Significance
A migration-based selection system is used to identify antibodies from combinatorial libraries that induce stem cells to both differentiate and selectively traffic to different tissues in adult animals. Significantly, a single agonist antibody induces microglia-like cells, which have the capacity to migrate to the brain and decrease amyloid beta deposition in the brain.
Keywords: antibody, stem cell, microglia, Alzheimer’s disease
Abstract
One goal of regenerative medicine is to repair damaged tissue. This requires not only generating new cells of the proper phenotype, but also selecting for those that properly integrate into sites of injury. In our laboratory we are using a cell-migration–based in vivo selection system to generate antibodies that induce cells to both differentiate and selectively localize to different tissues. Here we describe an antibody that induces bone marrow stem cells to differentiate into microglia-like cells that traffic to the brain where they organize into typical networks. Interestingly, in the APP/PS1 Alzheimer’s disease mouse model, these induced microglia-like cells are found at sites of plaque formation and significantly reduce their number. These results raise the intriguing question as to whether one can use such antibody-induced differentiation of stem cells to essentially recapitulate embryogenesis in adults to discover cells that can regenerate damaged organ systems.
Cell migration is central to the embryonic development and maintenance of all organisms. Despite its immense importance and much study, our knowledge of this process is still incomplete. For example, in the adult we still do not have a comprehensive understanding of which cells can migrate, where they go, and what sustains them in a differentiated state when they reach their destination.
Recently, we have developed a method that has the potential of allowing us to approach this problem. The method is based on the selection of intracellular antibodies from combinatorial libraries that modulate cell fates. One can test the effects of up to 108 different antibodies and, because the selection system is autocrine-based where the target and the antibody are in the same cell, each cell is a reporter system unto itself (1–9). We have previously used this method to select for antibodies that differentiate or transdifferentiate stem cells and even turn cancer cells into those with alternative phenotypes (10). Ultimately, its success is a function of the robustness of the section system. Protection from cell death, generation of a fluorescent reporter during signal transduction, or formation of a unique cellular morphology are examples of selection systems that have been used successfully (1–9). One important aspect of the method is that, once a functional antibody is found, it can then be used to identify its target, thereby giving insight into the pathway involved in the generation of the phenotype. In this respect the process is highly analogous to forward genetics except that the system and its de-convolution operate at the protein level.
We reasoned that our method should be ideal for the study of cell migration. Unlike previous selections where one must separate induced from uninduced cells, in a migration-based selection the end point of the experiment is when the cell population of interest is detected in a different location. Thus, rather than isolating phenotypically interesting cells from a mixture, one studies populations that, for physiological reasons, are enriched by self-separation. Here, we show that this method combined with adoptive transfer protocols can be used to isolate antibodies that induce hematopoietic stem cells (HSCs) from bone marrow cells to differentiate and then selectively migrate to different tissue compartments including the brain. Since the cells that migrate to the brain have many of the characteristics of microglia, we refer to them as microglia-like cells. This is because often the term “microglia” is reserved for yolk-sac–derived resident brain macrophages, and we want to avoid confusion.
If we add control of migration to the already known ability of intracellular antibodies to induce differentiation of cells, we can begin to think about reconstitution of organ systems in vivo.
Results
Selection of Antibodies in Vivo That Regulate Cell Migration.
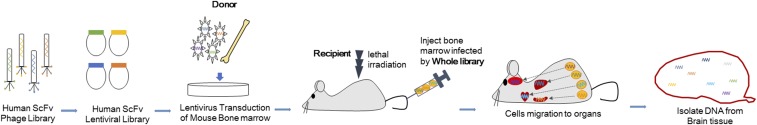
An in vivo selection scheme was developed (Fig. 1) to identify antibodies that cause differentiation of human stem cells into cell types capable of migration to specific tissues in the body, such as the brain where migratory cells are thought to be important in Alzheimer’s and Parkinson’s diseases. Genes obtained from a human single-chain fragment variable (ScFv) phage library were used to create a human ScFv lentiviral intracellular combinatorial antibody library with 108 unique antibody clones. In this system, the antibodies were expressed on the cell surface using a previously reported methodology (8). Total bone marrow cells were harvested from mice, infected in vitro with the lentiviral library, and then transplanted into lethally irradiated mice. After 7 d, brains of mice perfused with PBS were harvested to extract genomic DNA, which was subjected to PCR to amplify and sequence human ScFv sequences that were integrated into the genome of migrating cells. Different organs were assessed for cells that carried antibody genes from the library incorporated into their genome (Fig. S1).
Fig. 1.
Selection in vivo of an antibody that induces differentiation and migration of mouse HSCs. Scheme of the phenotype selection in vivo. Genes from a human ScFv phage library (108 members) were cloned into a lentiviral vector to make a lentiviral intrabody library in which antibody molecules are attached to the plasma membrane and displayed on the cell surface. Total mouse bone marrow cells were infected with the antibody library in vitro and transplanted into lethally irradiated C57BL/6J mice. The system is autocrine-based because each cell has a different antibody and the putative target. After 7 d, the mouse brains were harvested and analyzed by PCR to identify any antibody genes in cells that traffic to the brain.
