Significance
Peroxisome proliferator-activated receptor-γ (PPARγ) is a transcription factor that plays a central role in the formation of adipose tissue. We show that phosphorylation of a single amino acid of PPARγ alters the response of cells to DNA damaging agents, including multiple types of chemotherapy. Noncanonical agonist PPARγ ligands that block PPARγ phosphorylation sensitize a variety of cancer cell types to these chemotherapeutic agents in vitro and in vivo. We show that PPARγ interacts with the tumor-suppressor p53 in a manner dependent on PPARγ phosphorylation at S273. These data strongly suggest that noncanonical agonist PPARγ ligands, which lack many of the known side effects of classic agonists, should be explored for clinical use in combination with traditional chemotherapy for a variety of malignancies.
Keywords: PPARγ, DNA damage, lung cancer, chemotherapy
Abstract
The peroxisome-proliferator receptor-γ (PPARγ) is expressed in multiple cancer types. Recently, our group has shown that PPARγ is phosphorylated on serine 273 (S273), which selectively modulates the transcriptional program controlled by this protein. PPARγ ligands, including thiazolidinediones (TZDs), block S273 phosphorylation. This activity is chemically separable from the canonical activation of the receptor by agonist ligands and, importantly, these noncanonical agonist ligands do not cause some of the known side effects of TZDs. Here, we show that phosphorylation of S273 of PPARγ occurs in cancer cells on exposure to DNA damaging agents. Blocking this phosphorylation genetically or pharmacologically increases accumulation of DNA damage, resulting in apoptotic cell death. A genetic signature of PPARγ phosphorylation is associated with worse outcomes in response to chemotherapy in human patients. Noncanonical agonist ligands sensitize lung cancer xenografts and genetically induced lung tumors to carboplatin therapy. Moreover, inhibition of this phosphorylation results in deregulation of p53 signaling, and biochemical studies show that PPARγ physically interacts with p53 in a manner dependent on S273 phosphorylation. These data implicate a role for PPARγ in modifying the p53 response to cytotoxic therapy, which can be modulated for therapeutic gain using these compounds.
The peroxisome proliferator activator receptor-γ (PPARγ) is an orphan nuclear receptor that is essential for the development of adipocytes (1) and is the target for the thiazolidinedione (TZD) class of antidiabetic agents (2). In addition to its role in metabolism, PPARγ is mutated or overexpressed in certain human cancers (3–6). Despite initial excitement regarding the role of PPARγ ligands in cancer therapy, they were not effective as single agents in advanced epithelial malignancies (7, 8). As an alternative approach, our group has demonstrated that TZDs potently sensitize a variety of cancer cells to the cytotoxic effects of carboplatin (9, 10). The mechanism was thought to be via inhibition of metallothionein gene expression, although other mechanisms were not ruled out (11). While these data suggested that PPARγ ligands may play an important role in cancer therapy, the use of these drugs has declined dramatically due reports concerning toxicity, many of which are now known to have been potentially overstated (12).
Recent data from our group has shown that the pleiotropic effects of PPARγ ligands can be chemically separated into two distinct activities. One relates to the ability of ligands to act as canonical agonists of the nuclear receptor on peroxisome proliferator response elements, which leads to adipogenesis. The second relates to the allosteric inhibition of phosphorylation of the Ser273 (serine 273, S273) residue of PPARγ by a variety of kinases, including CDK5 (13) and ERK1/2 (14). Novel noncanonical agonist ligands (NALs) that only inhibit this phosphorylation event retain much of the antidiabetic activity of TZDs. Intriguingly, many of the known side effects of TZDs, including weight gain, fluid retention, and bone loss are correlated with the agonist properties of TZDs rather than their effect on S273 phosphorylation (15, 16). These data suggest that many of the effects of TZDs previously attributed to their effects as agonists may instead be due to their inhibition of S273 phosphorylation, and raise the question of whether S273 phosphorylation may control other previously unappreciated aspects of PPARγ biology.
The increasing prevalence of metabolic disease and cancer has led to a growing recognition of the mechanistic links shared by these two different diseases (17). The shared biology of obesity, diabetes, and cancer suggest that therapies developed for metabolic disease may be useful in cancer treatment (18, 19) or vice versa (14). One potential application of these therapeutics is to increase the efficacy of cytotoxic treatments in cancer (10, 20, 21). It is critical to note that despite rapid advances in both targeted therapy and immunotherapy, the majority of cancer patients will receive either chemotherapy or radiotherapy during the course of their disease. Thus, enhancing the efficacy of cytotoxic therapy remains a crucial goal for cancer patients.
Here we demonstrate that that PPARγ is phosphorylated in response to DNA damage; this phosphorylation can be inhibited by NALs. We show that inhibition of phosphorylation of PPARγ using chemical or genetic approaches results in dramatic sensitization of cells to DNA-damaging agents. S273 phosphorylation alters the association of PPARγ with the tumor suppressor p53 and impacts its function, which is required for the sensitizing effects of PPARγ ligands. These data suggest that PPARγ plays a more direct role in the cellular response to DNA damaging agents than has been previously demonstrated, and offer a therapeutic approach that can be combined with traditional cancer therapies.
Results
PPARγ Is Phosphorylated on S273 in Response to Carboplatin.
