiple somatic cells into MMCs (4, 5). In rice, this precise specification requires the MMC-expressed TAPETUM DETERMINANT-LIKE 1A (OsTDL1A) peptide ligand and its receptor MULTIPLE SPOROCYTE 1 (MSP1), whose expression is limited to somatic cells surrounding the MMC (6, 7). The maize ortholog of OsTDL1A, MULTIPLE ARCHESPORIAL CELLS 1 (MAC1), also plays a role in suppressing excessive MMC formation (8–10). MAC1 encodes a secreted protein and is preferentially expressed in the MMC (10), supporting the lateral inhibition model. Moreover, the putative RNA helicase gene MNEME (MEM) in Arabidopsis is expressed specifically in the MMC and inhibits neighboring somatic cells from acquiring MMC identity (11).
Intercellular signaling among somatic cells to restrict MMC specification, without involving the cell that eventually becomes the MMC, is only beginning to be understood. We recently showed that the epidermal layer (L1)-expressed TEX1 protein plays an important role in this process by promoting the biogenesis of TAS3-derived transacting siRNAs (ta-siRNAs), which repress the expression of ARF3 (12). Specifically, the expansion of ARF3 expression into lateral epidermal cells from the medio domain of ovule primordia in a TAS3 ta-siRNA–insensitive mutant led to the formation of supernumerary MMCs (12). This finding suggested that intercellular signaling among somatic cells in the ovule primordia is critical for restricting MMC fate to a single somatic cell, but the molecular mechanisms at work in the surrounding somatic cells are still far from clear.
The Arabidopsis cytochrome P450 gene KLU (also known as KLUH/CYP78A5) is thought to generate a mobile signal that promotes the growth of leaves and floral organs in a non–cell-autonomous manner (13, 14). In developing ovules, KLU is required for female meiosis and maternal control of seed size (15, 16). KLU is preferentially expressed in the inner integument, which is located at the proximal end of ovule primordia, opposite to the MMC along the proximal–distal axis (15, 16). The present findings show that KLU functions non–cell-autonomously in restricting MMC specification to a single cell. Along with the ATP-dependent chromatin remodeling complex SWR1, KLU activates the expression of the transcription factor (TF) gene WRKY28, previously unknown to play a role in MMC specification. The deposition of the histone variant H2A.Z by the SWR1 complex at WRKY28 is dependent on KLU, suggesting that the KLU-derived signal is required for the recruitment of SWR1 to WRKY28. Furthermore, we show that WRKY28 is required to prevent multiple somatic cells from differentiating into MMCs. We therefore uncover a mechanism in which KLU-expressing proximal somatic cells in ovule primordia repress MMC fate in distal cells through WRKY28 activation in these distal cells.
Results
The SWR1 Complex and KLU Genetically Interact to Prevent Supernumerary MMC-Like Cells.
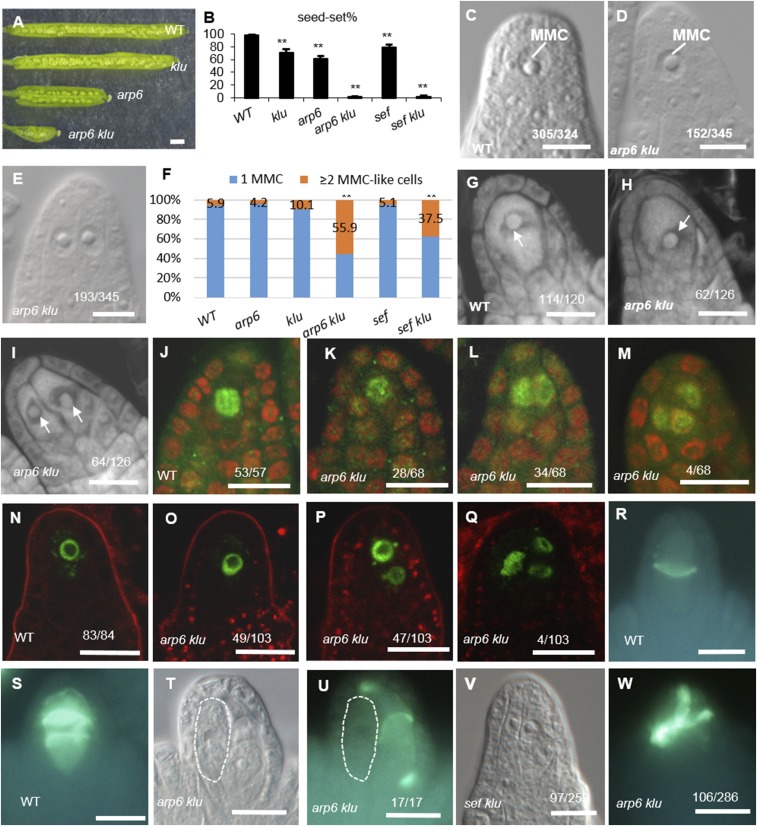
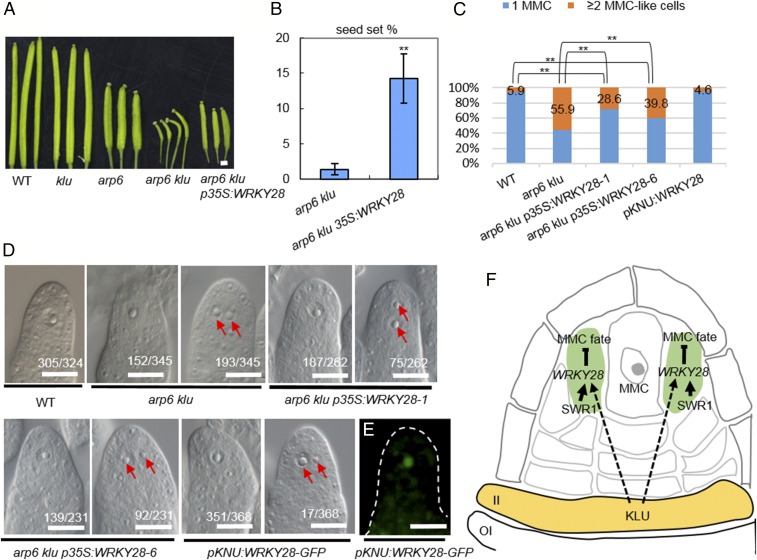
Previously, we showed that mutants in ACTIN-RELATED PROTEIN 6 (ARP6), a subunit gene of the chromatin remodeling SWR1 complex, are defective in chromosome pairing and organization during female meiosis I and have reduced seed set (17). We observed similar defects in a klu mutant, including impaired homolog pairing and recombination during female meiosis I and reduced fertility (16), which prompted us to generate the arp6 klu double mutant to investigate the genetic relationship between ARP6 and KLU in reproduction. Fertility was dramatically reduced in arp6 klu plants compared with WT, klu, and arp6 (Fig. 1 A and B). Reciprocal crosses between WT and the arp6 klu double mutant revealed that the low fertility of arp6 klu plants was primarily caused by female reproductive defects (Table S1).