The Selected Antibody Induces Migration of Cells to the Brain.
As a preliminary experiment, to confirm that the integrated antibody genes induced bone marrow cells to migrate to the brain, the entire collection of antibody genes that were recovered from the migrating cell population were cloned into lentivirus vectors, which were then inserted into the genomes of fresh bone marrow cells from mice expressing the red fluorescent protein (mCherry). These donor mCherry+ bone marrow cells that then contained the selected antibody genes were then adoptively transferred into irradiated wild-type mice (Fig. S2). After 2 wk, brains were perfused, harvested, and analyzed for the presence of cells ubiquitously expressing mCherry. The brains contained many cells expressing mCherry, indicating that at least some members of the selected antibody population could induce cells to migrate to the brain (Fig. S2).
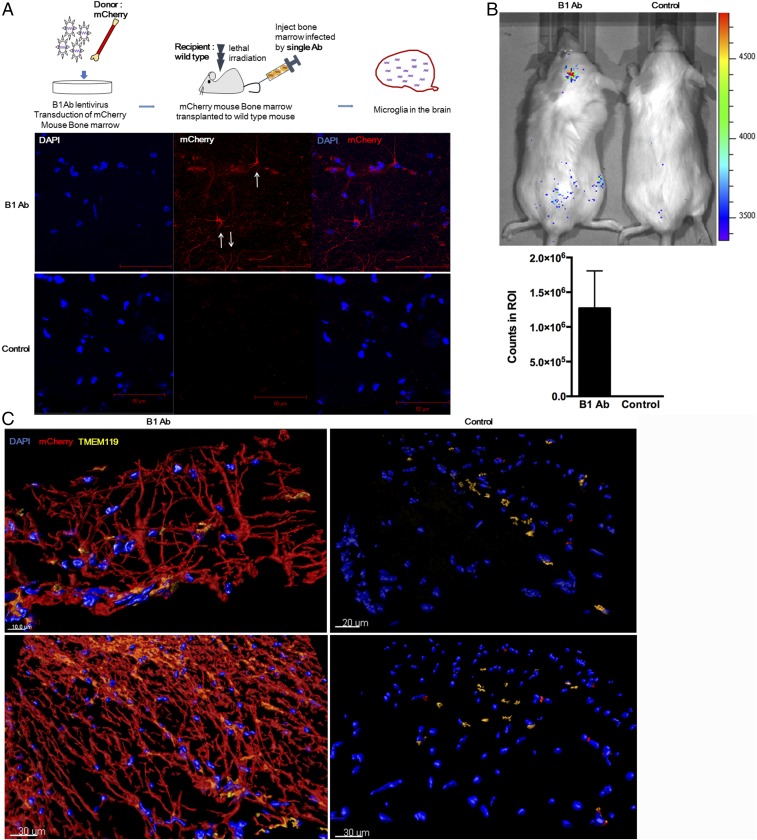
To investigate whether a single antibody from this population could induce cell migration from the bone marrow to the brain, the B1 gene alone was cloned into a lentivirus vector that was then used to infect total bone marrow cells freshly harvested from mice that ubiquitously expressed the monomeric mCherry protein. The B1 gene was selected for study because the four antibody gene sequences that were recovered from the cells that had migrated to the brain gave that gene the highest copy number. Antibody B1 was seen six times in cells that migrated to the brain but was not present in cells that migrated to other organs (Fig. S1). In antibody selections, because the numbers of input sequences are so high, selection of repeated sequences is of special significance. These mCherry+ cells with the single lentivirus-encoded B1 antibody integrated into their genome were then transplanted into lethally irradiated wild-type mice (Fig. 2A). After 1 wk, brains were perfused and prepared for immunofluorescence histochemistry. Donor mCherry+ cells infected with lentivirus expressing the B1 Ab migrated to the brain (Fig. 2A) and also stained positive for the microglia marker TMEM119 (Fig. S3A). A comprehensive analysis of whole-brain sections from treated and control mice showed that significantly more mCherry+ signal (63,270 versus 4,104 fluorescent units) was detected in the hippocampus, substantia nigra, and hypothalamus in mice, the bone marrow of which was infected with lentivirus encoding the B1 Ab (Fig. S3B).
Fig. 2.
