Significance
Structure–function aspects of dynamic, membrane-associated proteins are difficult to mechanistically define. In this study, computational and biophysical methods are applied to identify a ubiquitin-interaction domain essential for the activity of a bacterial cytotoxin, ExoU. Introduction of mutations that diminish or improve ubiquitin interaction alters the biochemical and biological activities of the toxin. This strategy and these verification procedures may be useful where traditional NMR or crystallographic technologies to analyze protein–protein interactions are limited. ExoU is a member of a family of newly discovered ubiquitin-activated enzymes encoded by several bacterial pathogens. Understanding the details of intracellular enzyme activation will be critical to the development of inhibitors aimed at reducing tissue damage during infection by a variety of organisms.
Keywords: computational modeling, DEER, continuous-wave spectroscopy, ubiquitin-binding domain, Rosetta
Abstract
ExoU is a type III-secreted cytotoxin expressing A2 phospholipase activity when injected into eukaryotic target cells by the bacterium Pseudomonas aeruginosa. The enzymatic activity of ExoU is undetectable in vitro unless ubiquitin, a required cofactor, is added to the reaction. The role of ubiquitin in facilitating ExoU enzymatic activity is poorly understood but of significance for designing inhibitors to prevent tissue injury during infections with strains of P. aeruginosa producing this toxin. Most ubiquitin-binding proteins, including ExoU, demonstrate a low (micromolar) affinity for monoubiquitin (monoUb). Additionally, ExoU is a large and dynamic protein, limiting the applicability of traditional structural techniques such as NMR and X-ray crystallography to define this protein–protein interaction. Recent advancements in computational methods, however, have allowed high-resolution protein modeling using sparse data. In this study, we combine double electron–electron resonance (DEER) spectroscopy and Rosetta modeling to identify potential binding interfaces of ExoU and monoUb. The lowest-energy scoring model was tested using biochemical, biophysical, and biological techniques. To verify the binding interface, Rosetta was used to design a panel of mutations to modulate binding, including one variant with enhanced binding affinity. Our analyses show the utility of computational modeling when combined with sensitive biological assays and biophysical approaches that are exquisitely suited for large dynamic proteins.
Opportunistic pathogens, such as Pseudomonas aeruginosa, are increasingly problematic in nosocomial settings. Intrinsic resistance to a wide variety of antibiotics makes P. aeruginosa particularly difficult to treat (1, 2). A major virulence determinant of P. aeruginosa is expression of a type III secretion system (T3SS). This system is used to inject effector enzymes directly into the cytoplasm of host cells. Up to four different enzymes can be delivered to the host cytosol by P. aeruginosa (3–6). Enzymatic activities generally result in cytoskeletal changes and cytotoxicity, aiding P. aeruginosa in colonization and evasion of the innate immune system. In vitro, the enzymatic activities of the effectors are undetectable unless a cognate eukaryotic protein cofactor is present in the reaction (3, 7–9). These noncovalent protein–protein interactions between enzyme and cofactor could be potential therapeutic targets. Our studies use the effector, ExoU, and its interaction with its cognate cofactor, ubiquitin, as a model system to study the mechanisms mediating enzyme activation within a eukaryotic environment.
ExoU is a 74-kDa patatin-like phospholipase (10–12). Upon interaction with ubiquitin or ubiquitylated proteins, ExoU cleaves membrane phospholipids, resulting in host cell lysis (8, 13). ExoU is one of several bacterial effectors known to interact with ubiquitin or ubiquitin-dependent pathways (8, 14–17). Additionally, ubiquitin-activated ExoU orthologs are encoded in the genomes of divergent gram-negative species (16). We have utilized electron paramagnetic resonance (EPR) spectroscopy in conjunction with site-directed spin labeling (SDSL) to analyze ExoU structure (18) and dynamics in response to the addition of ubiquitin, membrane substrates, or both (19, 20). Our analyses suggest that ExoU undergoes major conformational changes in the presence of both ubiquitin and membrane substrates not found with either component alone (19, 20). Additionally, ubiquitin has been shown to bind ExoU in the absence of substrate, suggesting a ubiquitin-binding interface is accessible in the apoenzyme state (8, 21). However, previous attempts to analyze the ExoU ubiquitin complex by cocrystallization and NMR in our group have been unsuccessful to date.
Most structural models of protein–protein interactions (PPIs) are determined using X-ray crystallography and are limited by the ability to obtain a crystal. Other techniques include NMR for small complexes (≤80 kDa) and electron microscopy for large complexes (>110 kDa). To date, solved structures of PPIs remain in the minority of known PPIs (22–24). Given the importance of PPIs (25–27), there has been strong interest in developing in silico techniques for protein structure modeling to improve the breadth and rate of protein structure determination (28–30). While techniques are constantly improving, they are currently limited by sampling the available conformational space (29). Similarly, low-resolution techniques like SDSL and EPR can give useful structural information under conditions incompatible with conventional techniques (i.e., presence of membrane lipids), but are not sufficient for high-resolution structural determination on their own. The use of integrative modeling techniques, combining sparse structural data with computational modeling, has been shown to significantly increase modeling accuracy and efficiency (31–33). The Rosetta protein modeling suite is particularly amenable to integrative modeling (34–37), and has been used in conjunction with SDSL EPR data to create high-resolution models of protein structures (38).