Fig. 1.
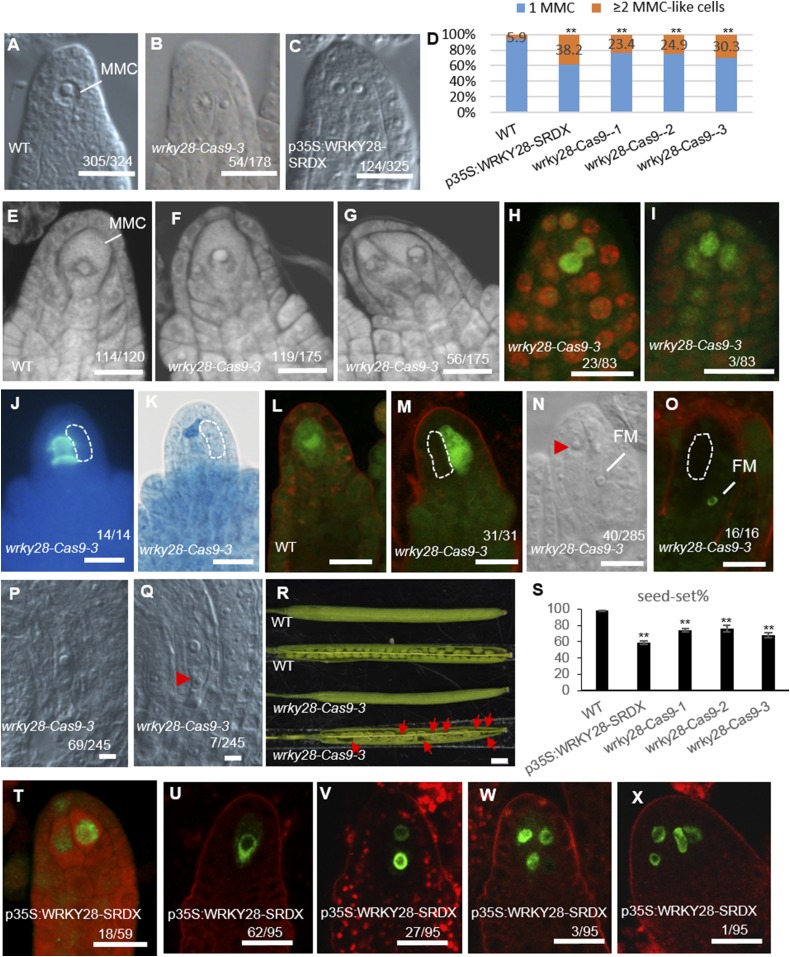
Supernumerary enlarged cells form in arp6 klu ovules. (A) Opened siliques of WT and the klu, arp6, and arp6 klu mutants. (B) Quantification of seed-set percentage. Data are means ± SD (n = 10; **P < 0.01 by t test). (C) Premeiotic WT ovule with a single MMC. (D and E) Premeiotic arp6 klu ovules showing one MMC (D) or two enlarged cells (E). (F) Quantification of aberrant MMC specification (**P < 0.01 by Pearson’s χ2 test). (G–I) Confocal sections of premeiotic WT (G) and arp6 klu (H and I) ovules stained by propidium iodide. Arrows point to the enlarged cells. (J–M) AGO9 immunolocalization in premeiotic WT (J) and arp6 klu (K–M) ovules. Green and red signals correspond to AGO9 localization and propidium iodide signal, respectively. (N–Q) Signal corresponding to pKNU:KNU-Venus expression in premeiotic WT (N) and arp6 klu (O–Q) ovules. (R, S, U, and W) Callose deposition in WT (R and S) and arp6 klu (U and W) ovules. (T) Differential interference contrast (DIC) image shows the morphology of the ovule shown in U; an abnormally enlarged cell adjacent to the MMC is outlined by the white dashed line. (V) Premeiotic sef klu ovule showing two enlarged MMC-like cells. Numbers in the panels denote the frequencies of the phenotypes shown. (Scale bars: A, 1 mm; C–E and G–W, 10 µm.)
To identify the cause of the female reproductive defects, we first examined MMC specification in arp6 klu ovules. Similar to WT ovules, arp6 and klu single mutant ovules have only one MMC at the distal end of ovule primordia (17). We found that 94.1% (n = 324) of premeiotic WT ovules (Columbia ecotype, stage 2-I to 2-II) contained one enlarged cell, which is typically regarded as the MMC in such assays (18) (Fig. 1C). This rate is comparable to what has previously been reported for Columbia ecotype WT ovules (18). In contrast, a single enlarged cell was seen in only 44.1% (n = 345) of premeiotic arp6 klu ovules (Fig. 1D). The remaining 55.9% had more than one enlarged cell (Fig. 1E), and this percentage was significantly higher than in arp6 (4.2%, n = 358) or klu ovules (10.1%, n = 276; Fig. 1F). The phenotype of multiple MMC-like cells in Arabidopsis varies by ecotype and developmental stage (18). Because arp6, klu, and WT control are all in the Columbia background, and all ovules were scored at the same developmental stages (2-I to 2-II) in this assay, the observed differences in the number of MMC-like cells are attributable to the arp6 and klu mutations rather than differences in ecotype or developmental stage.
To examine the MMC specification defects in arp6 klu ovules, we stained the ovules with propidium iodide, a method used to score MMCs in ovule primordia (18). Confocal imaging of WT ovules revealed a single enlarged hypodermal cell at the distal end of ovule primordia (Fig. 1G). In contrast, 50.8% (n = 126) of arp6 klu ovules contained more than one enlarged cell (Fig. 1 H and I). To determine whether the enlarged cells were somatic cells, we analyzed the localization of AGO9 protein. In WT ovules, AGO9 accumulates in cytoplasmic foci in somatic cells at the distal end of ovule primordia (19, 20) but exhibits nuclear localization in the MMC (18). In a whole-mount immunolocalization assay of premeiotic WT ovules, AGO9 localized to the nucleus in the MMC (Fig. 1J). In premeiotic arp6 klu ovules, AGO9 similarly accumulated in the nucleus in the supernumerary MMC-like cells (Fig. 1 K–M). Moreover, the expression of the MMC marker gene KNUCKLES (KNU; pKNU:KNU-Venus) (21), although specific to the MMC in WT (Fig. 1N), was detected in multiple enlarged MMC-like cells in arp6 klu ovules (Fig. 1 O–Q). These findings indicated that these enlarged cells in premeiotic arp6 klu ovules were distinct from the surrounding somatic cells and had acquired molecular characteristics of MMCs.