An agonist antibody regulates cell migration. (A) Scheme of the in vivo migration of cells from the bone marrow to the brain. From a pool of the antibody genes from all cells that had migrated to brain, a single antibody gene (B1 Ab) that was the most abundant was inserted into lentiviral vectors and used to infect total mCherry+ mouse bone marrow cells. After 3 d, these cells were transplanted into lethally irradiated wild-type C57BL/6J mice. After 1 wk, the mice were perfused with PBS followed by 2% PFA before harvesting the brains and sectioning the frozen Optimal Cutting Temperature compound blocks for immunofluorescence histochemistry. Brain sections (10 μm) were stained with DAPI and anti-mCherry antibodies and then analyzed by confocal microscopy. Significantly more mCherry+ cells were identified in the treated tissues compared with controls (untreated mCherry+ cells), suggesting that mCherry+ donor cells migrated from the bone marrow to the brain. White arrows indicate microglia-like cells. (Scale bars, 50 μm.) (B) Representative images of mice that received bone marrow from luciferase-expressing transgenic mice transduced with the lentiviral B1 Ab or no Ab (control). The mice were imaged 1 wk posttransplantation. Counts in brain ROI are shown below the image. Values are the mean ± SD for n = 5 mice. (C) Showing representative sections from two different locations from brains of mice treated with B1 Ab mCherry+ cells (Left) or untreated mCherry+ cells (Right). Tissue sections were stained with DAPI (blue), for mCherry-derived bone marrow cells (red), and the antibody to the microglia marker TMEM119 (orange). The confocal 3D volume reconstructed images were obtained using IMARS software. (Scale bars, 10, 20, and 30 μm.)
We used in vivo bioluminescence imaging to confirm that the integrated antibody genes induced bone marrow cells to migrate to the brain. For this experiment, fresh bone marrow cells from luciferase-expressing transgenic mice (luc+) were infected with lentivirus encoding the B1 Ab, adoptively transferred into irradiated wild-type mice, and imaged after 1 wk. Donor luc+ cells infected with the B1 Ab migrated to the brain (Fig. 2B) and appear as extensive microglia-like networks as observed in the 3D images (Fig. 2C).
Currently, it is thought that, in adoptive transfer experiments involving irradiation, some white blood cells are able to migrate into the brain due to a compromised blood–brain barrier (BBB) as a result of inflammation caused by the irradiation. To study the role of irradiation, mice were irradiated with or without a lead helmet and analyzed 2 wk later by histochemistry. No mCherry+ cells were seen in the brain when mice were wearing a lead helmet during irradiation (data not shown). This result is in agreement with the studies of others (11–13), such as Mildner et al. (11), who showed that shielding the brain from irradiation before adoptive transfer of cells did not lead to a significant invasion of monocyte-derived microglia into the brain as was observed in unshielded mice. Although we also see some migration of bone marrow cells to the brain in control mice, it is important to note that the efficiency was 15-fold lower than seen with cells exposed to the agonist antibody. This enhanced efficiency may be very important in a therapeutic setting where one may want to use as many cells as possible. At any rate, the results presented here describe a single agonist that induces microglia-like cells, which have the capacity to migrate to the brain.
Purified Antibody Differentiates Human and Murine Stem Cells into Microglia-Like Cells.
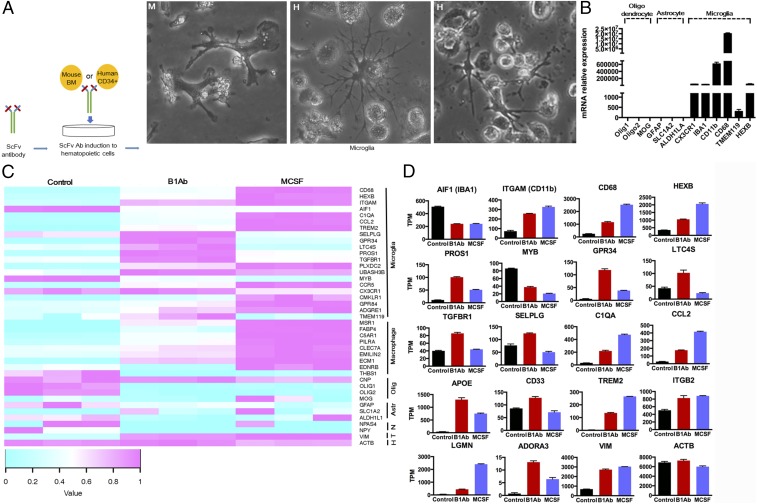
To determine if purified antibody could transform bone marrow cells, total mouse bone marrow or human CD34+ cells were incubated with the selected B1 Ab for 2 wk in vitro. The purified antibody induced both the mouse and human cells to differentiate into cells with a cellular morphology resembling microglia (Fig. 3A). To study the nature of the induced cells, mRNA expression levels were analyzed for specific oligodendrocyte, astrocyte, and microglia genes by qRT-PCR. The cells induced by purified B1 Ab expressed mRNA for the microglia markers CX3CR1, IBA1, CD11b, CD68, TMEM119, and HEXB, but failed to express mRNA for the established oligodendrocyte (Olig1, Olig2, and MOG) and astrocyte (GFAP, SLC1A2, and ALDH1LA) gene markers (Fig. 3B).
Fig. 3.