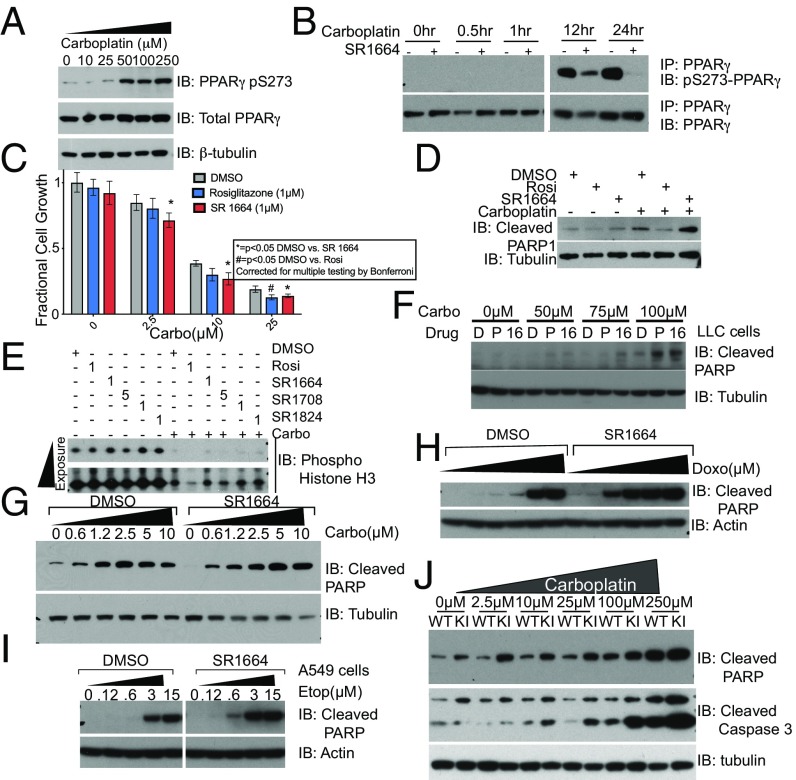
To investigate the role of phosphorylation of PPARγ in the response to DNA damage, we assessed whether S273 phosphorylation occurs in cancer cells upon treatment with carboplatin. A549 cells, which have been shown to be sensitized to carboplatin by TZDs (10), were treated with increasing concentrations of carboplatin for 24 h. Western blotting of whole-cell lysates prepared from these cells using an antibody specific for the S273 phosphorylated form of PPARγ (13) revealed a very robust dose-dependent increase in phosphorylated PPARγ (Fig. 1A).
Fig. 1.
(A) Dose-dependent phosphorylation of PPARγ on S273 with carboplatin treatment. (B) Time course of PPARγ phosphorylation as assessed by IP of PPARγ from lysates of A549 cells after treatment with 50 µM carboplatin treated with DMSO or 1 µM SR1664 shows phosphorylation can be inhibited by NALs. IB, immunobot. (C) A549 cells treated with increasing concentrations of carboplatin in the presence or absence of either rosiglitazone or SR1664 show equivalent effects on total cell number at 24 h. (D) Increased markers of apoptotic cell death with cotreatment of NALs and carboplatin as assessed by immunoblot for cleaved PARP1 (E) NALs do not further suppress phospho-Histone H3, a mitotic marker indicative of cell cycle progression. (F and G) Treatment of other cancer types, including LLC (F) and MDA-MB-468 cells, a triple-negative breast cancer cell line (G), show similar increased production of cleaved PARP when treated with a combination of SR1664 and carboplatin. (H and I) A549 cells cotreated with doxorubicin (H) or etoposide (I) with SR1664 shows an increase in apoptosis. Panels in I were taken from the same blot. (J) Treatment of wild-type (WT) and knock-in (KI) cells with carboplatin demonstrates a dramatically increased sensitivity of knock-in cells to the cytotoxic effects of carboplatin.
We then examined the dynamics of phosphorylation status of PPARγ after carboplatin treatment. PPARγ was immunoprecipitated from A549 lysates at the indicated times and analyzed by immunoblotting with the pS273 phospho-specific antibody. By 8 h there was a striking accumulation of phosphorylated PPARγ, which continued at 24 h posttreatment. As in adipose cells, coincubation of the cells with the NAL SR1664 (16) dramatically reduced the phosphorylation of PPARγ (Fig. 1B). These data suggest that PPARγ is indeed phosphorylated in cancer cells in response to carboplatin, and this phosphorylation can be inhibited by NALs.
Inhibition of S273A Phosphorylation with Noncanonical Agonist PPARγ Ligands Results in Increased Cell Death in Response to Multiple Genotoxic Agents.
We tested the functional consequences of blocking the phosphorylation of PPARγ using NALs in A549 cells treated with carboplatin. SR1664 significantly increased the cytoxic effects of carboplatin. Two-way ANOVA showed a significant interaction of the drug treatment with carboplatin treatment. (Fig. 1C) (P = 0.0009). This effect was also seen with the partial agonist MRL-24 and NAL SR1824 (Fig. S1 A and B). These experiments indicate that agonism of PPARγ is dispensable for the ability of TZDs to sensitize these cancer cells to the cytotoxic effects of carboplatin.
We assessed the relative contributions of apoptosis and growth inhibition by cell cycle arrest to the reduction in total cell numbers. A549 cells treated with rosiglitazone and NALs with and without carboplatin showed a dramatic increase in cleaved poly(ADP-ribose) polymerase 1 (PARP1) (Fig. 1D), a key marker of apoptosis. Similarly, analysis of cDNA prepared from the mRNA of these cells showed a significant increase in p53 upregulated modulator of apoptosis (PUMA) mRNA, a key mediator of apoptosis (Fig. S1C). Interestingly, apoptosis was significantly higher in cells treated with the NALs compared with rosiglitazone (Fig. 1D).
We next examined the induction of cell cycle arrest by studying the phosphorylation of histone H3, a key mitotic marker (Fig. 1E). As expected, carboplatin significantly induces cell cycle arrest. Rosiglitazone further suppresses H3 phosphorylation in comparison with the DMSO control, consistent with the ability of TZDs to induce cell cycle arrest in adipose cells (22). Contrastingly, cells treated with NALs do not show any further suppression of H3 phosphorylation. These data suggest that, compared with agonist ligands (TZDs), NALs preferentially cause apoptotic cell death in response to carboplatin, possibly due to their lack of effect on inhibiting cell cycle progression.