Here, we use RosettaEPR (34) to incorporate intermolecular double electron–electron resonance (DEER) distance measurements and perform global docking of monoubiquitin (monoUb) to ExoU with the RosettaDock application (39). Additional DEER distance measurements, continuous-wave (CW) EPR, and mutagenesis studies are used to test this model. Validation of the model is supported by both disruption and enhancement of the binding interface using Rosetta-generated predictions. Cumulatively, we develop and validate a model of the ExoU–monoUb interaction and show that RosettaDock can be used in conjunction with DEER distances to perform discovery-based global docking of large protein–protein complexes with low binding affinity. Ideally, this approach demonstrates a simple but effective way to determine high-resolution structures for PPIs that is complementary to conventional structural techniques.
Results
Global Docking of ExoU and MonoUb.
A C-terminal region of ExoU (amino acids 480 to 683) has been shown to be important for ubiquitin interaction (21). Previous work, based on structural homology to isopeptidase T, suggested that the C-terminal four-helix bundle of ExoU (amino acids 580 to 687) was a potential binding site for monoUb. Mutagenesis, solid-phase binding studies, and manual docking with DEER distance restraints indicated that the location of the binding domain likely included contacts between ExoU residues located at Q623, K611, Y619, and D625 (21). Subsequent CW EPR analyses of 41 individually labeled sites in the C-terminal region, however, failed to identify any change in spin label mobility upon the addition of monoUb alone (19). Further, power saturation studies in the presence of oxygen and nickel ethylenediaminediacetic acid as relaxation agents indicated that several of the identified residues were likely interacting with or intercalated within the membrane bilayer in the holo state (19). Using structural homology as a starting point limited the sampling space and biased the previous output models. Overall, our data suggest that a monoUb-binding interface is located within the C terminus but unlikely to be associated with the four-helix bundle of ExoU (19, 21).
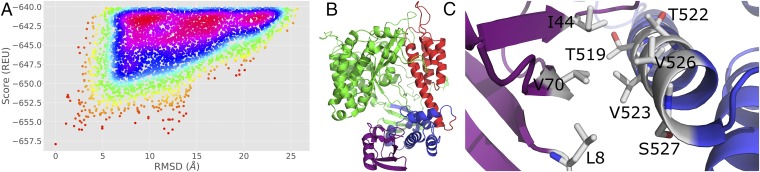
Published DEER distances were used as docking restraints for the RosettaDock (39) global docking protocol via the RosettaEPR scoring module (34). Approximately 30,000 decoy models of the ExoU [Protein Data Bank (PDB) ID code 3TU3] and ubiquitin (PDB ID code 1UBQ) interaction were generated. Ubiquitin was docked to a near–full-length ExoU to avoid biasing the output models. The model with the lowest Rosetta energy unit score was used as a reference structure to calculate the Cα root-mean-square deviation (rmsd) to each model. Score vs. rmsd was plotted for the top 5,000 models revealing a “binding funnel,” suggesting convergence of the data to a low-energy structure (Fig. 1A). The lowest-energy model (Fig. 1 B and C) depicts the interaction of the ubiquitin with α-helix 18 (11) in the bridging domain of ExoU (amino acids 480 to 580). In this model, the hydrophobic patch of ubiquitin, a known hotspot of ubiquitin binding (40–42), interacts with two solvent-exposed valines, V523 and V526, as well as the methyl groups of two threonines, T519 and T522. The best scoring model possesses good constraint satisfaction as determined by using the Rosetta cst info function with a theoretical best score of −1 (SI Appendix, Fig. S1). The best scoring model was relaxed using the Rosetta fast relax application and used as a working model to make predictions.
Fig. 1.
Modeling of the ExoU–monoUb interaction. (A) Score (Rosetta energy unit; REU) vs. rmsd plot of the top 5,000 scoring models. A positive correlation suggests convergence toward a low-energy structure. (B) Best scoring complex. ExoU catalytic domain (green), bridging domain (blue), and membrane localization domain (red) in complex with ubiquitin (purple). (C) The ubiquitin I44 hydrophobic patch interacts with helix 18 in the ExoU bridging domain.
CW EPR Scan of the Bridging Domain.
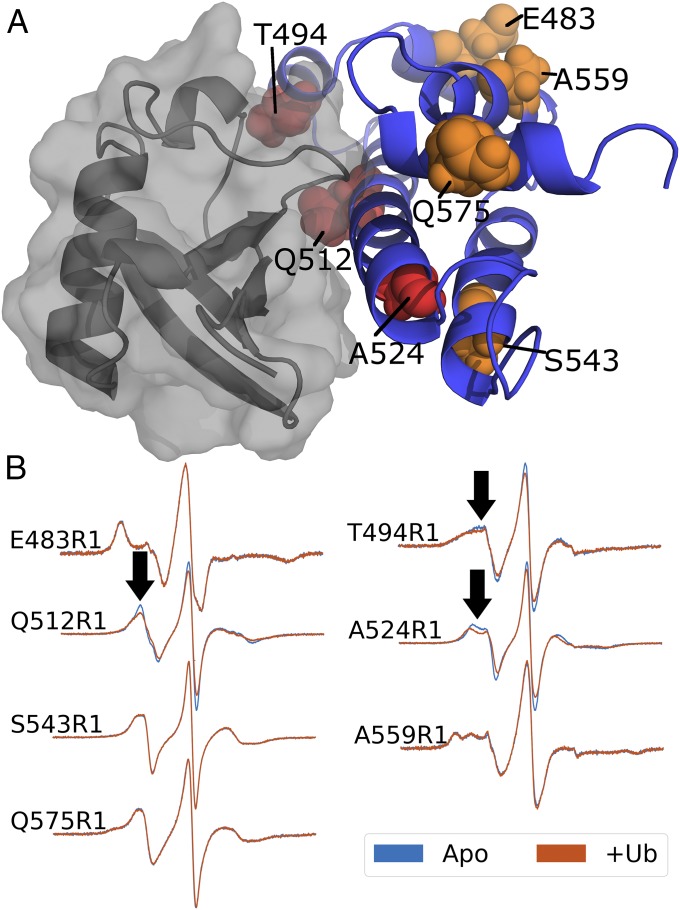
