Significance
Gut microbes positively affect the physiology of many animals, but the molecular mechanisms underlying these benefits remain poorly understood. Recent studies indicate that gut bacteria reduce oxygen levels in the mosquito gut, which serves as a growth signal. Here, we report that transduction of a bacteria-induced low-oxygen signal requires mosquito-encoded hypoxia-inducible transcription factors (HIFs). Our results further indicate that HIFs activate several processes with essential growth and metabolic functions. These findings can potentially be used to disrupt mosquito development into adults that transmit human diseases.
Keywords: insect, microbiota, growth, metabolism, molting
Abstract
Gut microbes positively affect the physiology of many animals, but the molecular mechanisms underlying these benefits remain poorly understood. We recently reported that bacteria-induced gut hypoxia functions as a signal for growth and molting of the mosquito Aedes aegypti. In this study, we tested the hypothesis that transduction of a gut hypoxia signal requires hypoxia-induced transcription factors (HIFs). Expression studies showed that HIF-α was stabilized in larvae containing bacteria that induce gut hypoxia but was destabilized in larvae that exhibit normoxia. However, we could rescue growth of larvae exhibiting gut normoxia by treating them with a prolyl hydroxylase inhibitor, FG-4592, that stabilized HIF-α, and inhibit growth of larvae exhibiting gut hypoxia by treating them with an inhibitor, PX-478, that destabilized HIF-α. Using these tools, we determined that HIF signaling activated the insulin/insulin growth factor pathway plus select mitogen-activated kinases and inhibited the adenosine monophosphate-activated protein kinase pathway. HIF signaling was also required for growth of the larval midgut and storage of neutral lipids by the fat body. Altogether, our results indicate that gut hypoxia and HIF signaling activate multiple processes in A. aegypti larvae, with conserved functions in growth and metabolism.
Gut-dwelling microbes positively affect the physiology of many animals, including insects (1, 2). However, the mechanisms through which gut microbes exert their beneficial effects remain unknown in most species (1, 2). Mosquitoes are important insects because they vector pathogens that cause disease in humans and other vertebrates. Recent studies indicate that gut microbes also strongly affect mosquito biology across all life stages. Acquisition and transmission of vertebrate pathogens only occurs through blood feeding by adult females (3), with several studies providing evidence that the gut microbiota affects permissiveness to infection (4–6). Recent evidence also indicates that mosquitoes require a gut microbiota to develop into adults (4, 6).
All mosquitoes hatch from eggs into larvae that are aquatic and feed primarily on detritus (6, 7). Most species also molt through four instars before metamorphosis into adults (3). First instars hatch with no gut microbes but rapidly acquire a gut microbiota from the environment by feeding (6). Comparative studies indicate that most gut community members are gram-negative aerobic and facultatively anaerobic bacteria, with diversity varying greatly within and between mosquito species as a function of collection site and other factors (8–16). Aedes aegypti is a broadly distributed mosquito that serves as the primary vector of the viruses that cause Dengue fever, yellow fever, and Zika syndrome (17, 18). A. aegypti larvae reared under conventional (nonsterile) laboratory conditions harbor a relatively simple gut microbiota of ∼100 bacterial species when fed a nutritionally complete diet (12). Axenic larvae with no gut microbiota consume food like conventional larvae but do not grow and die as first instars after several days (12). However, several community members and Escherichia coli K-12, which is not a community member, can individually colonize the gut to produce monoxenic, gnotobiotic larvae (i.e., larvae colonized by a single known species of bacteria) and rescue development (12, 16). Field-collected A. aegypti and several other species exhibit the same defects under axenic conditions, while development into adults is rescued if larvae are inoculated with different gut community members or E. coli (12, 16). Overall, these results indicate that A. aegypti and other mosquitoes require living microbes in their gut to become adults, but development does not depend on a particular species or community of microbes.
We recently used A. aegypti larvae fed a nutritionally complete diet and E. coli as a one-host/one-microbe model to characterize what bacteria in the gut provide that mosquito larvae require for development (19). Within each instar, mosquitoes and other insects grow by consuming nutrients until achieving a critical size, which stimulates a rise in titer of the hormone 20-hydroxyecdyone (20E) that stimulates molting (20, 21). Comparing gnotobiotic larvae inoculated with wild-type E. coli with conventionally reared first instars indicated both exhibit low midgut oxygen levels during growth to critical size, which is followed by 20E release and molting to the second instar at ∼24 h posthatching (19). In contrast, axenic first instars or gnotobiotic first instars inoculated with a mutant defective for cytochrome BD oxidase (ΔcydB-ΔcydD::kan E. coli) consume food but do not exhibit reduced gut oxygen levels, which correlates with minimal growth, no rise in 20E titer, and a failure to molt (19). These findings strongly suggested that aerobic respiration by bacteria reduces oxygen levels in the A. aegypti gut, and that hypoxia functions as a signal for growth and molting to the second instar. Additional experiments further supported that bacteria-induced gut hypoxia also functions as a signal for growth and molting in subsequent instars until pupation (19).
Some eukaryotic cells respond to fluctuating oxygen levels through conserved dimeric α/β hypoxia-induced transcription factors (HIFs) (22–24). HIFs are expressed under normoxia, but hydroxylation by a prolyl hydroxylase (PHD) usually targets HIF-α for degradation. In contrast, hydroxylation does not occur under hypoxia, resulting in HIF-α stabilization and activation of hypoxia-responsive genes (22–24). In this study, we tested the hypothesis that bacteria-induced gut hypoxia requires HIF signaling to function as a growth signal for A. aegypti larvae. Our results show that gut hypoxia and a PHD inhibitor stabilize HIF-α and stimulate growth, while normoxia and an HIF-α inhibitor disable growth. Our results further indicate HIF signaling is required for activation of downstream pathways and processes with essential growth functions.
Results
Bacteria-Induced Gut Hypoxia Stabilizes HIF-α in A. aegypti First Instars.
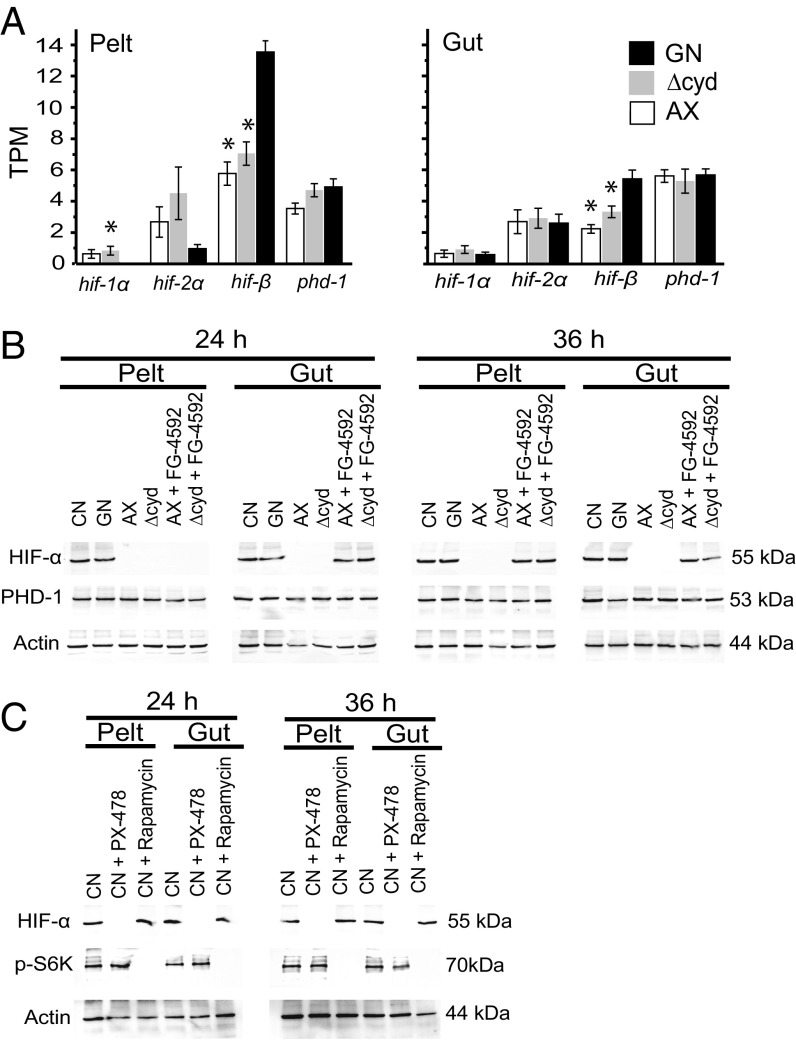
A. aegypti encodes conserved HIF pathway components, which include two HIF-α paralogs [hif-1α (AAEL001097) and hif-2α (AAEL015383)], HIF-β (AAEL010343), and PHD-1 (AAEL02798) (19). At the mRNA level, we assessed transcript abundance for these genes at 12 h posthatching in gnotobiotic first instars inoculated with wild-type E. coli, axenic first instars, and gnotobiotic first instars inoculated with ΔcydB-ΔcydD::kan E. coli (hereafter abbreviated Δcyd E. coli) (19). For each treatment, we examined the gut and remaining larval body (pelt), which primarily contained the fat body. Results indicated each gene was similarly expressed between treatments and tissues, with the exception of hif-β, which was more abundant in the gut and pelt of gnotobiotic larvae inoculated with wild-type E. coli (Fig. 1A). At the protein level, we generated one antibody designed to detect PHD-1 and a second antibody designed to detect both HIF-α paralogs. We also examined each treatment assessed at the mRNA level plus conventionally reared first instars that contained a mixed laboratory community of gut bacteria. Immunoblot analysis at 24 and 36 h posthatching detected PHD in the gut and pelt across all treatments (Fig. 1B). In contrast, HIF-α was detected in conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli, but it was not detected in axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli (Fig. 1B).