To determine whether one or all of the enlarged cells undergo meiosis, we analyzed callose deposition, a known cytological marker for MMCs undergoing meiosis (17). In WT stage 2-III/IV ovules, callose was deposited in transversely formed cell plates between daughter cells (Fig. 1 R and S). In arp6 klu ovules, transverse callose walls were detected in only one of the enlarged cells (Fig. 1 T and U), even at later developmental stages. Thus, despite the formation of multiple MMC-like cells in premeiotic arp6 klu ovules, only one differentiated further to undergo meiosis. Consistently, the supernumerary MMC-like cells adjacent to the functional megaspore in postmeiotic arp6 klu ovules did not express ANTIKEVORKIAN (AKV; Fig. S1H), a gene initially expressed in the functional megaspore and subsequently expressed in the developing female gametophyte (11). Taken together, our results show that multiple MMC-like cells acquired cytological and molecular characteristics of MMCs, but they did not all differentiate into fully functional MMCs.
To determine whether the function of ARP6 in repressing MMC-like cell fate applies to the SWR1 complex, we studied SERRATED LEAVES AND EARLY FLOWERING (SEF), another subunit of the SWR1 complex that physically interacts with ARP6 (22). Premeiotic ovule primordia of sef klu double mutants harbored supernumerary enlarged cells (Fig. 1V) much more frequently (37.5%, n = 259) than WT (5.9%, n = 324), sef (5.1%, n = 214), and klu (10.1%, n = 276) ovules (Fig. 1F). Furthermore, seed set was also reduced in sef klu plants (7.8%, n = 320) compared with WT (98.0%, n = 540), klu (70.6%, n = 428), and sef (69.2%, n = 351) plants (Fig. 1B). Taken together, our data suggest that the chromatin remodeling complex SWR1 and cytochrome P450 KLU genetically interact before meiosis to ensure that only one hypodermal somatic cell gains MMC characteristics.
In addition to multiple MMC-like cells, we detected other defects downstream of MMC specification in arp6 klu double mutant ovules. There was an increased number of ovules with meiotic defects [37.1% in arp6 klu (n = 286) vs. 0% in WT (n = 120); Fig. 1W], similar to the meiotic defects of arp6 (17) and klu (16) single mutants; a decreased number of ovules with a functional megaspore [45.6% in arp6 klu (n = 338) vs. 97.9% in WT (n = 329); Fig. S1 A–H]; an increased number of ovules with defective female gametogenesis [92.8% in arp6 klu (n = 292) vs. 0.8% in WT (n = 252); Fig. S1 I–M]; and a decreased number of ovules with a fully formed female gametophyte [7.2% in arp6 klu (n = 292) vs. 99.2% in WT (n = 252); Fig. S1 I and J]. The combined effects of these defects probably led to the reduced fertility of the arp6 klu double mutant (Fig. 1 A and B). Despite these pleiotropic phenotypes during reproduction, our study focused on the MMC specification defect, the earliest defect in the arp6 klu double mutant ovule.
WRKY28 Expression Is Decreased in arp6 klu Ovule Primordia.
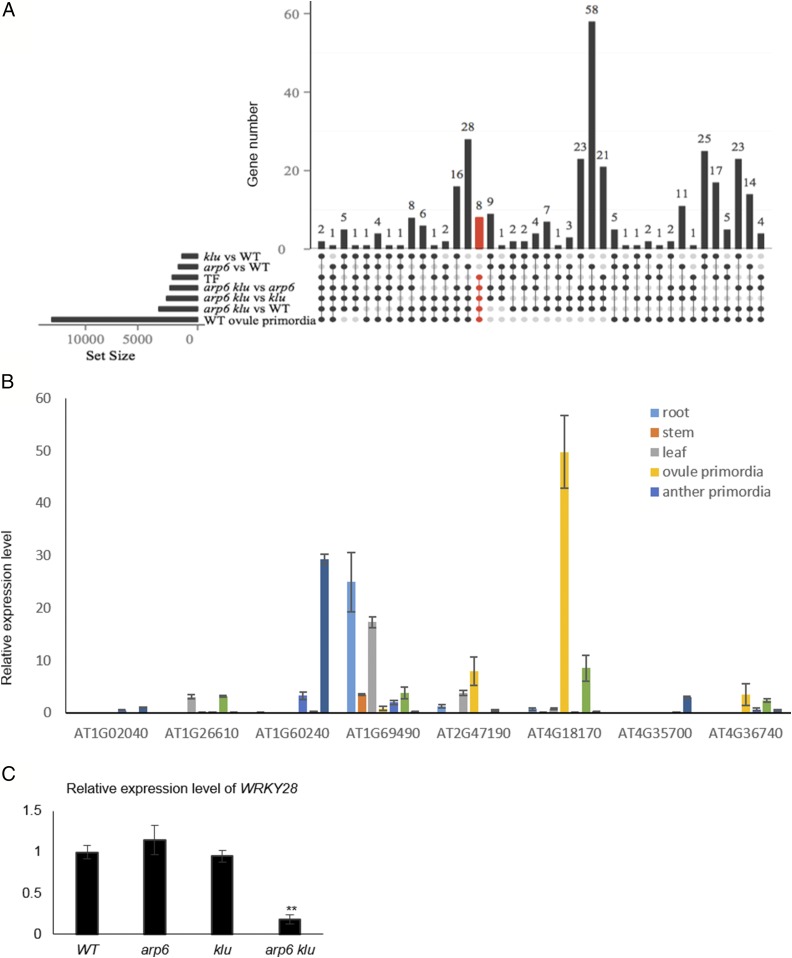
To gain insight into the mechanisms by which ARP6 and KLU cooperatively function in suppressing MMC specification, we identified genes with altered expression levels in arp6 klu ovules compared with WT and both single mutants. For RNA sequencing (RNA-seq), we extracted RNA from WT, arp6, klu, and arp6 klu flower buds. Because the MMC defect is specific to arp6 klu double mutant ovules, we screened for differentially expressed genes (fold change ≥ 2; P ≤ 0.05) in arp6 klu compared with WT, arp6, or klu that were not differentially expressed when comparing the arp6 or klu single mutants to WT. A total of 897 genes satisfied these criteria. To focus on genes potentially involved in megasporogenesis, we excluded genes that were not detected in a previously published RNA-seq dataset of ovule primordia undergoing megasporogenesis (16) and obtained 351 genes (Dataset S1) from the set of 897 genes.