Antibody-induced differentiation of HSCs into microglia-like cells. (A) Scheme of the hematopoietic stem cell differentiation by B1 Ab. In the presence of purified B1 antibody for 2 wk, total mouse bone marrow and human CD34+ cells differentiated into cells with the morphology of microglia. “H” indicates human CD34+ cells. “M” indicates mouse bone marrow. (Magnification: 100×.) (B) Human CD34+ cells treated with B1 or isotype control antibody were harvested after 2 wk to extract total RNA for qRT-PCR analysis. A distinct microglia mRNA expression profile was revealed when relative mRNA levels of oligodendrocyte (Olig1, Olig2, and MOG), astrocyte (GFAP, SLC1A2, and ALDH1LA), and microglia (CX3CR1, IBA1, CD11b, CD68, TMEM119, and HEXB) gene markers were compared by qRT-PCR. (C) Microglia gene profiling shown by RNA-sequencing analysis. Heat map and hierarchal clustering indicates that the microglia have a unique signature compared with the untreated human CD34+ cells and positive control of macrophages induced by treatment with M-CSF. The genes found to be highly expressed in the induced microglia were consistent with previous studies. In general, the induced microglia had lower expression of astrocyte (Astr), oligodendrocyte (Olig), and neuron (N) genes. T indicates target antigen. “H” indicates housekeeping gene. (D) A data summary of key microglia markers from RNA-seq replicates (n = 3) for genes expressed in transcripts per million (TPM).
RNA transcripts of human CD34+ cells treated with purified B1 Ab were also sequenced and compared with the profile of macrophages induced by treatment of human CD34+ cells with macrophage colony-stimulating factor (M-CSF) in vitro. RNA sequencing data from human CD34+ cells treated with B1 Ab or M-CSF were consistent with qRT-PCR results. To further identify transcripts that are expressed in microglia, we compared the results to expression data of previous reports (14–17). Notably, we found genes highly expressed in microglia—which include IGTAM, IBA1, TREM2, APOE, CD33, ITGB2, ADORA3, LGMN, PROS1, C1QA, GPR34, TGFBR1, SELPLG, HEXB, LTC4S, and CCL2—to be consistent with data published by other groups (Fig. 3 C and D and Dataset S1). Importantly, we also found that B1 Ab-induced microglia have a gene expression similar to human microglia. Among 52 genes, the most highly expressed are from human microglia [75% of the genes (39/52)], which is consistent with our data (Dataset S1).
To classify similarities and differences between the induced microglia and macrophages, we compared the top 10% of transcripts with the highest expression levels. Of the 3,996 total transcripts identified, 3,098 transcripts were shared between microglia and macrophages, 243 were unique to microglia differentiated with B1 Ab, and 312 were unique to macrophages differentiated with M-CSF (Fig. S4A). The most highly expressed genes that were expressed in both microglia and macrophages were ACP5, MMP9, APOC1, CTSL, COL6A2, CTSK, CYP27A1, and MSR1. The highly expressed genes unique to microglia included RPL3P4, FBP1, LIF, IL9R, SIGLEC6, MARCO, UTS2, CKAP4, and GPRC5C, whereas genes uniquely expressed in macrophages included RNASE1, LAIR2, PFKFB3, RNASE6, and GPR183 (Fig. S4B and Dataset S1). Of the highly expressed genes specific to microglia, 268 have been reported to be relevant to neuronal diseases such as Alzheimer’s, amyloidosis, tauopathy, dementia, inflammation of the central nervous system, and encephalitis (Dataset S1).
Identification of a Novel Target.
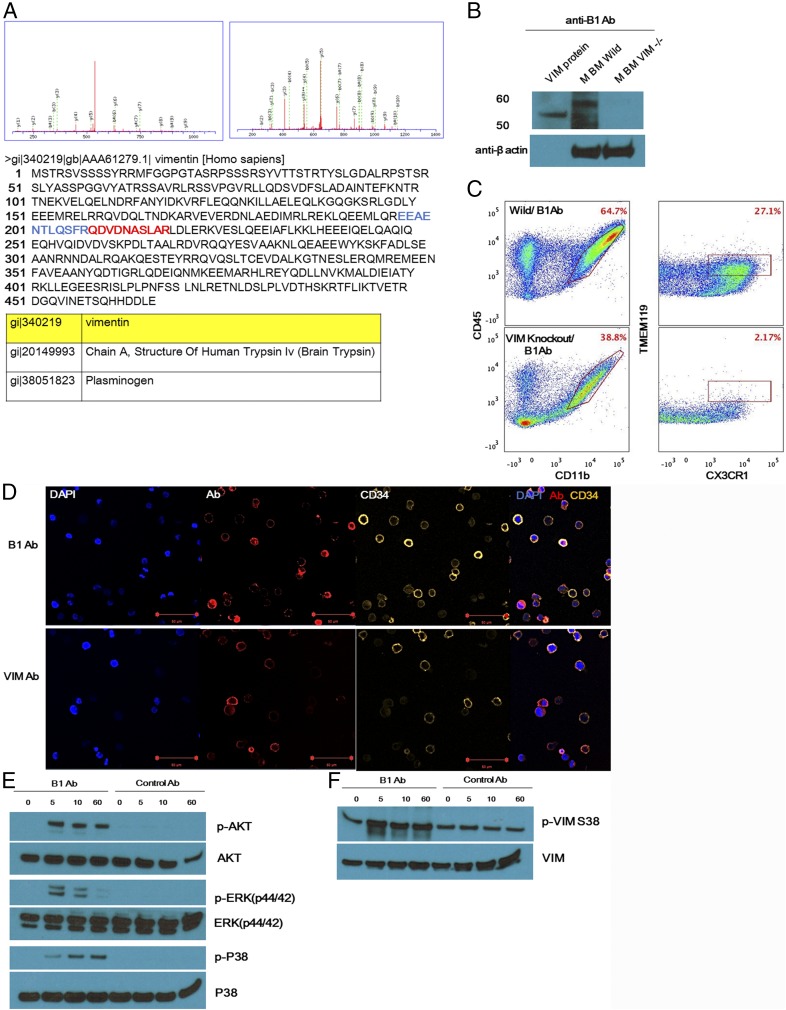