We then examined whether other cell types that expressed PPARγ were also sensitized to the cytotoxic effects of carboplatin. We saw similar effects of these drugs in the mouse Lewis lung carcinoma (LLC) cell lines as coincubation of LLC cells with the TZD pioglitazone or SR1664 with carboplatin increased the accumulation of cleaved PARP1 (Fig. 1F). We also assessed the ability of NALs to sensitize MDA-MB-468 cells, a model of triple-negative breast cancer. These cells showed increased phosphorylation of S273 of PPARγ upon treatment with carboplatin (Fig. S1F), as well as increased apoptosis with SR1664 cotreatment (Fig. 1G), which is quantitated in Fig. S1D.
The ability of NALs to sensitize PPARγ-expressing cells is not universal. HCT116 cells, which express high levels of PPARγ (23), show no significant increased accumulation of cleaved PARP (Fig. S1E), and no increased phosphorylation of PPARγ (Fig. S1G), suggesting that the sensitization effect of NALs is not present in every cell type despite the presence of PPARγ protein.
We next asked whether the ability of these ligands to sensitize cancer cells to chemotherapeutic cytotoxicity represented a generalized response to DNA damaging agents. We found that A549 cells treated with SR1664 and increasing concentrations of the anthracycline doxorubicin (Fig. 1H) and the topoisomerase II inhibitor etoposide (Fig. 1I) showed an increased accumulation of cleaved PARP1 compared with DMSO-treated controls. Contrastingly, cotreatment of A549 cells with the microtubule-stabilizing cytotoxic paclitaxel (Fig. S1H) did not result in increased cell death. This differential sensitization suggests that inhibition of the phosphorylation of PPARγ genetically sensitizes cells to cytotoxic agents that work directly by damaging the DNA, rather than drugs that are cytotoxic through other mechanisms.
Genetic Inhibition of PPARγ S273 Phosphorylation Mimics the Effects of Noncanonical Agonist PPARγ Ligands on Cell Death.
To verify that these results were specifically due to on target effects of inhibition of PPARγ phosphorylation, we used shRNA to generate A549 cells with low levels of PPARγ. Treatment of these cells with SR1664 and carboplatin shows that PPARγ is required for the increased apoptotic cell death (Fig. S1I).
To more precisely interrogate the importance of S273 phosphorylation of PPARγ, we took a genetic approach using mice bearing a Ser273→Ala knock-in mutation. We generated immortalized fibroblasts from the brown adipose tissue of these mice (Fig. S1J) and treated them with increasing doses of carboplatin. At each dose, from 2.5 µM to 250 µM, there is significantly increased accumulation of both cleaved PARP1 and cleaved Caspase 3 (Fig. 1J). These effects are especially striking at 2.5 µM, 10 µM, and 25 µM carboplatin, where there is no significant increase in PARP1 accumulation in the wild-type cells (quantitated in Fig. S1K). Thus, abolishing the phosphorylation of PPARγ by mutation of Ser273 to Ala is sufficient to confer a greatly increased sensitivity to apoptotic cell death induced by cytotoxic drugs. This was also true for fibroblasts generated from a separate body site (Fig. S1L). Consistent with our data from NAL treatment, we confirmed that these cells are also sensitized to other DNA damaging agents, such as etoposide (Fig. S1M) and doxorubicin (Fig. S1N), but not taxol (Fig. S1O).
Identification of a Core Gene Set Affected by Inhibition of PPARγ S273 Phosphorylation upon Treatment with Carboplatin.
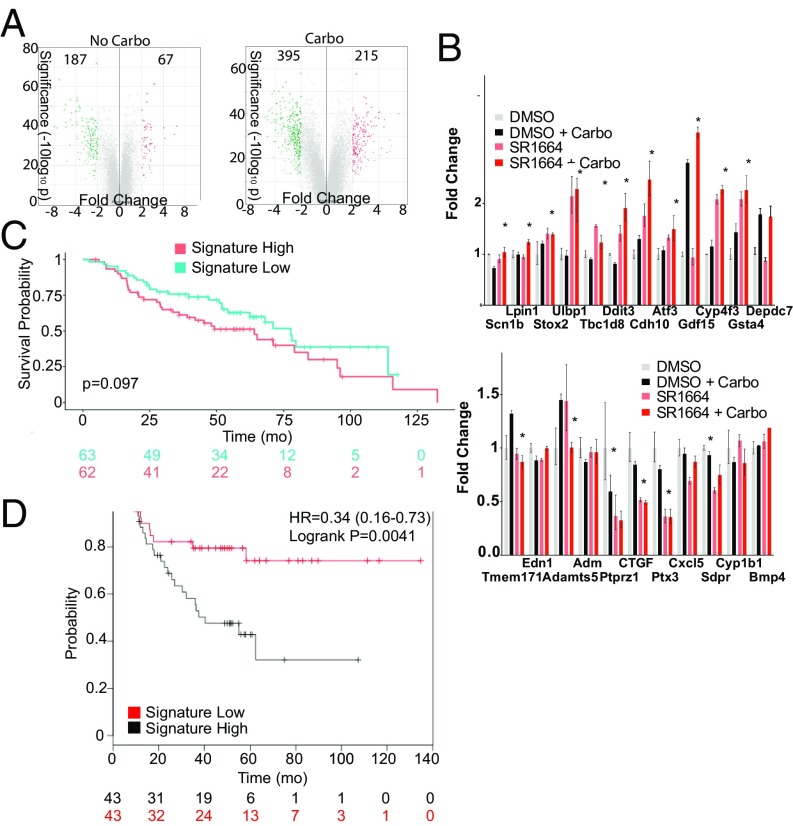
To assess the cellular response induced by inhibiting the phosphorylation of PPARγ, we examined global gene expression using Affymetrix arrays (Fig. 2A). Upon treatment with carboplatin, 395 genes were down-regulated in mutant cells and 215 genes up-regulated, consistent with the idea that inhibition of S273 phosphorylation results in a profound change in the transcriptome of the cells.