In parallel to in silico docking, we performed an SDSL scan of the bridging domain of ExoU. If ubiquitin interacts with ExoU near a labeled cysteine residue, the rotational freedom of the spin label will be restricted by quaternary contacts resulting in a broadening of the CW spectrum upon the addition of ubiquitin. Forty-one sites previously tested in the C-terminal four-helix bundle revealed no such change in their CW spectra in the presence of ubiquitin alone (19), supporting our hypothesis that ubiquitin does not interact with ExoU within the four-helix bundle. To extend this screen, seven ExoU variants were constructed in the bridging domain. Sites were selected to maximize coverage with at least one labeling site in each helix. Three of the seven sites in the bridging domain demonstrate broadening of spectral peaks upon the addition of ubiquitin (Fig. 2B). Two of these sites, Q512R1 and A524R1, are located within α-helix 18 and the third, T494R1, is within α-helix 17. Comparison with the initial RosettaDock model reveals ubiquitin binding near all three SDSL sites (Fig. 2A), suggesting the observed spectral changes are a result of new contacts limiting the motion of the spin labels. The remaining four sites show no response to the addition of ubiquitin (Fig. 2B). To control for structural variability introduced by SDSL, we performed activity assays on all seven SDSL variants. Under saturating conditions, all SDSL variants retain at least 70% WT enzyme activity (SI Appendix, Fig. S2). These data indicate that the overall tertiary fold of these ExoU derivatives is sufficient for ubiquitin interaction and catalysis.
Fig. 2.
SDSL scan of the ExoU bridging domain. (A) Cartoon model of the ExoU bridging domain in complex with ubiquitin (surface). SDSL sites are shown as spheres. Orange residues show no change in CW spectra upon addition of ubiquitin. Red residues indicate sites that show line broadening upon the addition of ubiquitin. (B) CW EPR spectra of seven SDSL sites screened in the presence (red) and absence (blue) of ubiquitin. Arrows indicate small but significant changes in CW spectra. Also, note the increased width of the center lines, reflected in the decreased amplitudes of the normalized spectra.
Model Testing with DEER.
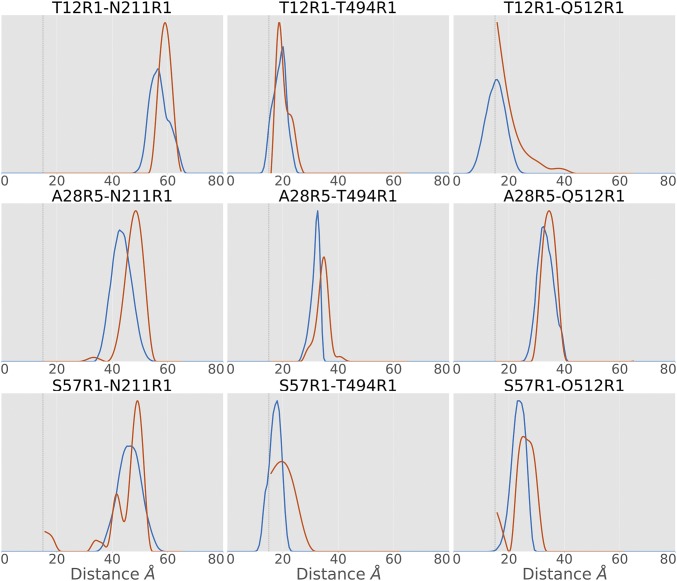
Based on our model, specific distances between ExoU and ubiquitin can be predicted. Using mtsslWizard (43, 44), spin label conformer ensembles were modeled into three sites for both ExoU and monoUb and nine distance distributions between pairs were calculated. Each pair was experimentally tested using SDSL and DEER to collect distance distributions (Fig. 3 and SI Appendix, Fig. S3 and Table S1). All nine tested pairs have significant overlap (>40%) between the experimental and the predicted distance distributions (Fig. 3 and SI Appendix, Table S1) and excellent agreement in identification of the most probable distance (SI Appendix, Table S1). All populations were validated for significance using DEERconstruct (45). One pair, T12R1–Q512R1, is predicted to have a significant portion of the distance distribution below 15 Å (Fig. 3). While the distance distribution below 15 Å cannot be determined using DEER analysis (46), the distribution above 15 Å closely resembles that of the predicted distribution. At least one peak from eight of the nine distance distributions is within a single SD of the predicted peak (SI Appendix, Table S1), suggesting strong agreement of the data with the model. The remaining distance, A28R5 to N211R1, is only 5 Å larger than predicted and shares 49% overlap with the predicted distribution. Differences observed between the predicted distance and experimental distance distributions likely arise from inherent rigidity of the in silico model, the lack of favorable conformer bias in mtsslWizard, and small conformational differences that may arise due to the presence of the spin label. Importantly, the shortest distances are in good agreement with the initial RosettaDock model and support the hypothesis that ubiquitin interacts with ExoU in the bridging domain.
Fig. 3.
Comparison of predicted distance distributions (blue) with experimental DEER distance distributions (red). Most experimental distances are in good agreement with predicted distance distributions. All experimental results share a significant population in agreement with the predicted distance distribution. Gray dashed lines indicate the lower limit of DEER.
Mutagenesis of the Predicted Binding Interface.