To identify the protein recognized by the B1 antibody, antibodies were produced recombinantly in Expi293F cells. Purified B1 antibody was incubated with mouse bone marrow, and immune complexes from cellular lysates were captured on a protein A/G column. Proteins that reacted with the antibody were identified by silver staining of SDS gels and mass spectrometry (MS). Three candidate proteins were identified above the background threshold (Fig. 4A). Vimentin (VIM) was one of the top hits and was confirmed to be the target antigen of the B1 Ab by Western blotting. The B1 Ab bound to purified VIM protein as well as to VIM from wild-type mouse bone marrow lysates, but did not bind to proteins in lysates from bone marrow obtained from VIM-deficient knockout mice (Fig. 4B). Bone marrow from wild-type and VIM knockout mice were incubated B1 Ab or commercial VIM Ab for 6 d. FACS analysis showed that microglia formation was increased by B1 Ab in wild-type mice. However, there was no induction of microglia in the VIM knockout mice (Fig. 4C). Interestingly, commercial VIM Ab did not induce microglia differentiation, indicating that our antibody had a unique binding mode because it was the product of selection for migration rather than simple binding (Fig. S5). Also, VIM was confirmed to be expressed on human CD34+ cells by immunofluorescence cytochemistry using B1 or commercial VIM antibodies (Fig. 4D). The amino acid sequence identity between mouse and human VIM is greater than 97%.
Fig. 4.
Identification of a novel antigen recognized by the B1 Ab. (A) Cell lysates of mouse bone marrow were incubated with the B1 Ab to bind antigens. Immunoprecipitated elutes were separated by SDS/PAGE and subjected to MS analysis. Nano-LC-MS/MS analysis identified three candidate hits as potential target antigens. The VIM peptides identified by the MS data are highlighted in blue and red. (B) The B1 Ab bound to commercial VIM protein and C57BL/6J mouse bone marrow lysates in Western blots. Importantly, no protein band was observed when the B1 Ab was blotted with bone marrow lysates from VIM-deficient mice (JAX stock 025692). (C) Bone marrow from wild-type and VIM knockout mice was incubated with B1 Ab for 6 d. Cells were then stained with antimicroglia markers such as anti-TMEM119 and anti-CX3CR1. FACS analysis showed that induction of microglia in vitro was increased by B1 Ab in wild-type mice but not in VIM knockout mice, again confirming that VIM is the target of the antibody. (D) Surface expression of VIM on human CD34+ cells was confirmed by confocal microscopy using DAPI, B1 or commercial VIM antibodies, or antibody against CD34. (Scale bars, 50 μm.) (E and F) Human CD34+ cells were treated with B1 Ab or control isotype antibody, and cell lysates were assessed by Western blotting using antibodies against nonphosphorylated and phosphorylated (p-) AKT, ERK, p38, and VIM S38.
To determine whether binding of B1 Ab leads to the activation of signaling pathways, human CD34+ bone marrow cells were treated with the B1 Ab, and cell lysates were assessed by Western blotting with antibodies against nonphosphorylated and phosphorylated (p-) AKT, ERK, and p38. Consistent with their known role in microglia differentiation, induction of p-AKT, p-ERK, and p-p38 was observed in the cells stimulated with B1 Ab, but not with an isotype control (Fig. 4E). In addition to activation of transcription factors after binding to VIM, the B1 Ab might be expected to induce phosphorylation of VIM itself. Therefore, CD34+ cells were activated with the B1 Ab, and the resulting VIM phosphorylation was determined by Western blot using an antibody that detects phosphorylation of VIM at serine 38. The treated cells showed a marked increase in VIM phosphorylation starting at 5 min (Fig. 4F).
The Induced Microglia-Like Cells Have an Antiinflammatory M2-Like Phenotype.