Fig. 2.
(A) Volcano plots of the comparison between wild-type and knock-in cell gene expression show a threefold increase in the number of differentially regulated genes. (B) A core set of genes regulated by S273 phosphorylation in multiple cell types upon carboplatin treatment was generated. Seventeen of a core set of 23 genes were similarly up-regulated (*P < 0.05) in A549 lung cancer cells by inhibition of PPARγ phosphorylation with the NAL SR1664 in the presence of carboplatin (χ2 P = 0.0218). (C) Kaplan–Meier plot of overall survival over time reveals a trend toward improved survival with high expression of the PPARγ S273A gene signature in the combined cohort of patients receiving adjuvant chemotherapy in the NIH Directors Challenge cohort and the UT Southwestern Lung SPORE cohort. (D) Kaplan–Meier analysis of a cohort of 86 ER−/PR− breast cancer patients using KMplot revealed a significant difference in recurrence-free survival based on expression of the gene signature.
We selected a group of genes that were at least threefold up-regulated with an ANOVA P value < 0.05 for validation. A total of 59 genes (excluding predicted genes and uncharacterized cDNAs) met these criteria and were analyzed (Fig. S2A). Forty of these were significantly (P < 0.05) regulated in separate experiments (χ2 P < 0.0063) and multiple others trended toward significance.
We then examined the expression of these genes in other cell types to generate a core signature of PPARγ phosphorylation inhibition after carboplatin treatment. A core set of 12 genes that were up-regulated in the S273A mutant and 11 genes that were down-regulated in the S273A mutant genes was generated based on their expression in multiple cell types with and without carboplatin treatment. Interestingly, most of the down-regulated genes [e.g., Ptprz1 (24), Edn1 (25), Adamts5 (25), Adm (26)] have been previously associated with chemotherapy resistance.
This gene set was assessed in A549 cells treated with SR1664 in combination with carboplatin (Fig. 2B). Ten of the 12 up-regulated genes were coordinately up-regulated in A549 cells treated with SR1664 and carboplatin. Seven of the 11 genes were appropriately down-regulated with SR1664 treatment with carboplatin, for a total of 17 of 23 genes appropriately regulated (χ2 P = 0.0218). The expression of these genes was also examined in MDA-MB-468 cells treated with SR1664 and carboplatin. We found a similar degree of regulation, although it did not reach significance by chi-square testing (Fig. S2B). This core gene set represents a gene-expression–based biomarker of the inhibition of PPARγ phosphorylation in response to carboplatin.
We hypothesized that this gene signature may reflect the sensitivity of tumors to cytotoxic chemotherapy. Using publicly available gene-expression datasets, we queried whether expression of the combined gene set correlated with the outcomes of patients treated with chemotherapy. Patients in the Director’s Challenge Consortium (27) who received adjuvant chemotherapy (n = 90) and the UT Lung SPORE cohort (n = 49) (28), two of the largest cohorts of lung cancer patients receiving adjuvant chemotherapy with available gene-expression data, were classified based on their expression of the genes in the signature. Notably, tissue was obtained before any chemotherapy. Kaplan–Meier analysis of overall survival in these two combined cohorts showed that patients with a greater than median signature score had a trend toward better survival than those who did not express the signature (P = 0.097) (Fig. 2C). These studies showed a similar trend when analyzed individually as well (P = 0.1 and P = 0.34) (Fig. S2C).
We also examined the gene signature in triple-negative breast cancer using the KMplot online tool (kmplot.com/analysis/index.php?p=background). Kaplan–Meier analysis of patients with estrogen receptor-negative/progesterone receptor-negative (ER−/PR−) breast cancers treated with chemotherapy showed that expression of the gene signature was associated with a significantly increased recurrence-free survival (median 58.15 mo vs. 21 mo) with a hazard ratio (HR) of 0.34 (P = 0.0041) (Fig. 2D). Interestingly, analysis of patients who did not receive chemotherapy shows that there was no difference in recurrence-free survival among the groups (Fig. S2D), suggesting that the gene signature does not simply reflect prognosis, but rather is predictive of chemotherapy response. A similar analysis of patients with lung cancer treated with chemotherapy showed a trend for improved outcomes with a HR of 0.27 (P = 0.0507) (Fig. S2E), although the analysis was limited by the small number of patients (n = 34). Of course, these analyses are limited due to the mixed clinical and pathologic features of these cohorts. However, these data suggest that low expression of the down-regulated genes and high expression of the up-regulated genes is associated with improved outcomes among patients receiving systemic chemotherapy.
Noncanonical Agonist PPARγ Ligands Synergize Effectively with Carboplatin in Vivo.