Fig. 1.
Detection of hypoxia pathway components in A. aegypti larvae. (A) Transcript abundance of hypoxia pathway genes in the gut and body (pelt) of first instars at 12 h posthatching. Treatments are gnotobiotic larvae inoculated with wild-type E. coli (GN), gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli (Δcyd), and axenic larvae (AX). Values are expressed in transcripts per kilobase million (TPM). For each gene, an asterisk above the bar indicates the treatment differed from GN, which served as the positive control (ANOVA followed by a post hoc Dunnett’s test, P < 0.05). (B) Immunoblots of pelt and gut extracts from conventional (CN) larvae, GN, AX, Δcyd, AX treated with FG-4592 (12 h posthatching), or Δcyd treated with FG-4592 (12 h posthatching). Samples were collected at 24 or 36 h and probed with antibodies to HIF-α, PHD-1, or actin (loading control). The molecular masses of HIF-α, PHD-1, and actin are indicated to the right of each blot. (C) Immunoblots of pelt and gut extracts from CN larvae and from CN larvae treated with PX-478 (12 h posthatching) or rapamycin (12 h posthatching). Samples were collected at 24 h and 36 h, and were probed with antibodies to HIF-α, p-S6K, or actin, with molecular masses of each indicated to the right of each blot.
These findings indicated that HIF-α was stabilized in conventional and gnotobiotic first instars inoculated with wild-type E. coli, which exhibit gut hypoxia, but was destabilized in axenic and gnotobiotic first instars inoculated with Δcyd E. coli, which do not. We thus assessed whether two small-molecule inhibitors altered HIF-α stability in A. aegypti: FG-4592, which is an oral, selective PHD inhibitor that stabilizes HIF-α in vertebrates under normoxia (25, 26), and PX-478, which is an oral selective inhibitor of HIF-α (25, 27). We also assessed the effects of rapamycin, which is a well-known inhibitor of the target of rapamycin C1 complex (TORC1) (28–30), because of (i) its known effects on delaying larval growth of insects (31) and (ii) the potential that TOR signaling can affect HIF stabilization (32). Previous results showed that adding FG-4592 (1 μM) to cultures at 12 h posthatching stimulated gnotobiotic first instars inoculated with Δcyd E. coli to grow, release 20E, and molt by 72 h (19). In the current study, immunoblotting further showed that FG-4592 stabilized HIF-α in the gut of axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli by 24 h posthatching and pelt by 36 h posthatching (Fig. 1B). Reciprocally, conventional first instars treated with PX-478 at 12 h posthatching remained viable over a range of concentrations but exhibited a dose-dependent reduction in molting to the second instar (Fig. S1). Larvae treated with the lowest dose of PX-478 that inhibited molting (50 μM) showed a loss of HIF-α by 24 h posthatching but exhibited no reduction in phosphorylated S6 kinase (p-S6K) (Fig. 1C), which is a well-known downstream target of TORC1 (28). Conventional first instars treated with rapamycin at 12 h posthatching also remained viable and exhibited a dose-dependent reduction in molting to the second instar within 72 h (Fig. S1). However, larvae treated with the lowest dose of rapamycin that delayed molting (50 μM) exhibited greatly reduced p-S6K in the gut and pelt but no reduction in HIF-α (Fig. 1C). We thus concluded that (i) FG-4592 stabilizes HIF-α at the protein level and rescues growth of A. aegypti larvae that exhibit gut normoxia, (ii) PX-478 reduces HIF-α and inhibits growth of conventional larvae that exhibit gut hypoxia, and (iii) rapamycin inhibits TOR signaling and delays growth of conventional larvae but does not affect HIF-α stabilization.
Gut Hypoxia and HIF Signaling in A. aegypti Larvae Affect Signaling Pathways with Growth and Metabolic Functions.
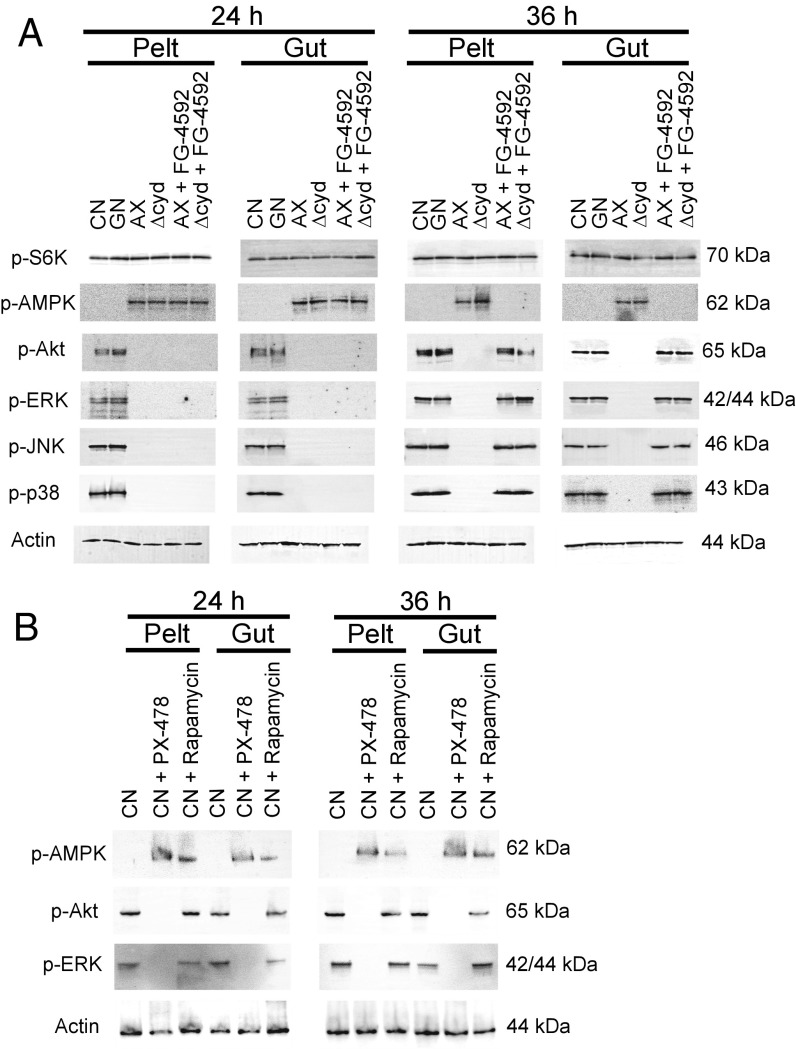
Signaling through TORC1 functions as a conserved nutrient-sensing pathway essential for growth of higher eukaryotes, including insects (28, 33). Two other pathways with conserved metabolic and growth functions are adenosine monophosphate-activated protein kinase (AMPK) signaling and insulin/insulin growth factor signaling (IIS). AMPK functions as a stress-activated energy sensor, which integrates multiple responses that enhance cellular resistance to stress (34, 35). The IIS pathway in A. aegypti and most other insects consists of a single insulin-like receptor that binds insulin-like peptides and activates two signaling cascades: a phosphatidylinositol 3-kinase (PI3K)/Akt branch with primarily metabolic functions and a mitogen-activated protein kinase (MAPK)/ERK branch with primarily growth functions (33, 36–39). We assessed whether activation of these pathways in conventional and gnotobiotic first instars inoculated with wild-type E. coli differed from axenic first instars or first instars inoculated with Δcyd E. coli by monitoring the phosphorylation status of specific proteins. S6K was used as a marker for TOR signaling as described above (28). Phosphorylation of AMPK was monitored directly (28), while Akt and ERK were used as established markers in mosquitoes for activation of the PI3K/Akt and MAPK/ERK branches of the IIS pathway (37–41). We also examined two other MAPK family members, c-Jun N-terminal kinase (JNK) and p38, because both have growth functions that can be activated by several pathways, including IIS, in other organisms (42–44).