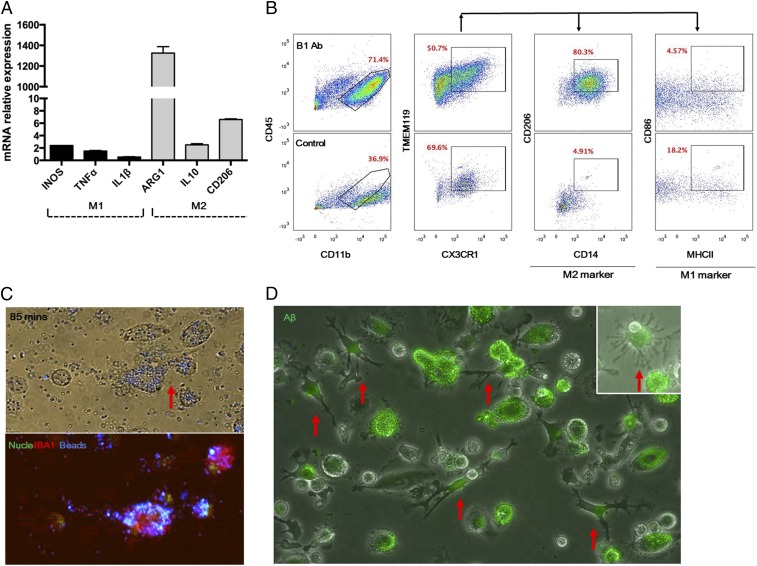
Polarization of the microglia is important because presumably one wants to induce those with antiinflammatory properties. To determine the nature of the microglia induced by the antibody, total mouse bone marrow cells were incubated with the selected B1 antibody for 2 wk in vitro, and specific M1/M2 marker gene mRNA and protein expression levels were analyzed by qRT-PCR and flow cytometry, respectively. Cells treated with B1 Ab up-regulated the M2 marker genes ARG1, IL10, and CD206, whereas expression of the M1 markers iNOS, TNFα, and IL1β remained low (Fig. 5A). Furthermore, flow cytometric analysis revealed that the majority of the induced CD45low-int CD11b+ cells stained positive for the microglial markers CX3CR1 and TMEM119 as well as the M2 markers CD36 and CD206, but negative for the M1 markers CD86 and MHCII (Fig. 5B). Together, these data suggest that the B1 Ab induced the mouse bone marrow HSCs to differentiate into microglia-like cells with M2 polarization.
Fig. 5.
B1 Ab-induced M2 polarization of differentiated microglia-like cells. (A) Mouse bone marrow cells were incubated with B1 Ab for 2 wk. Cells were then harvested to extract total RNA for qRT-PCR analysis. iNOS, TNFα, and IL1β were used as M1 gene markers, and ARG1, IL10, and CD206 were used for M2 markers. qRT-PCR suggested M2 polarization of the differentiated microglia with higher mRNA expression of M2 gene markers. (B) Flow cytometric analysis of the cells that were differentiated by the B1 Ab revealed that the cells were CD45low-intCD11b+TMEM119+CX3CR1+. Then the cells were gated further to find M1/M2 subpopulations using M1-specific antibodies against MHCII and CD86 and M2-specific antibodies against CD36 and CD206. The cells treated with B1 antibody expressed surface markers consistent with M2-polarized microglia. (C) A functional phagocytosis assay was performed on the cells that differentiated from human CD34+ cells. To assay function, we determined if the cells were capable of engulfing fluorescently labeled beads. After 85 min, the microglia-like cells had engulfed the beads, and the phagocytic cells stained positive for the microglia marker IBA1. “Nucle” indicates nucleus marker. “Beads” indicates DAPI-labeled beads. Red arrow indicates microglia-like cells. (Magnification: 100×.) (D) The Aβ peptide aggregation assay was conducted with the microglia that differentiated from human CD34+ cells. The microglia-like cells showed extensive uptake of fluorescent (HiLyte Fluor 488)-labeled Aβ peptides. Red arrows indicate microglia-like cells. (Magnification: 100×.)
The Induced Microglia-Like Cells Phagocytose the Amyloid Beta Peptide.
Since microglia are important phagocytic cells in the brain, functional phagocytic and amyloid beta (Aβ) peptide aggregation assays were performed on the microglia produced from the in vitro differentiation of human CD34+ cells by the B1 Ab. The induced microglia-like cells were incubated with fluorescently labeled beads and monitored by real-time fluorescence microscopy for engulfment of beads over time. Marked phagocytosis of the beads by the induced microglia-like cells was observed and was most notable after 85 min of incubation (Fig. 5C and Fig. S6). The phagocytic cells stained positive with the mouse microglia-specific marker IBA1 after fixation at 85 min (Fig. 5C). Active phagocytosis of the beads by the microglia-like cells was also followed in a time-lapse movie (Movie S1).
While, as noted above, it was important to establish that the induced microglia-like cells are phagocytic, in the therapeutic setting of Alzheimer’s disease the extension of this phagocytic function to the Aβ peptide is of central importance. Thus, we studied the ability of the microglia induced by the antibody to phagocytose Aβ (1–42). Indeed, the cells were strongly phagocytic for a fluorescent derivative of Aβ (Hilyte Fluor 488) (Fig. 5D).
The Induced Microglia-Like Cells Lower Aβ Deposition in the Brain.