Of these 351 genes, eight were TF genes (Fig. 2A) and therefore top candidates for genes with important regulatory roles. Among the eight candidate genes (Table S2), AT4G18170/WRKY28 (23) had the most enriched expression in ovule primordia with placenta (stage 2-I to 2-IV) relative to other tissues and organs, including mature ovule, anther primordia (stage 4–7) undergoing microsporogenesis, mature anther, leaf, root, and stem (Fig. 2B). WRKY proteins are plant-specific zinc-finger domain TFs implicated in mediating environmental and developmental responses (24). RNA-seq showed that WRKY28 transcript levels were significantly reduced in arp6 klu floral buds compared with WT, arp6, and klu floral buds (Table S2). To confirm the reduced expression of WRKY28 in arp6 klu floral buds, we excised ovule primordia with placenta (stage 2-I to 2-IV) and performed quantitative RT-PCR (qRT-PCR). This analysis showed a significant reduction in WRKY28 transcripts in arp6 klu ovule primordia compared with WT, arp6, and klu ovule primordia (Fig. 2C).
Fig. 2.
Identification of WRKY28 as a candidate gene downstream of ARP6 and KLU. (A) The left horizontal bar chart designates the following gene sets: differentially expressed genes between two genotypes, TF genes, and genes expressed in WT ovule primordia. The matrix diagram shows different combinations of overlap among these gene sets; the vertical bar chart indicates the gene number for a given combination. Dark/filled circles and light gray circles indicate gene sets included and excluded from a combination, respectively. For example, eight TF genes (marked in red) expressed in ovule primordia were differentially expressed in arp6 klu vs. arp6, klu, and WT, but not in klu vs. WT and arp6 vs. WT. (B) qRT-PCR analysis of the eight TF genes from A in different tissues. Data are means ± SD. (C) qRT-PCR analysis of WRKY28 mRNA levels in WT, arp6, klu, and arp6 klu ovules with placenta undergoing megasporogenesis. Data are means ± SD (n = 3; **P < 0.01).
H2A.Z Deposition at the WRKY28 Locus Requires ARP6 and KLU.
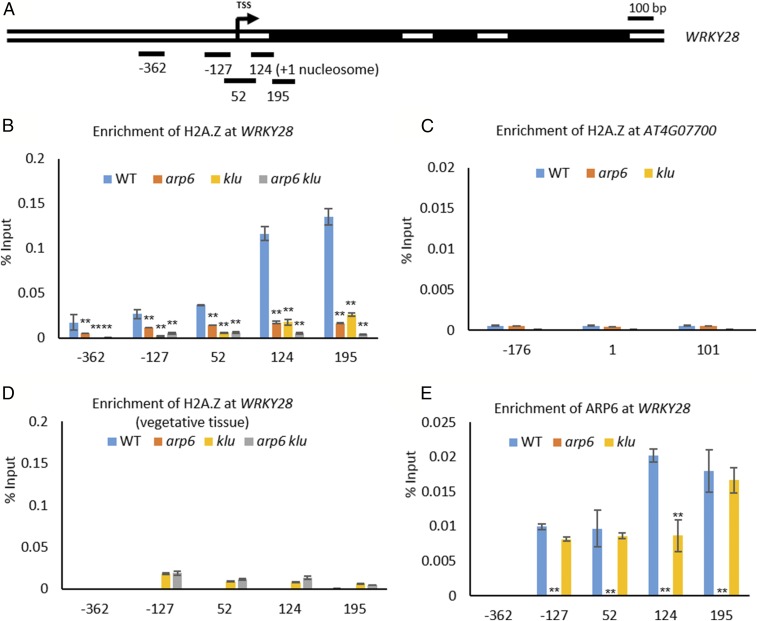
The highly conserved histone variant H2A.Z has been described as a “molecular rheostat” for the transcriptional control of gene expression, and it plays critical roles in the early development of multicellular organisms (25, 26). The replacement of H2A by H2A.Z in nucleosomes is mediated by the ATP-dependent chromatin remodeling complex SWR1 (27). Therefore, we hypothesized that ARP6 controls WRKY28 expression through the deposition of H2A.Z at the WRKY28 locus, similar to the role of ARP6 in regulating the expression of other genes (17, 28, 29).
To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) by using an H2A.Z antibody in WT and arp6 floral buds. In WT floral buds, we detected the highest level of H2A.Z occupancy downstream of the first nucleosome (+1) near the WRKY28 transcription start site (TSS) among the tested regions (Fig. 3 A and B). To confirm that the H2A.Z signals were in fact H2A.Z-specific, we included the gypsy-like transposon gene AT4G07700 (Fig. 3C), a gene previously shown to have H2A.Z-free nucleosomes, as a negative control (30). In WT floral buds, H2A.Z was not detected in any of the regions assayed in AT4G07700. As another negative control, we performed ChIP with the H2A.Z antibody on aerial nonreproductive tissues (i.e., lacking inflorescences or floral buds). No enrichment of H2A.Z was observed for any of the tested regions of WRKY28 in WT or any of the mutants (Fig. 3D), consistent with the low expression levels of WRKY28 in vegetative tissues (Fig. 2B). In arp6 floral buds, the enrichment of H2A.Z in the +1 nucleosome regions was greatly reduced (Fig. 3B). Taken together, these data suggest that the ARP6-containing SWR1 complex controls WRKY28 expression through H2A.Z deposition at the WRKY28 locus near the TSS.
Fig. 3.
KLU regulates H2A.Z deposition and ARP6 at WRKY28. (A) Gene diagram of WRKY28 with black boxes indicating exons and an arrow marking the TSS. Regions amplified by PCR primer sets are shown as black bars below the diagram; the numbers indicate the distance (in base pairs) to the TSS (designated as 0). (B) ChIP with polyclonal H2A.Z antibody to analyze H2A.Z enrichment at WRKY28 near the TSS in WT, arp6, klu, and arp6 klu floral buds (**P < 0.01 by Pearson’s χ2 test). (C) ChIP analysis for H2A.Z occupancy at the negative control gene At4g07700 in WT, arp6, and klu floral buds. (D) ChIP analysis for H2A.Z enrichment at WRKY28 in WT, arp6, klu, and arp6 klu aerial plant tissues lacking inflorescences. (E) ChIP with polyclonal ARP6 antibody to analyze ARP6 enrichment at WRKY28 in WT, klu, and arp6 floral buds (**P < 0.01 by Pearson’s χ2 test). (B–E) Values are means ± SD from two biological replicates.