The DEER distances acquired support the initial RosettaDock model, which predicts a testable interface for ubiquitin binding. We engineered four nonconservative mutations hypothesized to disrupt this interface. Three of these residues were selected based on predictions of an in silico alanine scan (ΔΔG > 1 kcal/mol) and the change in solvent-accessible surface area (ΔSASA > 50%) (SI Appendix, Table S2). The fourth mutation, T522D, was chosen due to a high predicted ΔΔG and proximity to the other sites. As a negative control, we constructed T554D, a mutation in the bridging domain, which is outside of the ubiquitin-binding interface in our model. To determine binding affinities, CW EPR spectra of A28R5 monoUb with different concentrations of ExoU variants were obtained. Complex formation significantly reduces the isotropic tumbling rate of the spin-labeled ubiquitin, allowing quantitation of bound and free populations (21). The resulting binding curves were fit using a single-site model to determine the dissociation constant (Kd; Table 1). WT ExoU binds A28R5 ubiquitin with low-micromolar affinity (122.8 ± 11.4 µM) typical of ubiquitin-binding domains. E520R ExoU, designed to disrupt a predicted electrostatic interaction, significantly decreases binding affinity (Kd 420.4 ± 92). T519D, T522D, and V523D mutations were all designed to disrupt interactions with the ubiquitin hydrophobic patch. Aspartate was chosen to maximize polarity. All three mutations dramatically reduce binding to the extent that little or no complex formation can be detected. In contrast, the control mutation exhibits little to no discernable effect on ubiquitin binding (Table 1 and SI Appendix, Fig. S4).
Table 1.
Binding constants and fit parameters for ubiquitin-binding-domain mutations in ExoU
| Sample | Kd, μM | R2 |
| WT ExoU | 122.8 ± 11.5 | 0.99 |
| T519D | >2,000 | N/A |
| E520R | 420.4 ± 92 | 0.98 |
| T522D | >2,000 | N/A |
| V523D | >2,000 | N/A |
| S527L | 31.9 ± 5 | 0.99 |
| T554D | 92.4 ± 16.7 | 0.99 |
N/A, not available.
An in silico mutagenesis screen was performed on all residues of ExoU within 8 Å of ubiquitin to assess the effect of amino acid substitutions on ubiquitin binding. A small number of substitutions were predicted to improve ubiquitin binding, such as S527L, providing a good validation test of the interface. ExoU S527L was constructed and tested for binding. A modest increase in affinity is observed, with a fourfold reduction in Kd (Table 1 and SI Appendix, Fig. S5) accompanied by an evident increase in the immobilized component of the CW EPR spectra (SI Appendix, Fig. S5). ExoU S527L immobilizes A28R5 ubiquitin at lower concentrations compared with other ExoU variants. These data strongly support the RosettaDock interface model given the unlikelihood of producing an off-target effect of improved ubiquitin binding. Furthermore, the ability of Rosetta to predict the S527L mutation is a strong testament to the overall accuracy of the model.
In addition to binding, we tested the effect of each substitution on enzymatic activity. Activity assays were conducted using a fluorogenic phospholipid analog, PED6, as described previously (47). MonoUb was titrated into reactions containing 10 nM ExoU or ExoU variant. Linear rates were calculated and used to fit activation curves (SI Appendix, Fig. S6 and Table S3). E520R has no discernable effect on ExoU activity. S527L demonstrates a modest improvement on the half-saturation constant, Kact. T519D, T522D, and V523D mutations, however, have a significant impact on Kact (SI Appendix, Fig. S6 and Table S3). To control for off-target effects, T519D, T522D, V523D, and S527L were tested for binding to SpcU, ExoU’s chaperone. All derivatives bind with similar affinities as WT ExoU and the T554D control, suggesting that these mutations are not disruptive to the overall structure of ExoU (SI Appendix, Fig. S7 and Table S4). While the reduction in activation is significant, it is not as dramatic as the effects on ubiquitin binding. The observed differences in the binding and activity data may be a result of the conditions in the in vitro phospholipase assay, which was optimized to measure activity of WT ExoU and contains ∼750 mM monosodium glutamate and up to 10,000-fold excess ubiquitin (SI Appendix).
Biological Effects of ExoU Ubiquitin-Binding Variants.
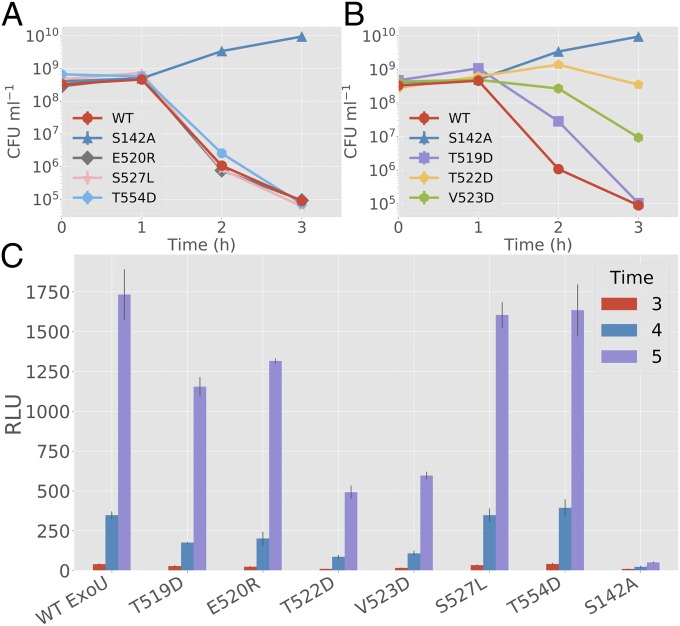
To test activity against biological membrane substrates, an Escherichia coli-based expression system was used to measure bacterial viability after coinduction of ExoU or ExoU variants and human monoUb (8). If a variant can be activated by ubiquitin in vivo, bacterial viability drops upon coinduction due to damage to the cytoplasmic membrane (16). In the absence of ubiquitin coexpression, ExoU and ExoU variants appear to be synthesized in similar amounts (SI Appendix, Fig. S8A). With ubiquitin coexpression, T519D, T522D, and V523D resulted in significantly lower toxicity compared with WT controls (Fig. 4 A and B). T522D and V523D had the most significant impact (P < 0.001 at 3 h post coinduction) in slowing bacterial death while T519D had delayed toxicity resulting in a significant difference at the 2-h time point but not at 3 h. The E520R and S527L mutations were not significantly different from parental ExoU in this assay. These results indicate that under physiological conditions and when monoUb is the only cofactor available, the bridging domain binding site is required for maximal activation.