We detected no differences across treatments in the phosphorylation status of S6K (Fig. 2A). In contrast, p-AMPK was only detected in axenic and gnotobiotic larvae inoculated with Δcyd E. coli, while phosphorylated Akt, ERK, JNK, and p38 were only detected in conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli (Fig. 2A). Since axenic and gnotobiotic larvae inoculated with Δcyd E. coli consume food (10), we reasoned that nutrient sensing through TOR is potentially operative despite larvae not growing. This interpretation was supported by analysis of conventional first instars that were reared without food (i.e., starved), which also do not grow or molt but exhibited reduced phosphorylation of S6K, Akt, and ERK, plus increased phosphorylation of AMPK (Fig. S2). We thus concluded that possible effectors of disabled growth in axenic larvae and gnotobiotic larvae inoculated with Δcyd E. coli included reduced signaling through the IIS pathway, reduced signaling through select other MAPKs, and increased signaling through the AMPK pathway. In contrast, reduced TOR signaling was only observed in conventional larvae that were starved or treated with rapamycin.
Fig. 2.
Gut bacteria and pharmacological manipulation of HIF-α affect the phosphorylation status of proteins in the TOR, AMPK, and IIS pathways plus select other MAPKs. (A) Immunoblots of pelt and gut extracts from conventional (CN) larvae, gnotobiotic larvae inoculated with wild-type E. coli (GN), axenic larvae (AX), gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli (Δcyd), AX with FG-4592 (12 h posthatching), or Δcyd treated with FG-4592 (12 h posthatching). Samples were collected at 24 h or 36 h and probed with antibodies to p-S6K, p-AMPK, p-Akt, p-ERK, p-JNK, p-38, or actin, with molecular masses of each protein target indicated to the right. (B) Immunoblots of pelt and gut extracts from CN larvae and CN larvae treated with PX-478 (12 h posthatching) or rapamycin (12 h posthatching). Samples were collected at 24 h and 36 h, and were probed with antibodies to p-AMPK, p-Akt p-ERK, or actin. The molecular masses of each protein are indicated to the right of each blot.
If the alterations exhibited by the IIS, MAPK, and AMPK pathways are related to gut normoxia and destabilized HIF-α, we hypothesized that FG-4592 should reverse these defects in axenic larvae or gnotobiotic larvae inoculated with Δcyd E. coli. Results strongly supported this prediction by showing that larvae treated with FG-4592 at 12 h posthatching showed increased phosphorylation of Akt, ERK, JNK, and p38 but reduced phosphorylation of AMPK by 36 h (Fig. 2A). Reciprocally, conventional larvae treated with PX-478 at 12 h posthatching exhibited increased phosphorylation of AMPK and reduced phosphorylation of Akt and ERK by 24 h, while larvae treated with rapamycin also showed increased phosphorylation of AMPK but no alteration in the phosphorylation state of Akt and ERK (Fig. 2B). Altogether, these results strongly supported that bacteria-induced gut hypoxia requires HIF signaling to stimulate growth, which correlates with activating the IIS pathway, activating select other MAPKs, and inhibiting the AMPK pathway.
Gut Hypoxia and HIF Signaling Promote Growth of the Larval Midgut.
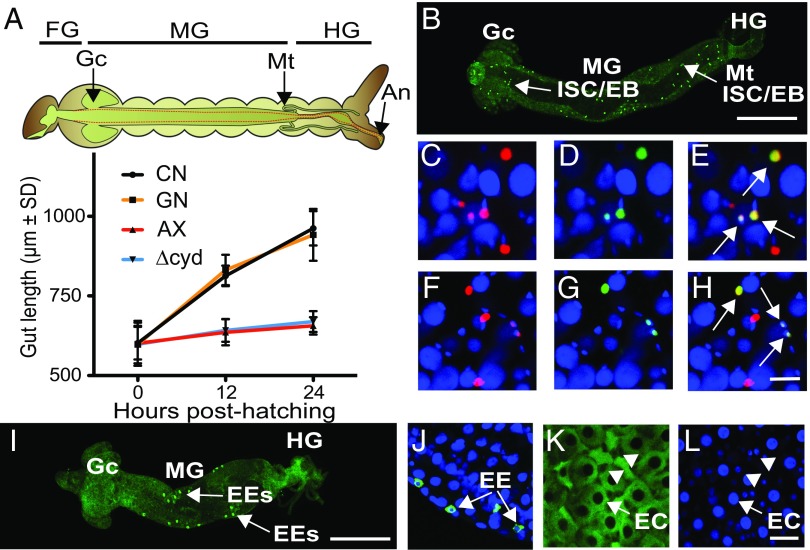
The midgut in conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli showed substantial growth after hatching as measured by length, whereas the midgut in axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli showed little growth (Fig. 3A). In adult-stage Drosophila, the midgut consists of four cell types (45, 46). Intestinal stem cells (ISCs) divide to produce one daughter ISC and one daughter enteroblast (EB) (47, 48). In turn, each EB differentiates without division into either an enteroendocrine cell (EE) or a nutrient-absorbing enterocyte (EC) (45–47). Several factors regulate ISC proliferation, including the IIS, p38, JNK, and JAK/STAT pathways (48–51). ISCs/EBs are also distinguished by their small size, small nuclei, and expression of a conserved Snail/Slug family transcription factor (escargot) (45–52). EEs are similarly small cells but do not divide and express several peptide hormones upon differentiation, including neuropeptide F (NPF) and tachykinin (TK) (50, 52). ECs, in contrast, are distinguished by their much larger size, endoreplicating nuclei, and function in uptake of nutrients like glucose (48–51).
Fig. 3.
Bacteria that induce hypoxia promote gut growth. (A) Gut length in conventional (CN) larvae, gnotobiotic larvae inoculated with wild-type E. coli (GN), axenic larvae (AX), and gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli (Δcyd) at hatching (0 h), 12 h posthatching, and 24 h posthatching. Above the graph is shown a schematic of the A. aegypti larval digestive tract with borders of the foregut (FG), midgut (Mg), and hindgut (HG) indicated. The midgut is demarcated by the gastric caeca (Gc) and Malpighian tubules (Mt). Gut length in the graph was measured from the Gc to the end of the HG at the anus (An). Gut length at 12 h and 24 h significantly differed between CN larvae and GN that exhibit gut hypoxia versus AX and Δcyd that do not (ANOVA followed by a post hoc Tukey–Kramer honest significant difference test, P < 0.05). (B) Low-magnification image of a gut from a CN larva at 24 h posthatching. The FG was removed, resulting in the MG and HG being oriented from left to right. PH3+ ISCs/EBs (bright green) are distributed over the length of the MG. (Scale bar: 200 μm.) (C–E) High-magnification images from the midgut of a 24-h posthatching CN larva. Small cells with nuclei that are Slg+ (C, red), PH3+ (D, green), and Slg+/PH3+ (E, yellow with arrows) are shown. (F–H) High-magnification images from the midgut of a 24-h posthatching GN showing Slg+, PH3+, and Slg+/PH3+ cells as labeled in C–E. All nuclei in C–H are counterstained with Hoechst 33342 (blue). (Scale bar: H, 10 μm.) (I) Low-magnification image of a gut from a CN larva at 24 h posthatching showing EEs that are NPF+ (bright green). (Orientation and scale bar as in B.) (J) High-magnification image of gut cells from a 24-h CN larva labeled with anti-NPF (green) and Hoechst 33342. Note that EEs exhibit a cytoplasmic NPF+ signal (arrows), while nuclei are blue. High-magnification images of the midgut from a 24-h CN larva showing large, columnar ECs (arrows) with cytoplasm labeled by 2-DG6P (K) and nuclei labeled with Hoechst 33342 (L) are shown. Note that ISCs/EBs or EEs with small nuclei (arrowheads) are present between ECs in K and L. (Scale bar: L, 20 μm.)