Chromatin remodeling complexes are recruited to specific target genes by gene-specific factors (31, 32). In light of the reduced WRKY28 expression in arp6 klu ovule primordia, we tested whether H2A.Z deposition at the WRKY28 locus is also dependent on KLU by performing ChIP with the H2A.Z antibody in WT, klu, and arp6 klu floral buds. H2A.Z enrichment at WRKY28 was greatly reduced in klu compared with WT and almost completely depleted in arp6 klu (Fig. 3B). To determine whether KLU is required for the recruitment of SWR1 to the WRKY28 locus, we performed ChIP with ARP6 antibody to examine ARP6 occupancy at WRKY28. In WT, we detected ARP6 at the +1 nucleosome position near the TSS (Fig. 3E), coinciding with the enrichment of H2A.Z (Fig. 3B). ARP6 enrichment was not detected in arp6 floral buds (Fig. 3E), indicating that the signals in WT truly corresponded to ARP6 occupancy. In klu floral buds, ARP6 enrichment at the +1 nucleosome of WRKY28 was reduced compared with WT (Fig. 3E). These findings are consistent with a role of KLU in recruiting ARP6 to WRKY28 to deposit H2A.Z.
Preferential Expression of WRKY28 in the Hypodermal Somatic Cells Surrounding the MMC Is Controlled by ARP6 and KLU.
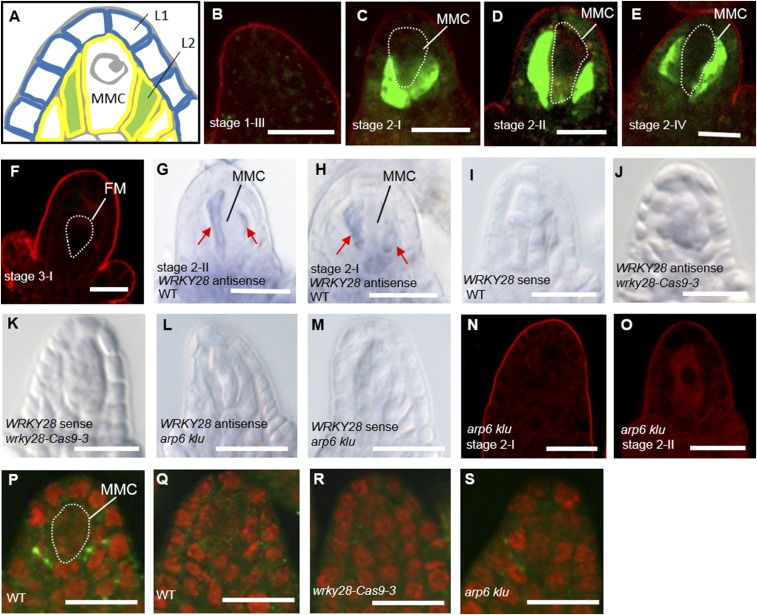
To analyze the expression pattern of WRKY28 in ovule primordia, we generated WRKY28 promoter-driven GFP (pWRKY28:GFP) lines in the WT background. Until stage 1-III, there was no GFP fluorescence in ovule primordia (Fig. 4B). The earliest GFP expression was detected exclusively in the hypodermal somatic cells flanking the developing MMC in the nucellus (Fig. 4A) at stage 2-I (Fig. 4C), and GFP expression in these cells persisted until stage 2-IV (Fig. 4 D and E). In postmeiotic stage 3-I ovules containing a teardrop-shaped functional megaspore, pWRKY28:GFP expression was not detected (Fig. 4F).
Fig. 4.
Cell-specific expression pattern of WRKY28 in ovules undergoing megasporogenesis. (A) Schematic representation of the distal structure of a premeiotic ovule with an MMC and the surrounding somatic cells (marked in green) in the hypodermal L2 cell layer (yellow outline), adjacent to epidermal L1 cells (blue outline). (B–F) pWRKY28:GFP expression in WT ovules from stage 1-III to stage 3-I. (G–M) In situ localization of antisense and sense WRKY28 mRNA in whole-mount WT (G–I), wrky28-Cas9-3 (J and K), and arp6 klu (L and M) ovules. (N and O) pWRKY28:GFP expression in arp6 klu ovules from stage 2-I to stage 2-II. No GFP signal was observed. Red signal corresponds to FM4-64 dye outlining the ovule. (P–S) WRKY28 (P) and preimmune serum (Q) immunolocalization in WT ovules and WRKY28 immunolocalization in wrky28-Cas9-3 (R) and arp6 klu (S) ovules. Green and red signals correspond to WRKY28 localization and propidium iodide signal, respectively. FM, functional megaspore. (Scale bars: 10 µm.)
Additional strategies confirmed the preferential expression of pWRKY28:GFP. By using ovule whole-mount in situ hybridization (33), we detected WRKY28 mRNA specifically in the hypodermal somatic cells surrounding the MMC (Fig. 4 G–I). As a negative control for the in situ hybridization experiment, we included a wrky28 mutant generated through CRISPR/Cas9 genome editing. Three independent wrky28-Cas9 mutant lines had nucleic acid deletions and/or insertions near the beginning of the WRKY28 ORF, leading to N-terminal truncation or premature translation termination (Fig. S2C). Western blotting using anti-WRKY28 polyclonal antibodies (Fig. S2D) showed reduced WRKY28 protein levels in these three lines, with the wrky28-Cas9-3 mutant having the greatest reduction (Fig. S2E). We therefore included the wrky28-Cas9-3 mutant for the in situ hybridization analysis and pistil qRT-PCR assay. The 2-nt deletion near the beginning of the WRKY28 ORF in wrky28-Cas9-3 (Fig. S2C) also caused reduced WRKY28 mRNA level as detected by the in situ hybridization (Fig. 4 J and K) and qRT-PCR experiments (Fig. S2H), probably because of nonsense-mediated mRNA decay in the wrky28-Cas9-3 line, indicating that the signals in the hypodermal somatic cells surrounding the MMC (Fig. 4 G and H) were specific to WRKY28 RNA. In addition, ovule whole-mount immunolocalization using anti-WRKY28 antibodies showed signals within the hypodermal somatic cells surrounding the MMC in WT (Fig. 4 P and Q) but not in wrky28-Cas9-3 ovule primordia (Fig. 4R). These results demonstrate the specific expression of WRKY28 in hypodermal somatic cells surrounding the MMC. To our knowledge, this is the first marker gene for cells surrounding the MMC in the hypodermal cell layer.