Fig. 4.
Biological activity of ExoU with bridging domain mutations. (A and B) Surrogate toxicity assays using E. coli strains coexpressing monoUb and ExoU variants. The y axis denotes colony-forming unit (CFU)/mL. (A) E520R, S527L, and T554D are compared with WT ExoU and S142A controls. No significant deviation from the parental ExoU control was detected (3-h time point). (B) T519D, T522D, and V523D are compared with WT ExoU and S142A controls. T522D and V523D demonstrate a significant difference (P < 0.001) from controls at 3 h. T519D shows a significant difference at hour 2 but not at hour 3, suggesting delayed toxicity. (C) Adenylate kinase release assays for HeLa cells infected with P. aeruginosa-expressing ExoU variants. S527L shows no significant deviation (P > 0.2) from WT and T554D controls at hour 5. Other mutations show a similar pattern to the E. coli dual-expression system, with T522D having the largest effect and E520R having the smallest. All experimental groups except for S527L are significantly different (P < 0.005) from both S142A and WT controls at hour 5. Error bars indicate SD about the mean of three independent data points. RLU, relative light unit.
To test the biological effect of ubiquitin-binding mutations in a mammalian host cell background, ExoU variants were transferred to the broad-host-range plasmid pUCP19 for delivery by the P. aeruginosa T3SS. Expression studies indicate that all variants are synthesized and secreted similarly from P. aeruginosa PA103∆exoUexoT::Tc grown under conditions of calcium limitation (SI Appendix, Fig. S8 B and C). Each strain was cocultured with HeLa cells, and cell death was measured over time using an adenylate kinase release assay (Lonza ToxiLight). HeLa cells cocultured with P. aeruginosa expressing T519D, T522D, and V523D ExoU variants showed significantly reduced mammalian cell killing compared with WT ExoU and control variants (Fig. 4C). These data implicate the importance of the bridging domain ubiquitin binding site for biological activity in human cells. S527L did not show increased killing compared with controls, likely due to the saturating concentrations of ubiquitin present in mammalian cells. Consistent with the bacterial dual-expression assays, T522D had the largest impact on ExoU cytotoxicity followed by V523D and T519D. Introduction of the S142A substitution into each ExoU variant reduced cell death to levels of an enzymatically inactive molecule, indicating that the residual phospholipase activity of each variant is responsible for cell death (SI Appendix, Fig. S8D). Cumulatively, both our in vitro and in vivo experiments of ExoU variants support the RosettaDock model of a predicted ubiquitin-binding interface being located in the bridging domain of ExoU.
Discussion
ExoU functions as a potent phospholipase toxin, only when it has been injected into mammalian cells and activated by a noncovalent interaction with ubiquitin or ubiquitylated proteins. Identifying this ubiquitin interface represents a critical step in understanding the activation mechanism and perhaps designing inhibitory drugs that may diminish cellular damage. The challenges to identifying the interface include a low-micromolar binding affinity for ubiquitin, and a complex that is likely too large for NMR (>80 kDa), too small for cryo-EM (<110 kDa), and too flexible for crystallography. In this manuscript, we describe a simple and effective approach that combines DEER distances with RosettaDock global docking to generate a model of an ExoU–monoUb complex. Mutations were designed to alter ubiquitin interaction and recombinant proteins tested in a variety of biophysical, biochemical, and biological analyses. Diminishing binding affinity at the interface impacted ExoU–ubiquitin association, enzymatic and biological activities supporting the model. Rosetta was further used to design a substitution that improved binding affinity and activation kinetics. Importantly, the use of EPR distance restraints allowed us to extend the size limit for global docking to a complex greater than 700 residues (48). Overall, this may be a useful strategy for identification of protein–protein interaction interfaces in addition to providing high-resolution structural information. It is important to emphasize that these approaches are complementary to conventional structural determination. DEER is particularly well suited for large flexible proteins, similar to ExoU, and protein complexes that can assume a number of conformational states (20). Distance distributions can be measured under a variety of conditions, including in the presence of phospholipids, that often confound NMR and crystallography. Moreover, DEER is highly amenable to the study of conformational changes, and can be used to identify subpopulations of structural ensembles.
Previous protein-deletion and mapping studies identified a large C-terminal region of ExoU that was involved in ubiquitin interactions (amino acids 480 to 687). Our data confirm the original mapping analysis but more accurately place the ubiquitin hydrophobic patch near an exposed α-helix in the ExoU bridging domain (amino acids 480 to 580). While eukaryotic ubiquitin-binding domains (UBDs) are often helical, there is no sequence homology between ExoU and known eukaryotic UBDs. Despite the lack of sequence homology, structural alignments of our model with other helical UBDs in complex with ubiquitin reveal a similar orientation (SI Appendix, Fig. S9). This structural homology further supports the ubiquitin binding model predicted by our analyses. We have identified, expressed, and tested ubiquitin activation of several PLA2 enzymes encoded in the genomes of a variety of gram-negative bacteria (16). Consistent with the variation observed in eukaryotic UBDs, this region is not well-conserved at the primary amino acid level in ExoU orthologs, but each likely maintains a structural homology that facilitates ubiquitin interaction.