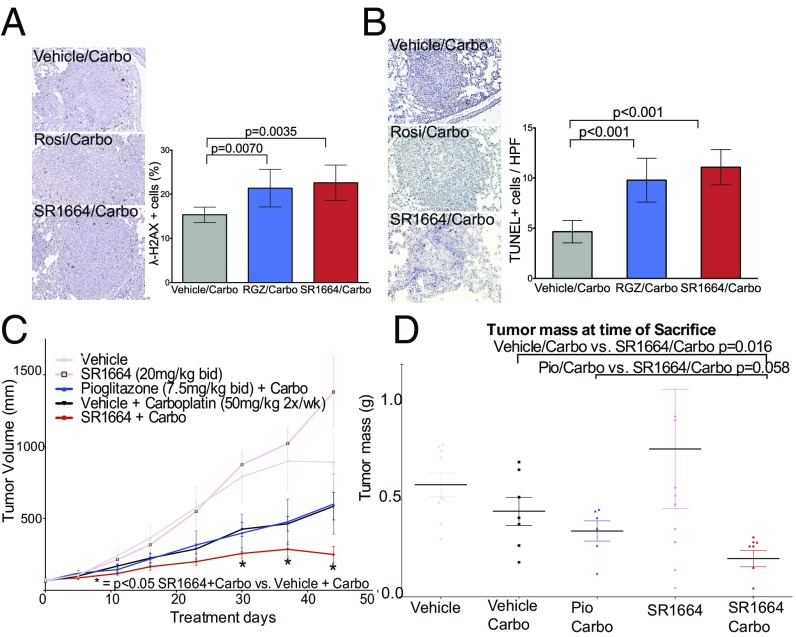
We next investigated whether inhibition of PPARγ phosphorylation could be a therapeutic target in vivo. We first examined short-term treatment of lung tumors in animals bearing a Lox-Stop-Lox mutant KRAS allele driven by inhaled adenoviral Cre (29). We treated animals with established lung tumors with carboplatin plus either rosiglitazone, SR1664, or vehicle for 2 d. Tumors were subjected to TUNEL staining for apoptotic cells, or immunohistochemistry for accumulation of γ-H2AX, a key marker of DNA damage. There was a significant increase in the number of γ-H2AX+ cells in animals treated both with rosiglitazone and with SR1664 when combined with carboplatin (Fig. 3A). There was also a significant increase in the number of TUNEL+ cells per field examined, increasing from 5% in controls to 10% with rosiglitazone and 12% for SR1664 (Fig. 3B) (P < 0.001). These data suggest that the inhibition of S273 phosphorylation of PPARγ is a bona fide therapeutic target, and that NALs can sensitize lung cancer cells to carboplatin in vivo.
Fig. 3.
(A and B) Aperio analysis of γ-H2AX staining (A) and TUNEL staining (B) in sections from tumors in lungs of inducible KRAS G12D allele activated with inhaled adenoviral Cre. An increase in labeled cells was seen with cotreatment of animals with both rosiglitazone and SR1664 compared with vehicle and carboplatin alone. (Magnification: 200×.) (C) Graph of xenograft volumes over time from a representative experiment. Tumors on animals treated with SR1664 and carboplatin were significantly smaller than those treated with vehicle and carboplatin. Tumors from animals treated with pioglitazone were not statistically smaller than those from control carboplatin animals, although this effect was due to a single outlier, as evidenced by the results when the data were transformed to a modified mean (Fig. S3A). (D) Tumor weights at the time of killing of mice from a separate experiment of nude mice with A549 cell xenografts treated with the indicated drugs (n = 7–10). There was a significant difference in tumor weight of xenografts in mice treated with SR1664 compared with those treated with vehicle and carboplatin (P = 0.016). The weights of tumors treated with SR1664 and carboplatin were lower than those treated with pioglitazone and carboplatin in a near significant trend (P = 0.058).
It was obviously important to investigate the effects of long-term therapeutic treatment of animals with these ligands. Tumor xenografts of A549 cells were grown in the flanks of nude mice and randomly assigned into treatment groups with vehicle, vehicle + carboplatin, pioglitazone, pioglitazone + carboplatin, SR1664, or SR1664 + carboplatin. Tumors from animals treated with SR1664 and carboplatin were significantly smaller than tumors from animals treated with vehicle and carboplatin alone. This trend was evident after about 2 wk of treatment, and became statistically significant by 30 d and remained so through the end of the experiment (Fig. 3C). These data were replicated in an independent experiment that showed essentially the same results (Fig. S3B). Tumor weights were measured from this second experiment (Fig. 3D) and confirmed that the SR1664/carboplatin group tumors weighed significantly less than those treated with vehicle/carboplatin (P = 0.016). Tumors from animals treated with SR1664/carboplatin trended toward being smaller than those from animals treated with pioglitazone/carboplatin (P = 0.058).
To verify that our treatment was affecting the S273 phosphorylation of PPARγ, we analyzed expression of the core gene set altered by inhibition of PPARγ phosphorylation in the presence of carboplatin (Fig. S4C). Sixteen of the 23 genes that were identified in our gene set were coordinately regulated in the appropriate direction (χ2 P = 0.06), suggesting that the xenografts were indeed responding to the effects of inhibition of PPARγ phosphorylation.
PPARγ Phosphorylation Plays a Role in the Response to DNA Damage.
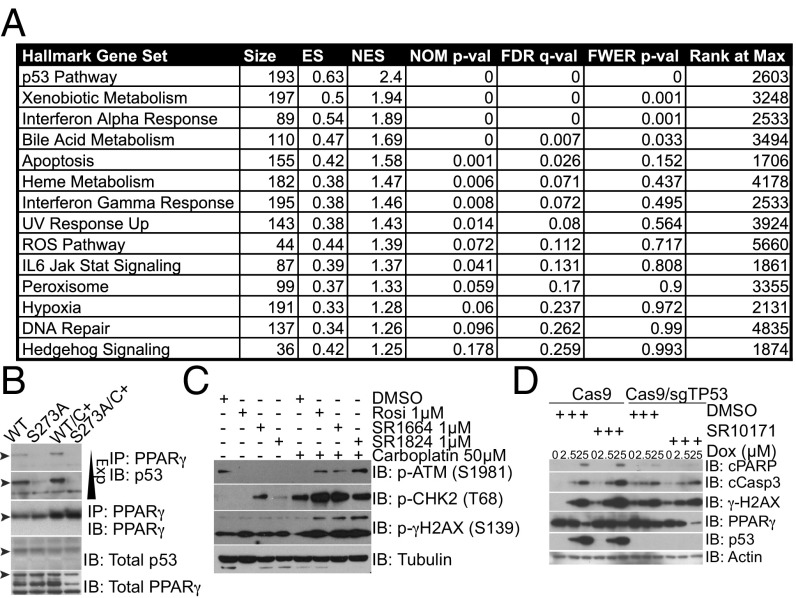
Although our prior data implicated metallothionein gene expression as a potential mechanism for the sensitization of TZDs to the effects of carboplatin, the broader effects of NALs with DNA damaging agents suggests a more general mechanism must be at work. Furthermore, treatment of A549 cells with NALs did not affect metallothionein gene expression (Fig. S4A). To assess for potential mechanisms of the increased sensitivity to genotoxic drugs, we performed gene set enrichment analysis (GSEA) using the microarray data generated above (Fig. 4A). The most enriched gene set associated with S273A mutation was the p53 pathway (Fig. S4C), along with several other DNA damage pathways. This analysis raises the intriguing possibility that the single amino acid change in the S273A knock-in mutants results in alteration of certain aspects of the DNA damage response.