We next examined WRKY28 expression in arp6 klu ovules. The signals of pWRKY28:GFP (Fig. 4 N and O), WRKY28 mRNA (Fig. 4 L and M), and WRKY28 protein (Fig. 4S) in the hypodermal somatic cells surrounding the MMC were greatly reduced in arp6 klu ovules compared with WT (Fig. 4 C–E, G, H, and P), indicating that the cell-specific expression of WRKY28 in ovule primordia is controlled by ARP6 and KLU.
Loss of WRKY28 Phenocopies the Defects of arp6 klu Ovule Primordia.
To test the hypothesis that WRKY28 functions downstream of ARP6 and KLU in restricting the MMC fate to a single cell, we first analyzed whether WRKY28 loss of function would lead to multiple MMC-like cells as observed in arp6 klu. Publicly available lines with transfer DNA (T-DNA) insertions in the WRKY28 locus are limited; none of the available T-DNA lines have insertions in the WRKY28 coding region, and one line with an insertion in the WRKY28 promoter did not have reduced WRKY28 mRNA levels (Fig. S2 A and B). We therefore generated wrky28 mutants to assess loss of function by using two different strategies, CRISPR/Cas9-induced mutations and the conversion of WRKY28 into a dominant repressor.
Ovules in three independent wrky28-Cas9 lines with reduced WRKY28 protein levels (Fig. S2 C–E) contained multiple enlarged MMC-like cells (Fig. 5 B, F, and G) at a much higher frequency than WT (Fig. 5 A, D, and E), showing that wrky28-Cas9 plants phenocopied the defects observed in arp6 klu. Additionally, the enlarged cells in premeiotic wrky28-Cas9-3 ovules exhibited nuclear AGO9 signal (Fig. 5 H and I), as observed in arp6 klu mutant ovules. Thus, WRKY28 represses ectopic MMC-like cell fate.
Fig. 5.
Ectopic MMC specification and aberrant female gametophyte development in mutants with disrupted WRKY28 function. (A) Premeiotic WT ovule with a single MMC. (B and C) Premeiotic wrky28-Cas9-3 (B) and p35S:WRKY28-SRDX (C) ovules with more than one enlarged cell. (D) Quantification of aberrant MMC specification (**P < 0.01 by Pearson’s χ2 test). (E–G) Confocal sections of premeiotic WT (E) and wrky28-Cas9-3 (F and G) ovules stained by propidium iodide. (H and I) AGO9 immunolocalization in premeiotic wrky28-Cas9-3 ovules. (J and K) Callose deposition (J) and ovule morphology (K) in wrky28-Cas9-3 ovules. (L and M) pDMC1:GFP expression in WT (L) and wrky28-Cas9-3 (M) ovules undergoing meiosis. (N) Postmeiotic wrky28-Cas9-3 ovule. (O) pAKV:H2B-YFP expression in a postmeiotic wrky28-Cas9-3 ovule. (P and Q) wrky28-Cas9-3 ovules at mature stage. (R) Siliques of WT and wrky28-Cas9-3. Red arrows point to aborted seeds. (S) Quantification of seed set in WT and wrky28 mutants. Data are means ± SD (n = 10; **P < 0.01). (T) AGO9 immunolocalization in premeiotic p35S:WRKY28-SRDX ovules. (U–X) Signal corresponding to pKNU:KNU-Venus in premeiotic p35S:WRKY28-SRDX ovules. Numbers denote the frequencies of the phenotypes shown. Abnormally enlarged cells are outlined by a white dashed line or indicated by a red arrowhead. FM, functional megaspore. (Scale bars: A–C, E–Q, and T–X, 10 µm; R, 1 mm.)
We next tested if WRKY28 functions as an activator of gene expression in ovules, similar to its reported function in other tissues (23). For this analysis, we examined the expression levels of ISOCHORISMATE1 (ICS1), whose expression is directly activated by WRKY28 in leaf protoplasts (23). In all three wrky28-Cas9 lines, ICS1 expression in the pistils of stage 9–11 flower buds was significantly reduced compared with WT (Fig. S2F).
In wrky28-Cas9-3 ovules undergoing meiosis, callose was detected only in the intermediate walls of a single MMC rather than in all enlarged cells (Fig. 5 J and K). We further examined these cells for the expression of a meiosis-related gene, AtDMC1, which is specifically expressed in MMCs undergoing meiosis (17) (Fig. 5L). In wrky28-Cas9 ovules, only one enlarged cell expressed pAtDMC1:GFP (Fig. 5M). In addition, the functional megaspore marker pAKV:H2B-YFP was not detected in the multiple enlarged cells adjacent to the functional megaspore in wrky28-Cas9-3 (Fig. 5O), consistent with the analysis of pAKV:H2B-YFP in arp6 klu (Fig. S1H).
Taken together, these results suggest that, even though multiple enlarged cells acquired MMC characteristics in wrky28-Cas9 ovules, only one cell fully differentiated into an MMC and underwent meiosis, similar to the observations in arp6 klu ovules (Fig. 1 J and K).
In addition to multiple enlarged cells, wrky28-Cas9-3 ovules more frequently contained abnormal female gametophytes (Fig. 5 N–Q) compared with WT (Fig. S1 A and I). This defect probably contributed to the reduced fertility of the wrky28-Cas9 lines (Fig. 5 R and S).
As a second strategy to probe the biological function of WRKY28, we converted WRKY28 into a transcriptional repressor by using the chimeric repressor silencing approach (34). WRKY28 was fused with an SRDX repressor domain, a 12-aa motif that converts TFs into dominant repressors (34), and the transgene was expressed under the 35S promoter. The SRDX line had reduced seed set (Fig. 5S) and contained multiple enlarged cells in ovule primordia more frequently (38.2%, n = 325) than WT plants (5.9%, n = 324; Fig. 5 A, C, and D). The enlarged cells in p35S:WRKY28-SRDX ovules also showed nuclear AGO9 localization pattern (Fig. 5T) and expressed the MMC marker gene KNU (pKNU:KNU-Venus; Fig. 5 U–X), indicating that the cells were distinct from the surrounding somatic cells and had acquired some of the molecular characteristics of MMCs. ICS1, a direct target of WRKY28, was expressed at a lower level in p35S:WRKY28-SRDX pistils than in WT (Fig. S2F), indicating that WRKY28-SRDX repressed ICS1 expression, whereas WRKY28 normally promotes ICS1 expression. The expression of WRKY28 paralogs in p35S:WRKY28-SRDX and wrky28-Cas9-3 pistils was comparable to their expression in WT (Fig. S2 G and H), indicating that the presence of multiple MMC-like cells in the WRKY28-SRDX and wrky28-Cas9 lines was not a result of misexpression of WRKY28 paralogs. Taken together, our results demonstrate that WRKY28 suppresses MMC fate in hypodermal cells surrounding the MMC.