Further support for our model came from an independent CW EPR scan of the ExoU bridging domain. Comparison of the results of this screen with our in silico docking model shows that the formation of newly formed quaternary contacts with ubiquitin are likely the cause of the shifts in the CW EPR spectra of T494R1, Q512R1, and A524R1 ExoU. An alternative explanation for this result is that new tertiary contacts are formed due to a conformational change caused by ubiquitin; however, previous biophysical studies of ExoU suggest that there is little to no detectable change in ExoU tertiary structure upon the addition of ubiquitin alone (19). While this screen was valuable in localizing this UBD of ExoU, the model was essential in determining the orientation and key residues of the interaction.
Orientation of ubiquitin with respect to ExoU in our model is supported by biophysical predictions made using mtsslWizard and verified by DEER. All nine distance distributions predicted were found to be within reasonable accuracy of the experimentally determined distances. Importantly, our model was developed using 11 previously published distance restraints (21), only one of which was under 30 Å. Long distances introduce ambiguity, while shorter distances tend to be more valuable. Our model allowed us to select and test residues that are predicted to be significantly closer in space, resulting in three distances under 20 Å, nearing the lower limit of detection using DEER.
In addition to structural predictions, our model allowed us to efficiently design high-impact substitutions using multiple in silico approaches. All three of the mutations directed to disrupt hydrophobic interactions with ubiquitin (T519D, T522D, V523D) resulted in a dramatic loss in binding and a measurable biochemical and biological impact. The strongest support for our model comes from a gain-of-affinity mutation, S527L. ExoU S527L demonstrated improved binding and catalytic activation by monoUb. This substitution had little effect on toxicity, however; given the high intracellular concentration of ubiquitin in eukaryotic cells (49) and the activation kinetics of ExoU with mono- and polyubiquitin, we suspect that both WT and S527L ExoU are saturated for activation in HeLa cells. Similarly, the isopropyl-d-1-thiogalactopyranoside–inducible E. coli expression system likely produces relatively high intracellular concentrations of ubiquitin, also leading to saturation of the limited quantities of toxic WT and S527L ExoU that can be expressed before membrane collapse. E520R had a significant but less dramatic effect relative to T519D, T522D, and V523D, suggesting that interactions with the ubiquitin hydrophobic patch dominate binding. This effect is more evident in the activity of E520R ExoU, which does not deviate from WT activity both in vitro and in E. coli. Interestingly, E520R did show reduced activity in the eukaryotic infection system, but had the least effect.
Similarly, it was surprising that ExoU UBD mutations, which appeared to abolish binding, had less of an effect on ExoU activation both in vitro and in vivo. ExoU is known to undergo a large conformational change upon activation (19, 20). A conformational change may expose an additional ubiquitin binding site, improve binding affinity, or improve substrate association. The existence of multiple UBDs is consistent with the previous observation that ExoU binds and is activated significantly better by polyubiquitin species (16). Future studies will seek to further explore the synergistic effects of the substrate and cofactor on conformational changes and activation. Despite the presence of residual activity, ExoU cytotoxicity was significantly impaired upon the introduction of UBD mutations, suggesting that disruption of the ExoU–ubiquitin interaction at this single site could be a useful therapeutic target. While this site has crude structural homology to eukaryotic UBDs, the lack of sequence homology may be exploited for the development of a unique therapeutic lacking off-target effects on other ubiquitin-binding proteins.
Materials and Methods
Protein–Protein Global Docking.
An ExoU model (19) generated from crystal structure PDB ID code 3TU3 (11) was used for all ExoU in silico docking. A monoUb crystal structure (PDB ID code 1UBQ) was used for all ubiquitin docking. Both monomers were prepared for docking by running the Rosetta clean_pdb.py script and relaxed with the Rosetta relax application using flags listed in SI Appendix, Procedure S1. Ten decoys were made, and the lowest-scoring relaxed structure was chosen for docking. Relaxed ExoU and ubiquitin structures were saved in the same pdb file ∼20 Å apart and randomly oriented. DEER distance restraints published by Anderson et al. (21) were entered into a constraints file as AtomPair constraints using the RosettaEPR Score function (34). ExoU and monoUb were docked with the Rosetta docking protocol application (39) using flags described in SI Appendix, Procedure S2. The resulting silent file was scored using flags listed in SI Appendix, Procedure S3, which takes into account the Rosetta and RosettaEPR score. Complex_20145 was selected as the best scoring model and used as a reference structure to plot score vs. rmsd of the top 5,000 decoys.
Predicting DEER Distances Using MtsslWizard.
MTSSL (methanethiosulfonate spin label) conformations were modeled into the complex using the PyMOL plugin mtsslWizard (43, 44) with “painstaking” as the speed parameter and “tight” as the van der Waals (vdW) restraints parameter. In the event that a site could not be modeled using these parameters, the vdW restraint parameter was set to “loose.” Distance distributions were measured using the “measurement” mode of mtsslWizard and saved to a file for graphical representation.
In Silico Prediction and Selection of Binding Mutations.