No markers have previously been generated to distinguish cell types in the mosquito midgut, although prior results do support that midgut growth during the larval stage is associated with an increase in cell size and number (53, 54). We therefore produced an A. aegypti anti-Slug (Slg) antibody, which we used in conjunction with previously generated NPF and TK antibodies (55, 56), a commercially available anti-phosphohistone H3 (PH3) antibody that marks dividing cells, and 5-ethynyl-2′-deoxyuridine (EdU) labeling. In conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli, we identified small cells distributed over the length of the midgut that were Slg+ or Slg+/PH3+, which strongly suggested they were ISCs/EBs (Fig. 3 B–H). Pairs of Slg+/PH3+ cells were also commonly observed in close proximity to one another, which was consistent with recent division of an ISC into a daughter ISC and EB (Fig. 3 C–H). Other small cells located in the midposterior midgut were NPF+/TK+/Slg−/PH3−, which identified them as differentiated EEs (Fig. 3 I and J and Fig. S3A). The most abundant cells over the length of the midgut were Sig−/PH3−/NPF−/TK−. These cells were classified as ECs on the basis that they were much larger than ISCs/EBs and EEs, absorbed the glucose analog 2-deoxyglucose-6-phosphate (2-DG6P) (Fig. 3K), and had large endoreplicating nuclei that incorporated EdU (Fig. 3L and Fig. S3B).
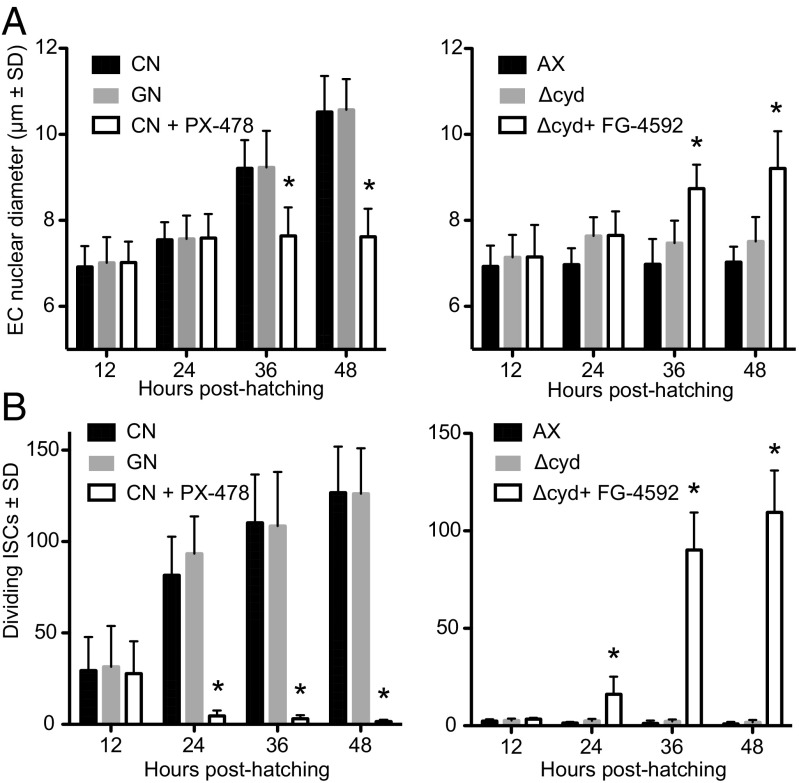
EC size, as estimated by nuclear diameter, was larger by 36 h posthatching in conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli than in axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli (Fig. 4A). Since only ISCs/EBs were PH3+, we used PH3 labeling to ask whether the abundance of proliferating ISCs also differed between treatments. Results showed that the midguts of conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli contained far more PH3+ cells than axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli (Fig. 4B). Together, these data strongly suggested bacteria-induced gut hypoxia promotes endoreplication of ECs and proliferation of ISCs. To test whether HIF signaling was required as an upstream activator of these responses, we compared the effects of FG-4592 on axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli versus PX-478 on conventional first instars and gnotobiotic first instars with wild-type E. coli. Strikingly, FG-4592 significantly increased EC nuclear diameter and the number of PH3+ ISCs/EBs in axenic first instars and gnotobiotic first instars with Δcyd E. coli, while PX-478 significantly reduced EC nuclear diameter and the number of PH3+ ISCs/EBs in conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli (Fig. 4).
Fig. 4.
Gut bacteria and pharmacological manipulation of HIF-α affect EC size and ISC proliferation. (A) EC size as measured by nuclear diameter in conventional (CN) larvae, gnotobiotic larvae inoculated with wild-type E. coli (GN), or CN larvae treated with PX-478 (Left), and in axenic larvae (AX), gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli (Δcyd), or Δcyd treated with FG-4592 (Right). Samples were measured from 12 to 48 h posthatching. An asterisk above a bar at a given time point indicates the treatment significantly differed from that of CN larvae (Left) or Δcyd (Right), which served as controls (analysis at each time point by ANOVA followed by a post hoc Dunnett’s test, P < 0.05). (B) Dividing ISCs as measured by PH3 labeling in CN larvae, GN, or CN larvae treated with PX-478 (Left), and AX, Δcyd, or Δcyd treated with FG-4592 (Right). An asterisk above a bar at a given time point indicates the treatment significantly differed from that of CN larvae (Left) or Δcyd (Right) as in A.
Gut Hypoxia and HIF Signaling also Promote Neutral Lipid Storage in the Fat Body.
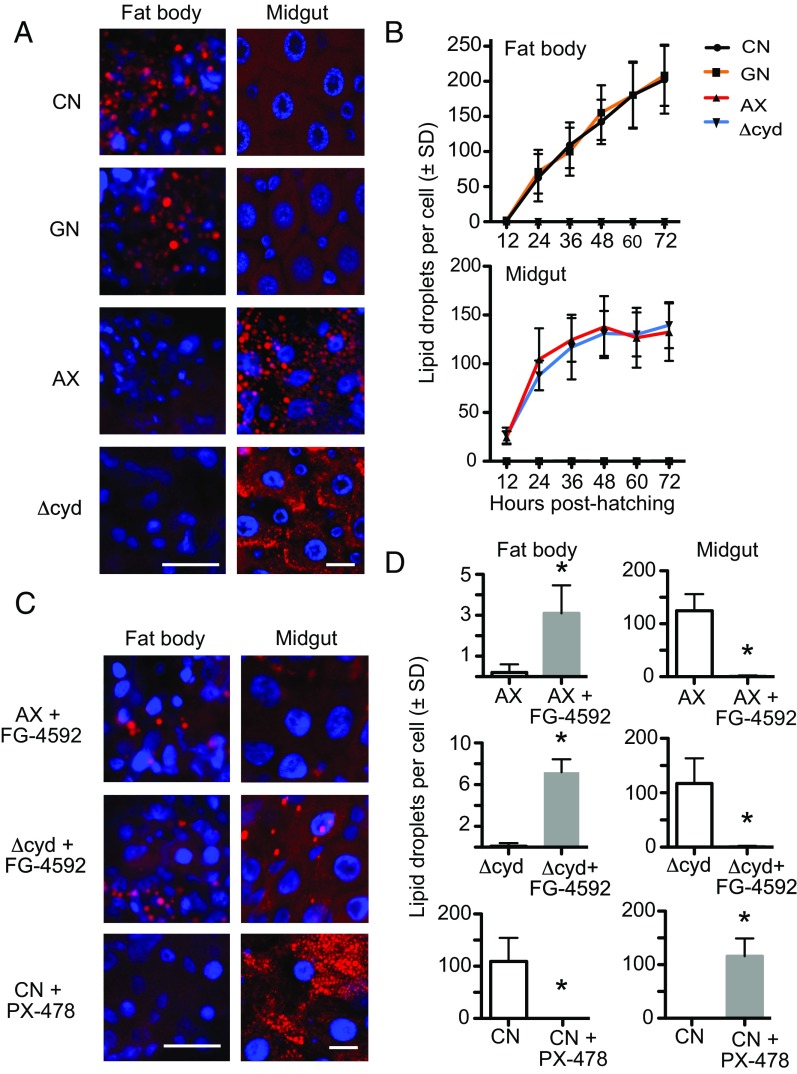
Under normal conditions, ECs absorb nutrients from digested food, which is resynthesized into triglyceride and other neutral lipids (57–60). Lipoproteins then transport these neutral lipids to the fat body, where they serve as the primary form of stored energy for growth (57–59). Lipid storage in the fat body also requires activation of the IIS pathway (33, 37, 60, 61). First-instar A. aegypti larvae hatch with minimal neutral lipid stores in the fat body (62). However, we hypothesized that activation of the IIS pathway in conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli should correlate with increased lipid stores as larvae grow, while reduced signaling through the IIS pathway in axenic larvae and gnotobiotic larvae inoculated with Δcyd E. coli should correlate with lower lipid stores since larvae do not grow. Because of the very small size of first instars (<1 mm), we quantified neutral lipid stores in fat body cells (adipocytes) by counting lipid droplets after Nile red staining (59). Results strongly supported predictions by showing that neutral lipid rapidly accumulated in fat body cells of conventional larvae and gnotobiotic larvae inoculated with wild-type E. coli, whereas no increase in stored lipid occurred in axenic larvae and gnotobiotic larvae inoculated with Δcyd E. coli (Fig. 5 A and B). Unexpectedly, however, ECs exhibited the opposite pattern, with few neutral lipid droplets detected in the fat body of conventional larvae and gnotobiotic larvae inoculated with wild-type E. coli, while an abundance of lipid droplets was detected predominantly in ECs of the anterior midgut of axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli (Fig. 5 A and B and Fig. S4 A–D). These outcomes were also clearly distinct from those of starved conventional larvae, which, as expected, showed no accumulation of neutral lipids in either the midgut or fat body (Fig. S4 E–H).