WRKY28 Overexpression Partially Complements the MMC Specification Defects in arp6 klu Ovules.
To test whether WRKY28 functions downstream of ARP6 and KLU to inhibit the formation of multiple MMC-like cells in ovule primordia, we transformed arp6+/− klu−/− plants with a full-length WRKY28 genomic fragment under the control of the 35S promoter. We used arp6+/− klu−/− plants for transformation because fertility was severely compromised in double homozygous arp6 klu plants. In the T1 generation, six independent arp6 klu transformants carrying the p35S:WRKY28 transgene were obtained, and all had elongated siliques (Fig. 6A) and increased seed set (Fig. 6B and Table S3) compared with arp6 klu plants. We examined the MMC phenotype in the ovule primordia of two of the six arp6 klu p35S:WRKY28 lines. In both lines, the phenotype of multiple MMC-like cells in premeiotic ovules occurred less frequently (28.6%, n = 262; and 39.8%, n = 231) than in arp6 klu plants (55.9%, n = 345) but more frequently than in WT plants (5.9%, n = 324; Fig. 6 C and D). This partial complementation indicates that WRKY28 is one player functioning downstream of ARP6 and KLU to inhibit ectopic MMC formation in ovule primordia.
Fig. 6.
Partial complementation of arp6 klu MMC specification defects by WRKY28 overexpression. (A) Siliques of plants with the indicated genotypes. (Scale bar: 1 mm.) (B) Quantification of seed set in arp6 klu plants with and without the WRKY28 transgene. Values are means ± SD (n = 6; **P < 0.01). (C) Quantification of aberrant MMC specification in premeiotic ovules (**P < 0.01 by Pearson’s χ2 test). (D) DIC images of premeiotic ovules with the indicated genotypes. Numbers denote the frequencies of the phenotypes shown. Red arrows indicate multiple enlarged cells. (Scale bar: 10 µm.) (E) Signal corresponding to pKNU:WRKY28-GFP expression in a premeiotic ovule (WT background). (F) Proposed model for the coordinated action of KLU and SWR1 in suppressing ectopic MMC fate by promoting WRKY28 expression in the hypodermal somatic cells surrounding the MMC. II, inner integument; OI, outer integument.
As arp6 klu p35S:WRKY28 plants still generated a normal MMC, we hypothesized that ectopic expression of WRKY28 in the MMC is not sufficient to inhibit normal development of the MMC itself. To test this hypothesis, we specifically expressed WRKY28-GFP in the MMC by using the MMC-specific promoter KNU (Fig. 6E) (21). pKNU:WRKY28-GFP transgenic plants exhibited normal MMC specification (95.4% of ovules showing a single enlarged MMC, n = 368; Fig. 4 C and D) and normal seed set (97.9%, n = 658; compared with 98.0%, n = 540 in WT). These results suggest that ectopic WRKY28 expression in the MMC is not sufficient to suppress MMC fate. This finding, together with the specific expression of WRKY28 in hypodermal cells surrounding the MMC, suggests that the main function of WRKY28 is to suppress MMC fate in the somatic cells surrounding the MMC.
Discussion
Although several signaling pathways important for germ-line specification in plants—for example, the TPD1–MSP1 ligand-receptor signaling pathway and the small RNA-involved pathways, have been uncovered in many angiosperms including Arabidopsis, maize, and rice (7, 8, 12, 20, 35), the mechanisms underlying the restriction of the germ-line precursor to a single cell remain largely unknown. Here, we show that the TF gene WRKY28 is specifically expressed in hypodermal cells surrounding the MMC, cells that appear to have the potential to develop into MMCs (Fig. 6F). Disruption of WRKY28 function resulted in these cells acquiring the cytological and molecular characteristics of MMCs. We therefore uncovered a mechanism in which the local expression of WRKY28 in somatic cells surrounding the MMC suppresses excessive MMC specification. This mechanism is different from that revealed in a recent study in which a group of redundant cyclin-dependent kinase inhibitors of the KIP-RELATED PROTEINs (KRPs) functions in ensuring a single MMC formation by preventing over-proliferation (i.e., additional mitotic divisions) of the already specified MMC (36). The excessive MMC cells in the krp4/6/7 triple mutants often appeared similar in size and were located side by side or one on top of another of the already specified MMC (36); however, supernumerary MMC-like cells in the wrky28 and arp6 klu mutants were rather randomly located, and these cells differed in size. Moreover, the excessive MMC-like cells in the wrky28 and arp6 klu mutants are positioned in the place of the hypodermal cells surrounding the MMC, indicating that loss of WRKY28 function in these cells may have led the cell to become enlarged and adopt some characteristics similar to that of MMC. We further showed that the chromatin remodeling complex SWR1 (29, 37–39) mediates the incorporation of the histone variant H2A.Z at WRKY28 to promote its expression. We found that H2A.Z deposition at WRKY28 by SWR1 is dependent on the Arabidopsis cytochrome P450 gene KLU. As ARP6 occupancy at WRKY28 is reduced in klu mutants, it is likely that KLU helps recruit SWR1 to the WRKY28 locus. Intriguingly, KLU is expressed preferentially in the inner integument of the ovule (22), which lies opposite the cells expressing WRKY28 along the proximal–distal axis. These observations suggest that the regulation of WRKY28 expression by KLU is not cell-autonomous. KLU is known to play an important role in determining plant organ size in a non–cell-autonomous manner (20, 22). The KLU-dependent signal has been proposed to have a range of activity beyond individual organs and flowers and to be distinct from classical phytohormones (21). We speculate that the same KLU-dependent mobile signal provides positional information to repress MMC fate in somatic cells. If so, how the MMC escapes this suppression is unknown.
Taken together, our findings elucidate a previously unknown mechanism at work in somatic cells surrounding the germ-line precursor to prevent ectopic germ-line formation. This mechanism involves the TF WRKY28 that suppresses MMC fate, the chromatin remodeling complex SWR1 that promotes WRKY28 expression through H2A.Z deposition, and, presumably, a KLU-dependent mobile signal that helps recruit SWR1 to the WRKY28 locus (Fig. 6F).
Materials and Methods
Detailed descriptions of the study materials and methods are provided in SI Materials and Methods.
Materials and Growth Conditions.
WT Arabidopsis thaliana (Col-0 ecotype), arp6 (Garlic_599_G03), klu (SALK_024697C), and sef (CS822749) were grown under 16 h light/8 h dark at 22 °C.
Histological Analysis.