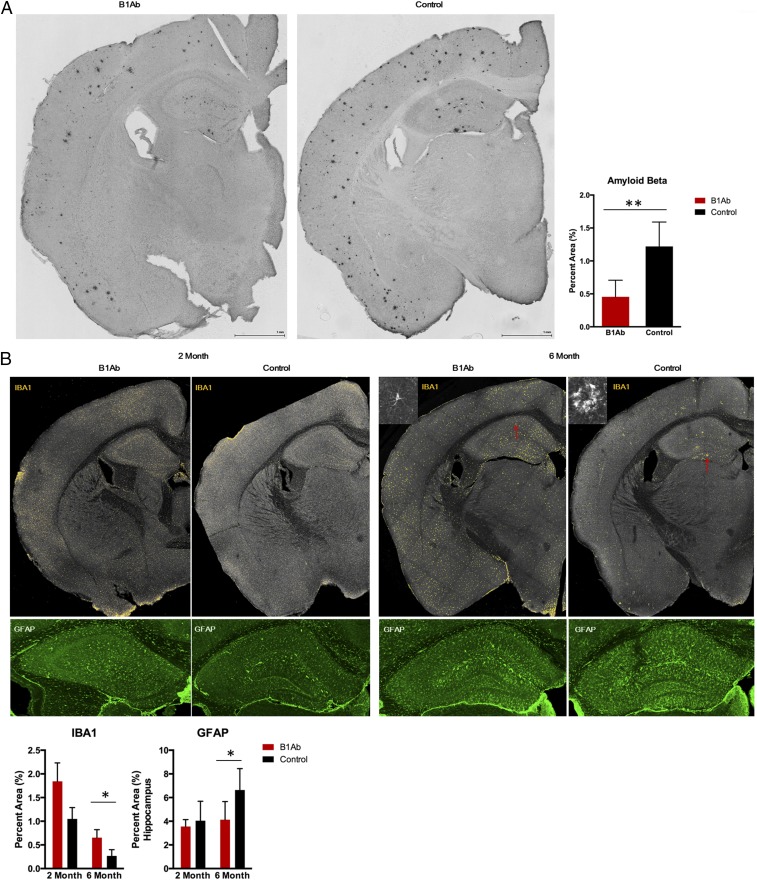
We next investigated whether this antibody could be used to lower Aβ plaques in the APP/PS1 Alzheimer’s disease mouse model, whereby the mice develop Aβ plaques and Alzheimer’s disease by 6 mo of age. The gene for the B1 antibody was inserted into the genome of fresh bone marrow cells from wild-type mice. Then these donor wild-type bone marrow cells were adoptively transferred into irradiated APP/PS1 8-wk-old mice. Brains were removed 10 d or 5 mo after adoptive transfer. The brains were perfused and prepared for immunofluorescence histochemistry. At the 6-mo time point, the brains of B1 Ab-treated mice had a significant decrease in Aβ deposition compared with control mice. Aβ deposition was 60% lower than in control mice (Fig. 6A). In addition, at the 6-mo time point, animals treated with B1 Ab had more microglia and fewer astrocytes (Fig. 6B).
Fig. 6.
Bone marrow cell expressing B1 Ab protect APP/PS1 mice. (A) Plaque deposition showing a protective effect of B1 Ab treatment relative to control. Total mouse bone marrow cells were infected with lentivirus-encoded B1 Ab or untreated cells (control) and transplanted into lethally irradiated 8-wk-old APP/PS1 mice (7/group). At the 6-mo time point, significant differences between B1 Ab-treated and control mice are indicated by **P < 0.005 (Student’s t test). (Scale bar, 1 mm.) (B) Total wild-type C57BL/6J mouse bone marrow cells were infected with lentivirus-encoded B1 Ab or untreated cells (control) and transplanted into lethally irradiated 8-wk-old APP/PS1 mice. After 2 wk (2 mo old) and 5 mo (6 mo old) post transfer, the mice were perfused with PBS, and brains were harvested and fixed in 2% PFA. Brain sections (50 μm) were stained with IBA1 for microglia and GFAP for astrocytes and analyzed by confocal microscopy. Yellow fluorescent units (Top) of IBA1 signal from brain coronal sections obtained by confocal microscopy were quantified using ImagePro software. Staining of the hippocampus with GFAP (Bottom) is shown. Red arrows indicate microglia at a higher magnification. Significant differences between B1 Ab-treated and control mice are indicated by *P < 0.05 (Student’s t test).
Microglia-Like Cells Migrate to the Injured Brain in the Absence of Irradiation.