Fig. 5.
Gut bacteria and pharmacological manipulation of HIF-α affect lipid accumulation in the fat body and midgut. (A) Nile red staining of neutral lipids (red) in fat body adipocytes and midgut ECs from conventional (CN) larvae, gnotobiotic larvae inoculated with wild-type E. coli (GN), axenic larvae (AX), and gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli (Δcyd) at 36 h posthatching. Cell nuclei are counterstained with Hoechst 33342 (blue). (Scale bars: fat body and midgut images, 20 μm.) (B) Quantification of neutral lipid droplets per fat body adipocyte (Upper) or midgut EC (Lower) from CN larvae, GN, AX, and Δcyd from 0 to 72 h posthatching. For both tissues, CN larvae and GN significantly differed from AX and Δcyd by 24 h posthatching (ANOVA followed by a post hoc Tukey–Kramer honest significant difference test, P < 0.05). (C) Nile red staining of neutral lipids (red) in fat body adipocytes and midgut ECs from AX and Δcyd treated with FG-4592 or CN larvae treated with PX-478 at 36 h posthatching. (Scale bars as in A.) (D) Comparison of neutral lipid droplets per adipocyte or midgut EC in: AX versus AX + FG-4592 treated larvae, Δcyd versus Δcyd + FG-4592 treated larvae, and CN larvae versus CN + PX-478-treated larvae. Larvae were drug-treated at 12 h posthatching, and samples were analyzed at 36 h posthatching. An asterisk above a bar indicates the FG-4592 or PX-478 treatment differed from that of the nontreated control (t test, P < 0.01).
The accumulation of neutral lipids in midgut cells of axenic and gnotobiotic first instars inoculated with Δcyd E. coli suggested these larvae absorbed some nutrients from the food they consumed, but defects in lipid biosynthesis and/or transport resulted in greatly reduced lipid stores in the fat body. An essential role for HIF signaling in neutral lipid transport to the fat body was also experimentally supported by results showing that PX-478 strongly reduced neutral lipid accumulation in the fat body, while, correspondingly, increasing neutral lipid accumulation in the midguts of conventional larvae and gnotobiotic larvae inoculated with wild-type E. coli (Fig. 5 C and D). Reciprocally, FG-4592 greatly reduced the accumulation of neutral lipids in the midguts of axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli (Fig. 5 C and D). The accumulation of neutral lipids in the fat bodies of FG-4592–treated axenic first instars and gnotobiotic first instars inoculated with Δcyd E. coli was also significantly increased (Fig. 5 C and D). However, this increase was lower than occurs in conventional first instars or first instars inoculated with wild-type E. coli, which indicated the rescue effect of FG-4592 was only partial.
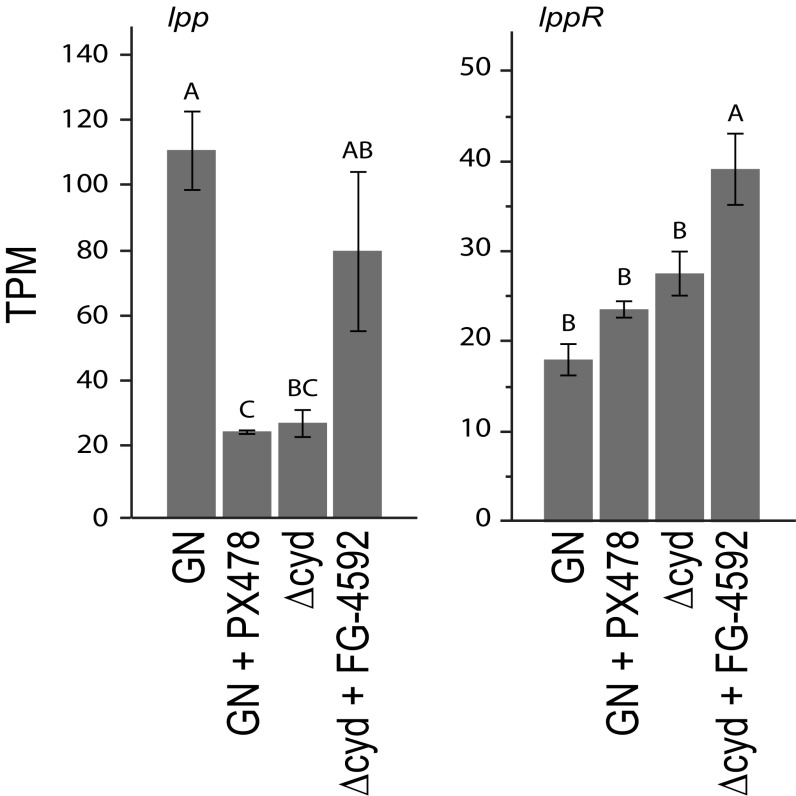
Lipophorin (Lpp) is the major neutral lipid carrier in A. aegypti and other insects, while lipid uptake by the fat body requires the Lpp receptor (LppR) (57–59). Expression data indicated that transcript abundance of lpp, but not lppR, was significantly lower in gnotobiotic first instars inoculated with Δcyd E. coli compared with gnotobiotic first instars inoculated with wild-type E. coli (Fig. 6). Notably, lpp transcript abundance was strongly reduced by PX-478 treatment of gnotobiotic first instars inoculated with wild-type E. coli, while FG-4592 treatment of gnotobiotic first instars inoculated with Δcyd E. coli increased transcript abundance of lpp, but not fully to the level of gnotobiotic first instars with wild-type E. coli (Fig. 6).
Fig. 6.
Transcript abundances of Lpp [apolipophorin 1,2 (AAEL009955)] (lpp; Left) and the LppR (AAEL018219) (lppR; Right) in pelts from gnotobiotic larvae inoculated with wild-type E. coli (GN), GN treated with PX-478 (12 h posthatching), gnotobiotic larvae inoculated with ΔcydB-ΔcydD::kan E. coli (Δcyd), or Δcyd treated with FG-4592 (12 h posthatching). Samples were collected at 12 h posthatching (GN, Δcyd) or 12 h posttreatment (GN + PX-478, Δcyd + FG-4592), with values expressed in transcripts per kilobase million (TPM). In each graph, bars with different letters significantly differ from one another (ANOVA followed by a post hoc Tukey–Kramer honest significant difference test, P < 0.05).