Cleared ovules, in situ hybridization, and callose staining were performed as previously described (40). Immunostaining of ovules and imaging of fluorescent signals (GFP, YFP, and antibody staining) were conducted as described in a previous study (40).
RNA-Seq Analysis.
RNA isolation and sequencing was performed as previously described (16). The RNA-seq datasets were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession no. SRP124412).
ChIP-qPCR.
ChIP-qPCR was performed as previously described (38). Primers used in this study are listed in Table S4.
Supplementary Material
Acknowledgments
We thank Dr. W. Yang for sharing the pAKV:H2B-YFP marker, R. Yadegari for providing the pDMC1:GFP marker, R. Deal and R. Meagher for sharing the H2A.Z and ARP6 antibodies, Dr. B. Sun for the pKNU:KNU-Venus line, and Z. Yang, L. Xia, and I. Lavagi for critical reading. This work was supported by National Natural Science Foundation of China (NSFC) Grants 31470284 (to Y.Q.), 31522009 (to Y.Q.), U1605212 (to Y.Q.), 31761130074 (to Y.Q.), 31600249 (to L.Z.), 31571264 (to H.Z.), and 31700278 (to Z.S.), Newton Advanced Fellowship NA160391 (to Y.Q.), and Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University Grant 1122yb035 (to L.Z.).
Footnotes
Conflict of interest statement: R.P. and reviewer D.Z. are co-editors on a 2017 special issue in Frontiers in Plant Science on Molecular and Cellular Plant Reproduction.
Data deposition: The data reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA) database, https://www.ncbi.nlm.nih.gov/sra (accession no.SRP124412).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716054115/-/DCSupplemental.
References
- 1.Yang WC, Shi DQ, Chen YH. Female gametophyte development in flowering plants. Annu Rev Plant Biol. 2010;61:89–108. doi: 10.1146/annurev-arplant-042809-112203. [DOI] [PubMed] [Google Scholar]
- 2.Grossniklaus U, Schneitz K. The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol. 1998;9:227–238. doi: 10.1006/scdb.1997.0214. [DOI] [PubMed] [Google Scholar]
- 3.Bachelier JB, Friedman WE. Female gamete competition in an ancient angiosperm lineage. Proc Natl Acad Sci USA. 2011;108:12360–12365. doi: 10.1073/pnas.1104697108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossniklaus U. Plant germline development: A tale of cross-talk, signaling, and cellular interactions. Sex Plant Reprod. 2011;24:91–95. doi: 10.1007/s00497-011-0170-3. [DOI] [PubMed] [Google Scholar]
- 5.Feng X, Zilberman D, Dickinson H. A conversation across generations: Soma-germ cell crosstalk in plants. Dev Cell. 2013;24:215–225. doi: 10.1016/j.devcel.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, et al. OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J. 2008;54:375–387. doi: 10.1111/j.1365-313X.2008.03426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nonomura K, et al. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell. 2003;15:1728–1739. doi: 10.1105/tpc.012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheridan WF, Avalkina NA, Shamrov II, Batygina TB, Golubovskaya IN. The mac1 gene: Controlling the commitment to the meiotic pathway in maize. Genetics. 1996;142:1009–1020. doi: 10.1093/genetics/142.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheridan WF, Golubeva EA, Abrhamova LI, Golubovskaya IN. The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics. 1999;153:933–941. doi: 10.1093/genetics/153.2.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CJ, et al. Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development. 2012;139:2594–2603. doi: 10.1242/dev.077891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt A, et al. Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol. 2011;9:e1001155. doi: 10.1371/journal.pbio.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Z, et al. The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr Biol. 2017;27:1597–1609 e2. doi: 10.1016/j.cub.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastasiou E, et al. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell. 2007;13:843–856. doi: 10.1016/j.devcel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson S, Stransfeld L, Adamski NM, Breuninger H, Lenhard M. KLUH/CYP78A5-dependent growth signaling coordinates floral organ growth in Arabidopsis. Curr Biol. 2010;20:527–532. doi: 10.1016/j.cub.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 15.Adamski NM, Anastasiou E, Eriksson S, O’Neill CM, Lenhard M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci USA. 2009;106:20115–20120. doi: 10.1073/pnas.0907024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, et al. Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J. 2014;80:615–628. doi: 10.1111/tpj.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Y, et al. ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell. 2014;26:1612–1628. doi: 10.1105/tpc.113.120576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodríguez-Leal D, León-Martínez G, Abad-Vivero U, Vielle-Calzada JP. Natural variation in epigenetic pathways affects the specification of female gamete precursors in Arabidopsis. Plant Cell. 2015;27:1034–1045. doi: 10.1105/tpc.114.133009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baroux C, Raissig MT, Grossniklaus U. Epigenetic regulation and reprogramming during gamete formation in plants. Curr Opin Genet Dev. 2011;21:124–133. doi: 10.1016/j.gde.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Olmedo-Monfil V, et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payne T, Johnson SD, Koltunow AM. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development. 2004;131:3737–3749. doi: 10.1242/dev.01216. [DOI] [PubMed] [Google Scholar]
- 22.March-Díaz R, García-Domínguez M, Florencio FJ, Reyes JC. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007;143:893–901. doi: 10.1104/pp.106.092270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Verk MC, Bol JF, Linthorst HJ. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 2011;11:89. doi: 10.1186/1471-2229-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian V, Fields PA, Boyer LA. H2A.Z: A molecular rheostat for transcriptional control. F1000Prime Rep. 2015;7:01. doi: 10.12703/P7-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: Essential contributions of histone variants to mammalian development and disease. Nat Rev Genet. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 28.Cai H, et al. ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol. 2017;214:1579–1596. doi: 10.1111/nph.14521. [DOI] [PubMed] [Google Scholar]
- 29.Dai X, et al. H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis. Mol Plant. 2017;10:1274–1292. doi: 10.1016/j.molp.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 32.Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hejátko J, et al. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc. 2006;1:1939–1946. doi: 10.1038/nprot.2006.333. [DOI] [PubMed] [Google Scholar]
- 34.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 35.Nonomura K, et al. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, et al. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science. 2017;356:eaaf6532. doi: 10.1126/science.aaf6532. [DOI] [PubMed] [Google Scholar]
- 37.Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell. 2007;19:74–83. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Sura W, et al. Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell. 2017;29:791–807. doi: 10.1105/tpc.16.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escobar-Guzmán R, Rodríguez-Leal D, Vielle-Calzada JP, Ronceret A. Whole-mount immunolocalization to study female meiosis in Arabidopsis. Nat Protoc. 2015;10:1535–1542. doi: 10.1038/nprot.2015.098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.