Fig. 4.
(A) Table from GSEA of the microarray gene-expression data generated from wild-type vs. S273A fibroblasts treated with carboplatin was performed using the Hallmark gene sets. (B) IP of endogenous PPARγ and immunoblots for p53 reveals a preferential association between wild-type PPARγ and p53 compared with the S273A mutant. (C) Western blotting of lysates harvested from A549 cells show accumulation of markers of the activated DNA damage response. (D) CRISPR/Cas9-mediated deletion of TP53 abrogrates the ability of NALs to sensitize A549 cells to doxorubicin.
Given the effects on the p53 gene set, we hypothesized that there may be a biochemical interaction between p53 and PPARγ. Immunoprecipitation (IP) of PPARγ from nuclear extracts of wild-type or S273A mutant fibroblasts followed by immunoblotting for p53 demonstrates that the wild-type PPARγ physically associates with p53 while the S273A mutant does not (Fig. 4B). This is true both in the presence and the absence of carboplatin, although there is increased association of p53 upon carboplatin treatment. There is no difference in the total levels of PPARγ, and no difference in nuclear accumulation of PPARγ (Fig. S4B). These data suggest that phosphorylation of PPARγ stabilizes the interaction of PPARγ and p53, and that mutant PPARγ that cannot be phosphorylated is not able to associate with p53 as efficiently. This differential interaction provides a potential mechanism whereby the blocking the phosphorylation of PPARγ with NALs reduces the interaction with p53 and potentiates apoptotic cell death.
Because p53 plays a central role in coordinating the DNA damage response, we next examined the effects of inhibition of S273 phosphorylation on markers of the DNA damage response. A549 cells treated with the NALs SR1664 and SR1824 showed increased accumulation of S1981 phospho-ATM (Fig. 4C) and increased phosphorylation of Chk2 T68, both markers of increased DNA damage signaling. Interestingly, increased accumulation of γ-H2AX, a marker of DNA double-strand breaks, was also seen. Thus, cells treated with NALs and genotoxic agents show an increased amount of unrepaired DNA damage.
We hypothesized that the interaction of p53 and PPARγ may play an important role in the ability of NALs to sensitize cancer cells to DNA damage. We examined the effects of NALs in combination with carboplatin in Calu-1 cells, which have deletion of p53 (Fig. S4F), as well as H2009 cells, which express mutant p53 (Fig. S4D). In both cell types, we fail to see an increase in the DNA damage marker γ-H2AX when cells are treated with both SR1664 and carboplatin. Ectopic expression of wild-type p53 in Calu-1 cells rescued the ability of SR1664 to sensitize cells to the DNA damage produced by carboplatin (Fig. S4F). These data suggest that the presence of wild-type p53 is required for the sensitizing effects of NALs.
To further investigate the role of p53 in the ability of NALs to sensitize cells to DNA damaging agents, we performed CRISPR/Cas9-mediated deletion of TP53 from A549 cells. Control cells transduced with Cas9 alone show robust increases in cleaved PARP and cleaved Caspase 3 when treated with the NAL SR10171 and doxorubicin compared with doxorubicin alone (Fig. 4D, lane 3 vs. lane 6). Contrastingly, cells depleted of p53 show no significant increase in accumulation of apoptotic markers or γ-H2AX phosphorylation when cotreated with SR10171 and doxorubicin. We also confirmed these results using shRNA-mediated knockdown of TP53 from A549 cells (Fig. S4E). These data suggest that p53 is required for the ability of NALs to sensitize cells to genotoxic agents.
Discussion
We have shown that NALs are able to sensitize cancer cells to DNA damaging agents in vitro and in vivo. Our data show that PPARγ is phosphorylated upon exposure to DNA damage, and inhibition of this phosphorylation results in increased DNA damage and tumor cell death. Taken together, these data indicate that PPARγ is a very promising target for cancer-directed therapy, and that modulation of S273 phosphorylation may provide a wider therapeutic window than conventional agonist ligands.
These data show that cell-autonomous effects of NALs cause increased apoptosis in response to DNA damaging agents, but the magnitude of the effects in vivo seem to be larger than the effects seen on fractional cell growth in vitro. It is possible that there may be additional effects of NALs on other cell types in the tumor microenvironment, including immune cells that may play a significant role in impacting tumor growth in vivo.
We have demonstrated that mutation of a single amino acid of PPARγ results in a profound change in the response of fibroblasts to a variety of DNA damaging agents. Our data suggest a model where phosphorylation of PPARγ is a cellular response to DNA damage, and this nuclear receptor then aids in the repair response via p53. Inhibition of phosphorylation by NALs disrupts the PPARγ/p53 interaction, resulting in increased DNA damage, which triggers apoptotic cell death. To the best of our knowledge, PPARγ has not been known to be involved in DNA repair or in the response to DNA damaging agents. At this point, we have not identified the kinase responsible for PPARγ phosphorylation. It is reasonable to suspect that one of any number of kinases involved in the DNA damage response (e.g., DNA-PK, ATM, or ATR) might be involved. However, the S273 site is not within a consensus motif for these PI3K family members, making this possibility less likely. However, this site can be phosphorylated both by ERK (14) and CDK5 (13), both of which can be activated by DNA damage (30, 31). Additional studies are needed to clarify which of these kinases play a role in PPARγ phosphorylation.