Discussion
Previous results indicated that bacteria-induced gut hypoxia functions as a signal that A. aeygpti larvae require for growth to a critical size, 20E release, and molting (19). Here, we tested the hypothesis that transduction of a bacteria-induced hypoxia signal requires hypoxia-inducible transcription factors. Our results establish that bacteria-induced gut hypoxia stabilizes HIF-α in first instars and that one pharmacological intervention, FG-4592, stabilizes HIF-α under gut normoxia, while another, PX-478, reduces HIF-α under gut hypoxia. Results further support that HIF signaling is required for activation of the IIS pathway and select MAPKs with known functions in regulating ISC proliferation in the midgut and neutral lipid storage in the fat body. Our rapamycin assays suggest that TOR signaling is also required for larval growth, but our results do not indicate that bacteria-induced gut hypoxia affects TOR pathway activation if larvae are provided a nutritionally complete diet.
Studies conducted in the vertebrate literature indicate the HIF pathway inhibitors we used are highly specific (25, 26), although some data do suggest prolonged treatment with PHD inhibitors can affect NF-κB transcription factors (63). Likewise, rapamycin is considered a highly specific TORC1 inhibitor, although prolonged exposure in vertebrate cell culture studies indicates it can also inhibit TORC2 activity (28). However, an important strength of our study is that results are not based on a single pharmacological intervention but, rather, on reciprocal interventions that either rescued growth of larvae that exhibit gut normoxia (axenic first instars and first instars inoculated with Δcyd E. coli) or inhibited growth of larvae that exhibit gut hypoxia (conventional first instars and gnotobiotic first instars inoculated with wild-type E. coli). Together, these treatments provide strongly corroborating evidence that bacteria-induced hypoxia stabilizes HIF-α in A. aegypti larvae and that HIF signaling is required for activation of several pathways with essential functions in growth and metabolism. While axenic larvae or gnotobiotic larvae inoculated with Δcyd E. coli fed a standard diet fail to grow like starved or rapamycin-treated conventional larvae, they also differ in exhibiting no loss of TOR signaling.
Gut bacteria produce many metabolites, including short-chain fatty acids (SCFAs) that have been implicated in systemic health of vertebrates and growth of some invertebrates (64–67). However, previous experiments showed that supplementing the diets of A. aegypti axenic first instars or gnotobiotic first instars inoculated with Δcyd E. coli with acetate or several other SCFAs has no rescue effect on growth or molting (19). Thus, our results support bacteria-induced hypoxia as a growth signal in A. aegypti, but do not identify an essential role for SCFAs in growth. In vertebrates, bacteria-derived lipopolysaccharide (LPS) has also been implicated in up-regulating HIF-1α expression through inflammatory responses mediated by Toll signaling, reactive oxygen species, and certain cytokines (68). Our finding that transcript abundance levels for HIF-β are lower in axenic larvae than gnotobiotic larvae inoculated with wild-type E. coli is consistent with a potential role for bacteria cell wall components in regulating HIF-β expression. On the other hand, our results also indicate that HIF-β transcript abundance is lower in gnotobiotic larvae inoculated with ΔcydB-ΔcydD:kan E. coli, which argues against bacterial cell wall components being the primary factor that up-regulates HIF-β expression in A. aegypti first instars that grow.
As previously noted, several factors modulate ISC proliferation in adult Drosophila, including the IIS pathway (50), EGF-activating JNK and ERK signaling (51, 52), Janus kinase signal transducers and activators of transcription (JAK-STAT) (49, 69, 70), and p38 MAPK (70). ISC proliferation also increases in response to indigenous and pathogenic bacteria in the gut, with evidence supporting a role for JNK and JAK-STAT signaling in bacteria-induced stimulation of ISCs (70–72). We did not examine JAK-STAT signaling, which is implicated in regulating proliferation of several stem cell populations in addition to ISCs (73), but our results are consistent with IIS, ERK, JNK, and p38 playing a potentially synergistic role in regulating ISC proliferation during larval growth of A. aegypti. However, our study implicates bacteria-induced gut hypoxia and HIF signaling as upstream activators of IIS, ERK, JNK, and p38. Interestingly, our study also implicates bacteria-induced gut hypoxia and HIF signaling in transport of neutral lipids from the midgut to the fat body during normal growth of A. aegypti larvae. The accumulation of neutral lipids in ECs that occurs in axenic larvae, gnotobiotic larvae inoculated with Δcyd E. coli, or conventional larvae treated with PX-478 is reminiscent of lipid accumulation in the midgut of Drosophila melanogaster adults after lethal infection by Vibrio cholera, which down-regulates the IIS pathway (66). It also resembles the effects of disabled TK production by midgut EEs, which is implicated in increased synthesis of neutral lipids by the midgut (74). Our finding that bacteria-induced gut hypoxia and HIF signaling activate the IIS pathway and lpp expression suggests a potentially important role for gut bacteria in activating normal transport of neutral lipids from the midgut to the fat body. However, that down-regulated HIF signaling in A. aegypti larvae stimulates lipid accumulation in the midgut also suggests a potential role for resident gut bacteria in either directly modulating lipid biosynthesis or indirectly modulating lipid biosynthesis through effects on TK production by EEs.
In summary, bacteria-induced gut hypoxia requires HIF signaling to function as a growth signal in A. aegypti. Given the evolutionary conservation of the HIF pathway and dependence of other mosquito species on gut microbes for growth (12, 16), we think it likely all members of the Culicidae rely on gut hypoxia as a developmental signal. Our results further suggest HIF signaling modulates several conserved signaling processes with essential functions in growth and metabolism. In vertebrates, HIFs directly regulate target genes, including transcription factors, through binding to promoter response elements (75–77). Thus, future studies on the repertoire of genes that mosquito HIFs activate will be necessary to elucidate how gut hypoxia up-regulates the IIS and other downstream pathways that larvae rely upon for development into adults.
Materials and Methods
Mosquitoes, Bacteria, and Culture Conditions.
The University of Georgia strain of A. aegypti was used in the study (19). Wild-type and mutant E. coli K12 cells were grown in Luria broth at 37 °C with 25 μg/mL kanamycin. The double mutant ΔcydB-ΔcydD::kan E. coli (referred to as Δcyd in this study) was constructed by P1 phage transduction as previously described (19). For experiments, wild-type E. coli and Δcyd E. coli cells were grown to midlog phase, followed by pelleting and resupension in water (wild-type) or M9 minimal medium (Δcyd). Conventional larvae were hatched from unsterilized eggs and individually reared at 26 °C in wells of 24-well culture plates (Corning) containing 1 mL of nonsterile double-distilled water and 100 μg of nutritionally complete diet consisting of rat chow, lactalbumin, and inactive torula yeast (1:1:1) (10). Axenic larvae were hatched from surface-sterilized eggs and individually reared in 1 mL of sterile water containing the above diet (100 μg) that had been sterilized by γ-irradiation (10). Gnotobiotic larvae were produced by placing individual axenic larvae in 1 mL of sterile water plus 100 μg of sterilized diet and 106 colony-forming units (cfu) of wild-type E. coli or 1 mL of M9 minimal medium containing 100 μg of sterilized diet and 106 cfu of Δcyd E. coli (19). Larger numbers of larvae for each condition were reared together in culture flasks (Corning) by linearly increasing the amount of water, food, and bacteria, as described above.
FG-4592, PX-478, and Rapamycin Treatment.
Stocks of FG-4592 (Selleckchem) and PX-478 (Selleckchem) were dissolved in water, while rapamycin (Selleckchem) was dissolved in ethanol. FG-4592 (1 μM) was added to cultures containing axenic or gnotobiotic larvae inoculated with Δcyd E. coli at 12 h (19). PX-478 (1–100 μM) or rapamycin (1–100 μM) was initially added to cultures containing conventional first instars, followed by determination of the proportion of larvae that molted to the second instars within 72 h. In subsequent assays, 50 μM PX-478 or rapamycin was added to cultures containing 1–300 conventional or gnotobiotic first instars inoculated with wild-type E. coli at 12 h posthatching.
RNA-Sequencing Analysis.