The ExoU–ubiquitin model was submitted to both the InterProSurf (curie.utmb.edu) and Robetta interface alanine scan (50, 51) servers. Interface residues identified from both results were compiled and compared (SI Appendix, Table S2). Sites were selected based on >50% ΔSASA (InterProSurf) and >1 kcal/mol predicted ΔΔG (Robetta). Disruptive mutations were selected to either disrupt predicted electrostatic interactions (E520R) or disrupt hydrophobic interactions (T519D, T522D, V523D). The gain-of-affinity mutation S527L was selected by running the Rosetta ddg_monomer application (50, 52) on the ExoU–ubiquitin complex, mutating each residue in SI Appendix, Table S2 to each other possible residue except cysteine. The ddg_monomer application was run using parameters described in SI Appendix, Procedure S4. S527L was selected as the most reasonable mutation with the largest reduction in the ddg_monomer score.
Continuous-Wave Electron Paramagnetic Resonance and Ubiquitin Binding Analyses.
Continuous-wave EPR was conducted at room temperature using an ELEXSYS-II E500 spectrometer (Bruker) equipped with a high-Q cavity operating at X band. All samples were prepared in 20 mM Tris, 150 mM NaCl, 20% glycerol (pH 7.5) buffer. ExoU with cysteine substitutions within the bridging domain was analyzed at a concentration of 50 μM in the presence or absence of 600 μM purified recombinant monoUb. Spectrometer acquisition parameters were set as follows: time constant, 5.12 ms; conversion time, 20.48 s; sweep time, 20.97 s; 100-kHz field modulation amplitude, 1.0 G; microwave power, 10-mW; and sweep width, 100 G. Spectra were collected as an average of 25 scans. Ubiquitin binding assays were conducted using 25 μM A28R5 ubiquitin and a similar procedure as described previously (21). Acquisition parameters for ubiquitin binding assays differed from the above by the following: time constant, 20.48 ms; conversion time, 40.96 s; sweep time, 41.94 s; and field modulation amplitude, 1.6 G. All CW EPR spectra were analyzed using MATLAB 2016 (MathWorks) and EasySpin (53) or Python 3 (www.python.org).
Double Electron–Electron Resonance Distance Distribution Analyses.
Four-pulse DEER spectroscopy (54) was carried out at ∼33 GHz on an ELEXSYS E580 spectrometer (Bruker BioSpin) equipped with an EN 5107D2 resonator and 10-W microwave amplifier. Samples at a final volume of 12 µL contained spin-labeled ExoU and spin-labeled ubiquitin each at ∼0.1 mM final concentration in 20 mM Tris, 150 mM NaCl (pH 7.5) buffer and 25% (vol/vol) perdeuterated glycerol (Sigma-Aldrich) as cryoprotectant. Samples in 1.1- × 1.6-mm glass capillaries (VitroCom) were flash-frozen by immersion in an acetone/dry ice bath, immediately placed in the sample resonator, and maintained at 80 K using an Oxford cryostat. Pump pulses (32 ns) were positioned at the low field maximum of the Q-band absorption spectrum, and π and π/2 observer pulses (32 and 16 ns, respectively) were positioned 50 to 56 MHz upfield, corresponding to the center field maximum. Signal averaging times ranged from 8 to 16 h. Dipolar evolution data were corrected assuming a homogeneous 3D background and analyzed by model free Tikhonov regularization using DeerAnalysis (46) version 2016.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) NIAID AI104922 (to D.W.F.) and NIGMS GM114234 (to J.B.F.). The National Biomedical EPR Center, Department of Biophysics, is supported by NIH Grant P41 EB001980. DEER instrumentation was supported by NIH Grants S10RR022422 and S10OD011937. Work in the J.M. laboratory is supported through NIGMS R01 GM080403 and R01 GM073151.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716861115/-/DCSupplemental.
References
- 1.McCarthy K. Pseudomonas aeruginosa: Evolution of antimicrobial resistance and implications for therapy. Semin Respir Crit Care Med. 2015;36:44–55. doi: 10.1055/s-0034-1396907. [DOI] [PubMed] [Google Scholar]
- 2.Poole K. Pseudomonas aeruginosa: Resistance to the max. Front Microbiol. 2011;2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yahr TL, Barbieri JT, Frank DW. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finck-Barbançon V, et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 6.Iglewski BH, Sadoff J, Bjorn MJ, Maxwell ES. Pseudomonas aeruginosa exoenzyme S: An adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DM, et al. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol. 2011;82:1454–1467. doi: 10.1111/j.1365-2958.2011.07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belyy A, et al. Actin activates Pseudomonas aeruginosa ExoY nucleotidyl cyclase toxin and ExoY-like effector domains from MARTX toxins. Nat Commun. 2016;7:13582. doi: 10.1038/ncomms13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato H, Frank DW. ExoU is a potent intracellular phospholipase. Mol Microbiol. 2004;53:1279–1290. doi: 10.1111/j.1365-2958.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 11.Halavaty AS, et al. Structure of the type III secretion effector protein ExoU in complex with its chaperone SpcU. PLoS One. 2012;7:e49388. doi: 10.1371/journal.pone.0049388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gendrin C, et al. Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog. 2012;8:e1002637. doi: 10.1371/journal.ppat.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato H, Feix JB, Frank DW. Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry. 2006;45:10368–10375. doi: 10.1021/bi060788j. [DOI] [PubMed] [Google Scholar]