These data suggest that this therapeutic strategy would be best adopted in p53 wild-type tumors, which accounts for ∼50% of human cancers. Our data also imply that expression of the S273A gene signature is associated with a trend toward improved outcomes after chemotherapy in multiple cancer types. Because these samples were taken before chemotherapy, the differences in gene expression among samples suggest that there may be phosphorylation occurring in tumors at baseline. We hypothesize that tumor inflammation, ongoing DNA damage, or other factors may result in phosphorylation of PPARγ in some tumors, which results in low expression of the gene signature. Direct interrogation of phosphorylation in tumors via better phospho-specific antibodies or mass spectrometry would also help clarify the extent of PPARγ phosphorylation in vivo. We believe that the patients most likely to show a synergy with NALs and chemotherapy would be those with tumors with a low signature score. Treatment with NALs might boost expression of the gene signature by inhibiting phosphorylation and sensitize those tumors to adjuvant chemotherapy. Of course, such an approach would need to be tested in a prospective manner.
To our knowledge, this work is unique in reporting an interaction between PPARγ and the tumor suppressor P53. We have shown via IP that wild-type PPARγ can interact with p53, while the S273A mutant is unable to bind. This interaction may be direct, or may be indirect in a larger protein complex. Further characterization of this interaction may yield insights into which aspects of p53 biology are specifically being affected by PPARγ. Interestingly, p53 has been show to play a role in adipose tissue inflammation (32), a scenario where PPARγ is also phosphorylated. Our observations raise the possibility that the PPARγ/p53 interaction may also play an important role in adipose tissue biology.
One intriguing aspect of this work is that the induction of markers of apoptosis is much greater with SR1664 compared with rosiglitazone. One potential explanation for this change is due to their differential effects on cell cycle progression. In general, cells confronted with genotoxic stress can either arrest at some stage of the cell cycle and attempt repair or initiate apoptotic cell death. One function of conventional agonist PPARγ ligands like TZDs is to cause cell cycle arrest (22) as part of its prodifferentiation program. The NALs do not appear to influence mitotic progression. Thus, cells treated with these drugs do not arrest the cell cycle, perhaps allowing for continued division in the face of DNA damage, triggering apoptotic cell death. Consistent with that theory, there was a trend toward increased tumor control in xenografts treated with SR1664 compared with TZDs, although this did not achieve statistical significance. Additional studies appropriately powered to detect these differences may provide further insight into the differential efficacy of these drugs.
In this study, we have shown activity of PPARγ ligands in combination with DNA damaging agents in nonsmall cell lung cancer as well as triple-negative breast cancer. However, PPARγ is expressed in a variety of other cancers, including 22% of colorectal cancer, (33) and 71% of pancreatic cancers (34). However, PPARγ has been largely overlooked as a potential therapeutic in cancer, likely due to the controversies surrounding the black box warning for rosiglitazone as well as failure in nonbiomarker driven clinical studies (35). Of course, not all tumor cells (e.g., HCT116 colorectal cells) are sensitized by NALs, which may result from any combinations of low expression levels of PPARγ, lack of phosphorylating kinases, poor drug penetration, or efflux pumps that limit effective drug concentrations. However, we believe that these data suggest that targeting PPARγ phosphorylation may be a valuable therapeutic approach applicable in combination with a wide variety of genotoxic agents directed toward many different cancer types.
Experimental Procedures
Detailed methods for reagents, antibodies, cell culture, qPCR, protein analysis, and microarray analysis are provided in SI Experimental Procedures.
Animal Experiments.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Beth Israel Deaconess Medical Center. Six- to 10-wk-old nude male mice were randomly assigned to treatment groups of: vehicle, vehicle + carboplatin (50 mg/kg, Monday, Wednesday, and Friday), pioglitazone (7.5 mg/kg, twice a day) + carboplatin, SR1664 (20 mg/kg), and SR1664 + carboplatin. Full methods are detailed in SI Experimental Procedures.
Microarray Analysis.
Full methods of microarray analysis are in SI Experimental Procedures. A gene set was defined as genes that were >threefold up-regulated with a significant P value (false-discovery rate P < 0.05). A refined gene set was generated from these genes with exclusion of genes that were not expressed across a wide variety of cells and tissues. GSEA was performed as described previously (36) using the Hallmark Gene sets defined in the MSigDB.
For analysis of clinical data, raw Affymetrix data and clinical data were downloaded from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) for the NIH Director’s Challenge’s study (27) and the UT Lung Spore Cohort (28), along with the clinical data. A gene signature score reflecting greater or less than median gene expression was defined. The association of signature classification with survival was analyzed using the Kaplan–Meier method in R studio. As an alternative approach, we used the online tool KMplot (kmplot.com/analysis/index.php?p=background) to analyze data for breast cancer (37).
Statistics.
Student’s test was used for single comparisons of mean values. Error bars represent ± SEM except when otherwise specified. A two-way ANOVA was used to compare multiple groups. A chi-square test was used to compare gene expression changes in the gene set. An asterisk (*) indicates P < 0.05 except when specified.
Supplementary Material
Acknowledgments
We thank Dr. Pere Puigserver, Dr. Rana Gupta, Dr. David Miyamoto, and Dr. Evan Rosen for helpful comments, as well as constructive comments from members of the B.M.S. laboratory. This work was supported by National Cancer Institute–Massachusetts General Hospital Federal Share Program NCI-C06-CA-059267 (to M.J.K.); Department of Defense Lung Cancer Research Program Career Development Award LC140129 (to M.J.K.); NIH Grants DK31405 (to B.M.S.) and DK107717 (to A.S.B.); and the JPB Foundation (B.M.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE107709).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717776115/-/DCSupplemental.