Cultures containing 300 axenic first instars, gnotobiotic first instars inoculated with wild-type E. coli, or Δcyd E. coli were reared for 12 or 18 h and dissected in RNase-free PBS (pH 7.4) to separate the gut and remaining body (pelt). Gnotobiotic first instars inoculated with wild-type E. coli were treated with PX-478, while gnotobiotic first instars inoculated with Δcyd E. coli were treated with FG-4592 at 12 h posthatching and similarly dissected at 24 or 36 h posthatching (12 h or 18 h posttreatment, respectively). Samples were then snap-frozen in RNase-free PBS and stored at −80 °C. Four biological replicates were performed for each treatment. Total RNA was extracted using a Qiagen RNAEasy Kit with DNA removed by two consecutive DNase treatments. Samples were then prepared for Illumina sequencing and run as single-end 75-nt reads on two high-throughput chips using the NextSeq platform. Additional details on library construction and data analysis are provided in SI Materials and Methods.
Detection of HIF-α and PHD-1.
Antibodies were commercially generated (Pacific Immunology) in rabbits to recognize proteins from hif-1α/-2α and phd-1. For a cross-reacting HIF-α antibody, a rabbit was immunized with one peptide corresponding to amino acids 292–313 (CDHEELREALNGRHHSPSELLK) that is identical in hif-1α/hif-2α plus a second peptide corresponding to amino acids 441–549 (KNVISKGQSETARYRFLAR) that is hif-1α–specific. For PHD-1, a second rabbit was immunized with peptides corresponding to amino acids 18–42 (HHQQRSQPQSPSPSSQQASPSSTSS-Cy) and 464–479 (DAEERESARLRYQDC). Gut and pelt homogenates were prepared for each of the treatments described for RNA-sequencing analysis plus conventional larvae and conventional larvae treated with PX-478. Gut and pelt samples were collected at 24 and 36 h posthatching in protease/phosphatase inhibitor mixture, with protein concentrations determined using Coomassie Plus Protein Reagent (ThermoFisher). Samples were resuspended in Laemmli buffer with mercaptoethanol (10 μM). Samples were electrophoresed (100 μg per lane) on 4–20% Tris⋅HCl gels (Biorad), followed by transfer to polyvinyl difluoride (PVDF; ThermoFisher). After blocking in 5% nonfat dry milk in PBS + 0.1% Tween 20 for 1 h, blots were probed with rabbit anti–HIF-α (1:5,000), anti-PHD (1:5,000), or anti-actin (1:1,000, A2103; Sigma–Aldrich), which was used as a loading control. Samples were then washed and probed with a peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000; Jackson), followed by visualization using a chemiluminescent substrate (Clarity Western ECL Substrate; Biorad) and Syngene imaging system.
Detection of Phosphorylated Proteins.
Gut and pelt homogenates were prepared, electrophoresed, and transferred to PVDF as described above. After blocking, membranes were probed with the following primary antibodies: phospho-p70 S6 kinase (1:1,000, 9209; Cell Signaling Technology); phospho-AMPK (1:1,000, 2531; Cell Signaling Technology); phospho-Drosophila Akt (Ser505) (1:1,000, 4054; Cell Signaling Technology); phospho-p44/42 MAPK (ERK1/2) (1:1,000, 9101; Cell Signaling Technology); phospho-SAPK/JNK (1:1,000 dilution, 9251; Cell Signaling Technology); phospho-p38 MAPK (1:1,000, 9211; Cell Signaling Technology); and actin (1:1,000, A2103; Sigma), which served as the loading control. After washing, samples were probed with secondary antibody and visualized as described above.
Gut Length.
Larvae from different treatments and time points posthatching were dissected as described above to collect the gut. Gut length was determined from the anterior end of the gastric caeca to the anus using a Zeiss LSM 710 confocal microscope and Zen 2011 software. Ten larvae per treatment and time point were measured, with data analyzed by ANOVA using JMP Pro-13 (SAS).
Immunocytochemistry and Neutral Lipid Staining.
An affinity-purified antibody against the product of the A. aegypti slg gene (AAEL008336) was commercially generated (Genescript) by immunizing a rabbit with peptides corresponding to amino acids 64–77 (IKAEDTLPTPPPSSC), 279–292 (CSILTEKSTSNSNIQ), and 309–322 (CGAPRYQCPDCGKSY). For immunocytochemistry, guts were fixed in 4% paraformaldehyde in PBS for 20 min (anti-PH3, anti-NPF, and anti-TK) or graded ethanol, followed by postfixation in 4% paraformaldehyde in PBS for 20 min (anti-Slg). Samples were washed three times in PBS, permeabilized in PBS + 0.2 Triton X-100 (PBT) for 1 h, and blocked in PBT + 5% goat serum (PBT-GS) 1 h. Samples were incubated overnight at 4 °C with primary antibodies diluted in PBT-GS, anti-Slg (1:25), rabbit anti-PH3 (1:250; Santa Cruz Biotechnology), rabbit anti-NPF (1:250) (54), and rabbit anti-TK (1:500) (55). Following washing in PBT-GS, samples were incubated for 2 h (anti-PH3, anti-NPF, and anti-TK) or overnight (anti-Slg) at 4 °C with a goat anti-rabbit Alexa Fluor 488 or 568 secondary antibody (1:2,000; Molecular Probes); washed; and, in some cases, counterstained with Hoechst 33342 (1:2,000; ThermoFisher). The cytoplasm of ECs was visualized by uptake of 2-DG6P using a commercially available kit (ab136956; Abcam). Samples were then slide-mounted in 1:1 PBS/glycerol and imaged by confocal microscopy. For EdU labeling, 1 mM EdU was added to larval cultures at different time points posthatching. After 12 h, larvae were dissected and guts were fixed in 4% paraformaldehyde for 20 min, washed, and permeabilized as described above. EdU was visualized using a Click-iT EdU Alexa Fluor 488 Imaging Kit (C10337; Molecular Probes), followed by slide mounting and confocal microscopy. All images were processed using Adobe Photoshop CS6. For lipid droplet staining, guts and pelts were fixed as described above, washed three times in PBS, and stained for 30 min with Nile red (100 μg/mL; Molecular Probes) or Bodipy (Molecular Probes). After washing two times in PBS for 5 min, samples were counterstained with Hoechst 33342, slide-mounted, and examined as described above. The nuclear diameter of ECs, lipid droplets per EC, and lipid droplets per adipocyte were quantified for 10 cells from 10 larvae per treatment and time point, with data subsequently analyzed by ANOVA.
Supplementary Material
Acknowledgments
We thank J. Veenstra for providing the anti-TK antibody and J. A. Johnson for assistance with rearing of mosquitoes. This work was supported by an Achievement Rewards for College Scientists (ARCS) Foundation Scholarship (to K.L.C.), National Science Foundation Graduate Research Fellowship 038550-04 (to K.L.C.), and National Institutes of Health Grants R01AI106892 and T32GM007103 (to M.R.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. PRJNA420687).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719063115/-/DCSupplemental.