- 14.Anderson DM, Frank DW. Five mechanisms of manipulation by bacterial effectors: A ubiquitous theme. PLoS Pathog. 2012;8:e1002823. doi: 10.1371/journal.ppat.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson DM, Feix JB, Frank DW. Cross kingdom activators of five classes of bacterial effectors. PLoS Pathog. 2015;11:e1004944. doi: 10.1371/journal.ppat.1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson DM, Sato H, Dirck AT, Feix JB, Frank DW. Ubiquitin activates patatin-like phospholipases from multiple bacterial species. J Bacteriol. 2015;197:529–541. doi: 10.1128/JB.02402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyson GH, et al. A novel phosphatidylinositol 4,5-bisphosphate binding domain mediates plasma membrane localization of ExoU and other patatin-like phospholipases. J Biol Chem. 2015;290:2919–2937. doi: 10.1074/jbc.M114.611251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer AW, et al. Structure and dynamics of type III secretion effector protein ExoU as determined by SDSL-EPR spectroscopy in conjunction with de novo protein folding. ACS Omega. 2017;2:2977–2984. doi: 10.1021/acsomega.7b00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tessmer MH, Anderson DM, Buchaklian A, Frank DW, Feix JB. Cooperative substrate-cofactor interactions and membrane localization of the bacterial phospholipase A2 (PLA2) enzyme, ExoU. J Biol Chem. 2017;292:3411–3419. doi: 10.1074/jbc.M116.760074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson MA, et al. Induced conformational changes in the activation of the Pseudomonas aeruginosa type III toxin, ExoU. Biophys J. 2011;100:1335–1343. doi: 10.1016/j.bpj.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson DM, et al. Identification of the major ubiquitin-binding domain of the Pseudomonas aeruginosa ExoU A2 phospholipase. J Biol Chem. 2013;288:26741–26752. doi: 10.1074/jbc.M113.478529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosca R, Pons T, Céol A, Valencia A, Aloy P. Towards a detailed atlas of protein-protein interactions. Curr Opin Struct Biol. 2013;23:929–940. doi: 10.1016/j.sbi.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Pache RA, Aloy P. Incorporating high-throughput proteomics experiments into structural biology pipelines: Identification of the low-hanging fruits. Proteomics. 2008;8:1959–1964. doi: 10.1002/pmic.200700966. [DOI] [PubMed] [Google Scholar]
- 24.Aloy P, Pichaud M, Russell RB. Protein complexes: Structure prediction challenges for the 21st century. Curr Opin Struct Biol. 2005;15:15–22. doi: 10.1016/j.sbi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Alberts B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 26.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 27.Duran-Frigola M, Mosca R, Aloy P. Structural systems pharmacology: The role of 3D structures in next-generation drug development. Chem Biol. 2013;20:674–684. doi: 10.1016/j.chembiol.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Janin J, et al. Critical Assessment of PRedicted Interactions CAPRI: A critical assessment of predicted interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 29.Soni N, Madhusudhan MS. Computational modeling of protein assemblies. Curr Opin Struct Biol. 2017;44:179–189. doi: 10.1016/j.sbi.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Aloy P, Russell RB. Structural systems biology: Modelling protein interactions. Nat Rev Mol Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 31.Melquiond ASJ, Bonvin AMJ. Data-driven docking: Using external information to spark the biomolecular rendez-vous. In: Zacharias M, editor. Protein-Protein Complexes: Analysis, Modeling and Drug Design. Imperial College Press; London: 2010. pp. 182–208. [Google Scholar]
- 32.Karaca E, Bonvin AM. Advances in integrative modeling of biomolecular complexes. Methods. 2013;59:372–381. doi: 10.1016/j.ymeth.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Mouradov D, Kobe B, Dixon NE, Huber T. Hybrid methods for protein structure prediction. In: Rangwala H, Karypis G, editors. Introduction to Protein Structure Prediction: Methods and Algorithms. John Wiley & Sons; Hoboken, NJ: 2010. pp. 265–277. [Google Scholar]
- 34.Hirst SJ, Alexander N, Mchaourab HS, Meiler J. RosettaEPR: An integrated tool for protein structure determination from sparse EPR data. J Struct Biol. 2011;173:506–514. doi: 10.1016/j.jsb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Y, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahraman A, et al. Cross-link guided molecular modeling with ROSETTA. PLoS One. 2013;8:e73411. doi: 10.1371/journal.pone.0073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bender BJ, et al. Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochemistry. 2016;55:4748–4763. doi: 10.1021/acs.biochem.6b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander N, Bortolus M, Al-Mestarihi A, Mchaourab H, Meiler J. De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure. 2008;16:181–195. doi: 10.1016/j.str.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gray JJ, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 40.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 41.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains—From structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagelueken G, Ward R, Naismith JH, Schiemann O. MtsslWizard: In silico spin-labeling and generation of distance distributions in PyMOL. Appl Magn Reson. 2012;42:377–391. doi: 10.1007/s00723-012-0314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagelueken G, Abdullin D, Schiemann O. mtsslSuite: Probing biomolecular conformation by spin-labeling studies. Methods Enzymol. 2015;563:595–622. doi: 10.1016/bs.mie.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Casey TM, Fanucci GE. Spin labeling and double electron-electron resonance (DEER) to deconstruct conformational ensembles of HIV protease. Methods Enzymol. 2015;564:153–187. doi: 10.1016/bs.mie.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeschke G, et al. DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 47.Benson MA, Schmalzer KM, Frank DW. A sensitive fluorescence-based assay for the detection of ExoU-mediated PLA(2) activity. Clin Chim Acta. 2010;411:190–197. doi: 10.1016/j.cca.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daily MD, Masica D, Sivasubramanian A, Somarouthu S, Gray JJ. CAPRI rounds 3–5 reveal promising successes and future challenges for RosettaDock. Proteins. 2005;60:181–186. doi: 10.1002/prot.20555. [DOI] [PubMed] [Google Scholar]
- 49.Kaiser SE, et al. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods. 2011;8:691–696. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein–protein complexes. Proc Natl Acad Sci USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 52.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 54.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole–dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.