References
- 1.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 3.Kroll TG, et al. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma [corrected] Science. 2000;289:1357–1360; erratum in (2000) 289:1474. doi: 10.1126/science.289.5483.1357. [DOI] [PubMed] [Google Scholar]
- 4.Sarraf P, et al. Loss-of-function mutations in PPAR gamma associated with human colon cancer. Mol Cell. 1999;3:799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- 5.Mueller E, et al. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 6.Mueller E, et al. Effects of ligand activation of peroxisome proliferator-activated receptor gamma in human prostate cancer. Proc Natl Acad Sci USA. 2000;97:10990–10995. doi: 10.1073/pnas.180329197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith MR, et al. Rosiglitazone versus placebo for men with prostate carcinoma and a rising serum prostate-specific antigen level after radical prostatectomy and/or radiation therapy. Cancer. 2004;101:1569–1574. doi: 10.1002/cncr.20493. [DOI] [PubMed] [Google Scholar]
- 8.Burstein HJ, et al. Use of the peroxisome proliferator-activated receptor (PPAR) gamma ligand troglitazone as treatment for refractory breast cancer: A phase II study. Breast Cancer Res Treat. 2003;79:391–397. doi: 10.1023/a:1024038127156. [DOI] [PubMed] [Google Scholar]
- 9.Girnun GD, et al. Regression of drug-resistant lung cancer by the combination of rosiglitazone and carboplatin. Clin Cancer Res. 2008;14:6478–6486. doi: 10.1158/1078-0432.CCR-08-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girnun GD, et al. Synergy between PPARgamma ligands and platinum-based drugs in cancer. Cancer Cell. 2007;11:395–406. doi: 10.1016/j.ccr.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J, Morley TS, Scherer PE. Inhibition of endotrophin, a cleavage product of collagen VI, confers cisplatin sensitivity to tumours. EMBO Mol Med. 2013;5:935–948. doi: 10.1002/emmm.201202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soccio RE, Chen ER, Lazar MA. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab. 2014;20:573–591. doi: 10.1016/j.cmet.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JH, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banks AS, et al. An ERK/Cdk5 axis controls the diabetogenic actions of PPARγ. Nature. 2015;517:391–395. doi: 10.1038/nature13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marciano DP, et al. Pharmacological repression of PPARgamma promotes osteogenesis. Nat Commun. 2015;6:7443. doi: 10.1038/ncomms8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi JH, et al. Antidiabetic actions of a non-agonist PPARγ ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandekar MJ, Cohen P, Spiegelman BM. Molecular mechanisms of cancer development in obesity. Nat Rev Cancer. 2011;11:886–895. doi: 10.1038/nrc3174. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin PJ, et al. Evaluation of metformin in early breast cancer: A modification of the traditional paradigm for clinical testing of anti-cancer agents. Breast Cancer Res Treat. 2011;126:215–220. doi: 10.1007/s10549-010-1224-1. [DOI] [PubMed] [Google Scholar]
- 20.Shackelford DB, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiralerspong S, et al. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol. 2009;27:3297–3302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altiok S, Xu M, Spiegelman BM. PPARgamma induces cell cycle withdrawal: Inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai Y, et al. Peroxisome proliferator-activated receptor-gamma contributes to the inhibitory effects of Embelin on colon carcinogenesis. Cancer Res. 2009;69:4776–4783. doi: 10.1158/0008-5472.CAN-08-4754. [DOI] [PubMed] [Google Scholar]
- 24.Fu F, et al. Expression of receptor protein tyrosine phosphatase ζ is a risk factor for triple negative breast cancer relapse. Biomed Rep. 2016;4:167–172. doi: 10.3892/br.2016.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd KL, Cree IA, Savage RS. Prediction of resistance to chemotherapy in ovarian cancer: A systematic review. BMC Cancer. 2015;15:117. doi: 10.1186/s12885-015-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Ayllon BD, et al. Cancer stem cells and cisplatin-resistant cells isolated from non-small-lung cancer cell lines constitute related cell populations. Cancer Med. 2014;3:1099–1111. doi: 10.1002/cam4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shedden K, et al. Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang H, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19:1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian B, Yang Q, Mao Z. Phosphorylation of ATM by Cdk5 mediates DNA damage signalling and regulates neuronal death. Nat Cell Biol. 2009;11:211–218. doi: 10.1038/ncb1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang D, et al. ERK activation mediates cell cycle arrest and apoptosis after DNA damage independently of p53. J Biol Chem. 2002;277:12710–12717. doi: 10.1074/jbc.M111598200. [DOI] [PubMed] [Google Scholar]
- 32.Minamino T, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 2009;15:1082–1087. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 33.Ogino S, et al. Colorectal cancer expression of peroxisome proliferator-activated receptor gamma (PPARG, PPARgamma) is associated with good prognosis. Gastroenterology. 2009;136:1242–1250. doi: 10.1053/j.gastro.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristiansen G, et al. Peroxisome proliferator-activated receptor gamma is highly expressed in pancreatic cancer and is associated with shorter overall survival times. Clin Cancer Res. 2006;12:6444–6451. doi: 10.1158/1078-0432.CCR-06-0834. [DOI] [PubMed] [Google Scholar]
- 35.Shaw AT, et al. 2012. Randomized phase 2 study of efatutazone in combination with carboplatin and paclitaxel as first-line therapy for metastatic nonsmall cell lung cancer (NSCLC). Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research (American Association for Cancer Research, Philadelphia), Abstract 4606.
- 36.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 37.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.