References
- 1.Sommer F, Bäckhed F. The gut microbiota–Masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 2.Engel P, Moran NA. The gut microbiota of insects–Diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 3.Clements AN. The Biology of Mosquitoes: Development, Nutrition, and Reproduction. Vol 1 Chapman & Hall; New York: 1992. [Google Scholar]
- 4.Hegde S, Rasgon JL, Hughes GL. The microbiome modulates arbovirus transmission in mosquitoes. Curr Opin Virol. 2015;15:97–102. doi: 10.1016/j.coviro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Tol S, Dimopoulos G. Influences of the mosquito microbiota on vector competence. Adv Insect Physiol. 2016;51:243–291. [Google Scholar]
- 6.Strand MR. The gut microbiota of mosquitoes: Diversity and function. In: Wikel S, Aksoy S, Dimopoulos G, editors. Arthropod Vector: Controller of Disease Transmission. Vol 1. Academic; San Diego: 2017. pp. 185–199. [Google Scholar]
- 7.Merritt RW, Dadd RH, Walker ED. Feeding behavior, natural food, and nutritional relationships of larval mosquitoes. Annu Rev Entomol. 1992;37:349–376. doi: 10.1146/annurev.en.37.010192.002025. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Gilbreath TM, 3rd, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One. 2011;6:e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol. 2012;21:5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- 10.Boissière A, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8:e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gimonneau G, et al. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect Genet Evol. 2014;28:715–724. doi: 10.1016/j.meegid.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 12.Coon KL, Vogel KJ, Brown MR, Strand MR. Mosquitoes rely on their gut microbiota for development. Mol Ecol. 2014;23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duguma D, et al. Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiol. 2015;15:140. doi: 10.1186/s12866-015-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muturi EJ, Bara JJ, Rooney AP, Hansen AK. Midgut fungal and bacterial microbiota of Aedes triseriatus and Aedes japonicus shift in response to La Crosse virus infection. Mol Ecol. 2016;25:4075–4090. doi: 10.1111/mec.13741. [DOI] [PubMed] [Google Scholar]
- 15.Coon KL, Brown MR, Strand MR. Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae) Parasit Vectors. 2016;9:375. doi: 10.1186/s13071-016-1660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol Ecol. 2016;25:5806–5826. doi: 10.1111/mec.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 18.Enserink M. INFECTIOUS DISEASES. An obscure mosquito-borne disease goes global. Science. 2015;350:1012–1013. doi: 10.1126/science.350.6264.1012. [DOI] [PubMed] [Google Scholar]
- 19.Coon KL, et al. Bacteria-mediated hypoxia functions as a signal for mosquito development. Proc Natl Acad Sci USA. 2017;114:E5362–E5369. doi: 10.1073/pnas.1702983114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nijhout HF, et al. The developmental control of size in insects. Wiley Interdiscip Rev Dev Biol. 2014;3:113–134. doi: 10.1002/wdev.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel KJ, Valzania L, Coon KL, Brown MR, Strand MR. Transcriptome sequencing reveals large-scale changes in axenic Aedes aegypti larvae. PLoS Negl Trop Dis. 2017;11:e0005273. doi: 10.1371/journal.pntd.0005273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 23.Dekanty A, et al. Drosophila genome-wide RNAi screen identifies multiple regulators of HIF-dependent transcription in hypoxia. PLoS Genet. 2010;6:e1000994. doi: 10.1371/journal.pgen.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward JPT. Oxygen sensors in context. Biochim Biophys Acta. 2008;1777:1–14. doi: 10.1016/j.bbabio.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Rabinowitz MH. Inhibition of hypoxia-inducible factor prolyl hydroxylase domain oxygen sensors: Tricking the body into mounting orchestrated survival and repair responses. J Med Chem. 2013;56:9369–9402. doi: 10.1021/jm400386j. [DOI] [PubMed] [Google Scholar]
- 26.Jain IH, et al. Hypoxia as a therapy for mitochondrial disease. Science. 2016;352:54–61. doi: 10.1126/science.aad9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masoud GN, Li W. HIF-1α pathway: Role, regulation, and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjedov I, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Q, Gilbert LI. S6 phosphorylation results from prothoracicotropic hormone stimulation of insect prothoracic glands: A role for S6 kinase. Dev Genet. 1994;15:332–338. doi: 10.1002/dvg.1020150404. [DOI] [PubMed] [Google Scholar]
- 30.Schreiber KH, et al. Rapamycin-mediated mTORC2 inhibition is determined by the relative expression of FK506-binding proteins. Aging Cell. 2015;14:265–273. doi: 10.1111/acel.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemirembe K, Liebmann K, Bootes A, Smith WA, Suzuki Y. Amino acids and TOR signaling promote prothoracic gland growth and the initiation of larval molts in the tobacco hornworm Manduca sexta. PLoS One. 2012;7:e44429. doi: 10.1371/journal.pone.0044429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Land SC, Tee AR. Hypoxia-inducible factor 1α is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J Biol Chem. 2007;282:20534–20543. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- 33.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 35.Salminen A, Kaarniranta K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res Rev. 2012;11:230–241. doi: 10.1016/j.arr.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Teleman AA, Chen Y-W, Cohen SM. Drosophila Melted modulates FOXO and TOR activity. Dev Cell. 2005;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Pakpour N, et al. Ingested human insulin inhibits the mosquito NF-κB-dependent immune response to Plasmodium falciparum. Infect Immun. 2012;80:2141–2149. doi: 10.1128/IAI.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strand MR, Brown MR, Vogel KJ. Mosquito peptide hormones: Diversity, production and function. Adv Insect Physiol. 2016;51:145–188. doi: 10.1016/bs.aiip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy SG, Hansen IA, Raikhel AS. Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2007;37:1317–1326. doi: 10.1016/j.ibmb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhara A, et al. Ovary ecdysteroidogenic hormone functions independently of the insulin receptor in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2013;43:1100–1108. doi: 10.1016/j.ibmb.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel KJ, Brown MR, Strand MR. Ovary ecdysteroidogenic hormone requires a receptor tyrosine kinase to activate egg formation in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2015;112:5057–5062. doi: 10.1073/pnas.1501814112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 43.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 44.Teng J-A, et al. The activation of ERK1/2 and JNK MAPK signaling by insulin/IGF-1 is responsible for the development of colon cancer with type 2 diabetes mellitus. PLoS One. 2016;11:e0149822. doi: 10.1371/journal.pone.0149822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 46.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 47.Apidianakis Y, Tamamouna V, Teloni S, Pitsouli C. Intestinal stem cells: A decade of intensive research in Drosophila and the road ahead. Adv Insect Physiol. 2017;52:139–175. [Google Scholar]
- 48.Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138:1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang H, Grenley MO, Bravo M-J, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc Natl Acad Sci USA. 2011;108:18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw RL, et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiher W, et al. Peptidomics and peptide hormone processing in the Drosophila midgut. J Proteome Res. 2011;10:1881–1892. doi: 10.1021/pr101116g. [DOI] [PubMed] [Google Scholar]
- 53.Trager W. Cell size in relation to growth and metamorphosis of the mosquito Aedes aegypti. J Exp Zool. 1937;76:467–489. [Google Scholar]
- 54.Ray K, Mercedes M, Chan D, Choi CY, Nishiura JT. Growth and differentiation of the larval mosquito midgut. J Insect Sci. 2009;9:1–13. doi: 10.1673/031.009.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanek DM, Pohl J, Crim JW, Brown MR. Neuropeptide F and its expression in the yellow fever mosquito, Aedes aegypti. Peptides. 2002;23:1367–1378. doi: 10.1016/s0196-9781(02)00074-8. [DOI] [PubMed] [Google Scholar]
- 56.Veenstra JA, Agricola HJ, Sellami A. Regulatory peptides in fruit fly midgut. Cell Tissue Res. 2008;334:499–516. doi: 10.1007/s00441-008-0708-3. [DOI] [PubMed] [Google Scholar]
- 57.Canavoso LE, Jouni ZE, Karnas KJ, Pennington JE, Wells MA. Fat metabolism in insects. Annu Rev Nutr. 2001;21:23–46. doi: 10.1146/annurev.nutr.21.1.23. [DOI] [PubMed] [Google Scholar]
- 58.Arrese EL, Soulages JL. Insect fat body: Energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palm W, et al. Lipoproteins in Drosophila melanogaster–Assembly, function, and influence on tissue lipid composition. PLoS Genet. 2012;8:e1002828. doi: 10.1371/journal.pgen.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown MR, et al. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2008;105:5716–5721. doi: 10.1073/pnas.0800478105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atella GC, Shahabuddin M. Differential partitioning of maternal fatty acid and phospholipid in neonate mosquito larvae. J Exp Biol. 2002;205:3623–3630. doi: 10.1242/jeb.205.23.3623. [DOI] [PubMed] [Google Scholar]
- 63.Eltzschig HK, Bratton DL, Colgan SP. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat Rev Drug Discov. 2014;13:852–869. doi: 10.1038/nrd4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell WR, Hoyles L, Flint HJ, Dumas ME. Colonic bacterial metabolites and human health. Curr Opin Microbiol. 2013;16:246–254. doi: 10.1016/j.mib.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Shin SC, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 66.Hang S, et al. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe. 2014;16:592–604. doi: 10.1016/j.chom.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci USA. 2017;114:4775–4780. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flück K, Fandrey J. Oxygen sensing in intestinal mucosal inflammation. Pflugers Arch. 2016;468:77–84. doi: 10.1007/s00424-015-1722-4. [DOI] [PubMed] [Google Scholar]
- 69.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: Activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park J-S, Kim Y-S, Yoo M-A. The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging (Albany NY) 2009;1:637–651. doi: 10.18632/aging.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang H, et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gregory L, Came PJ, Brown S. Stem cell regulation by JAK/STAT signaling in Drosophila. Semin Cell Dev Biol. 2008;19:407–413. doi: 10.1016/j.semcdb.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 74.Song W, Veenstra JA, Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mole DR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia X, Kung AL. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 2009;10:R113. doi: 10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schödel J, et al. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.