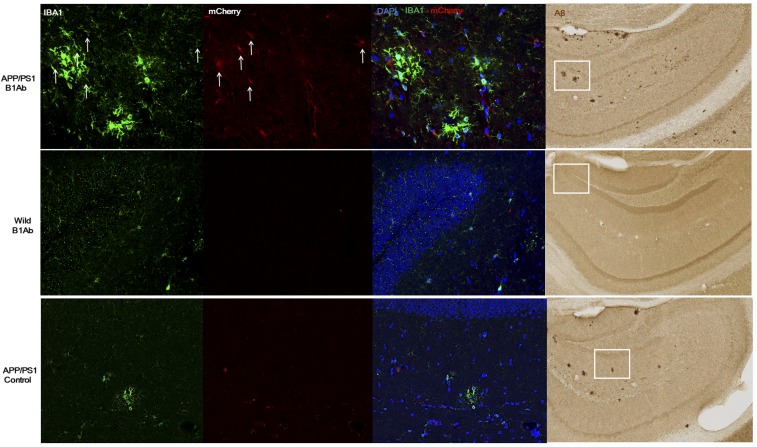
In the studies above, brain irradiation was used to increase the efficiency of the adoptive transfer. Thus, one could argue that irradiation was also necessary for migration of microglia to the brain, and thus our studies would not be applicable to other types of brain injury such as Alzheimer’s. Therefore, we carried out studies in aged APP/PS1 mice where bone marrow transfer was carried out without irradiation. mCherry+ mouse bone marrow cells treated with B1 Ab were transplanted into nonirradiated 8-mo-old APP/PS1 mice and C57BL6 wild-type mice. After 1 wk, brain sections were stained with DAPI, IBA1, anti-mCherry, and anti-Amyloid β antibodies. mCherry+ cells from B1 Ab-treated bone marrow in these mice significantly migrated into the brains of aged APP/PS1 mouse brains compared with controls such as aged APP/PS1 mice that were not treated with B1 Ab and aged wild-type mice (Fig. 7).
Fig. 7.
Microglia-like cells migrate to injured brain in the absence of irradiation. mCherry+ mouse bone marrow cells treated by B1 Ab were transplanted into nonirradiated 8-mo-old APP/PS1 and C57BL6 wild-type mice. After 1 wk, brain sections were stained with DAPI (blue), anti-IBA1 (green,), anti-mCherry (red), and anti-Aβ (brown). mCherry+ cells were identified in the B1 Ab-treated 8-mo-old APP/PS1 mice. However, neither wild-type mice nor nontreated mice showed significant migration of mCherry+ cells. The white boxes indicate the confocal images that correspond to the adjacent fluorescent images. Showing representative images from two mice in each group. (Magnification: Left, 20×; Right, 10×.)
Importantly, the mCherry+ cells were found adjacent to plaques in the hippocampus that already contained abundant endogenous microglia (Fig. 7). In sum, these studies suggest that the brain injury associated with Alzheimer’s disease is a permissive condition and/or driving force that allows bone marrow cells to migrate to the brain where they are found at sites of injury.
Discussion
To repair damaged tissues in the adult, one must induce cells of a desired phenotype that also properly integrate into the tissue of interest. While this might seem to be a formidable two-step event, it may be simpler if, as observed here, once certain phenotypes are induced, appropriate migration may be obligatory. This is similar to what happens in embryogenesis. Toward this end, migration-based selections from very large libraries of cells may be a powerful approach for generating those rare cells that have the appropriate phenotype as well as the ability to migrate and properly integrate into target tissues. If we can generalize these results to other systems, we may have a way to reconstitute damaged organ systems in the adult. Essentially, we want to recapitulate embryonic events. For example, one might want to find an antibody that recapitulates the embryonic events that cause cells to differentiate into insulin-producing cells that localize to islets in the pancreas.
In this study we have focused on the selection of an antibody agonist that induces bone marrow cells to differentiate into microglia-like cells that have the capacity to migrate to brain and function to reduce plaque in a murine model of Alzheimer’s disease. When selection from agnostic antibody libraries is used to generate cells with a particular phenotype, the question invariably arises as to exactly what cell has been induced. Here, because of the consistency and diversity of the accumulated data, we can conclude that the induced cells have characteristics of microglia, which are brain resident macrophages. Indeed, the cells have the morphology of microglia, are strongly phagocytic, and contain multiple markers associated with microglia. Perhaps the strongest evidence is that the cells migrate to the brain where they form networks typical of microglia.
The identification of VIM as the target aligns with a growing body of evidence suggesting that, in addition to its more mechanical role as an intermediate filament protein involved in polar morphology, VIM also plays a role in cell attachment, migration, and cell signaling (18–20). In general, VIM is considered to be a cytoplasmic intermediate filament protein, but more recent evidence has shown that VIM is also located on the cell surface. For example, VIM is located on the cell surface of metastatic hepatocarcinoma cells (21) and serves as a cell-surface receptor for enterovirus 71 on a variety of cell lines (22). This surface localization is relevant to our studies using the lentivirus library to generate microglia that migrate to the brain because the antibodies are displayed on the plasma membrane where they can bind to membrane-associated proteins on the cell (8). Moreover, we have shown that exogenous soluble antibody added to stem cells elicits the same differentiation program. Indeed, there is a certain parallelism with the role that VIM plays in the chemotactic migration of macrophages to tissues that are sites of infection in the periphery and the putative migration of microglia to sites of brain injury (23–26). In both cases the source of the cells may be the bone marrow. It will be interesting to learn whether the injured brain generates unique chemotactic factors that direct migrating cells to sites of injury.
We do not as yet completely understand how an antibody agonist for VIM induces cell differentiation. VIM is also a scaffold for assembly and regulation of signal transduction pathways (19). For example, VIM filament assembly is required for the β3-adrenergic receptor-mediated activation of the ERK map kinase (27) and ERK-mediated epithelial–mesenchymal transition of cancer cells (28). The role of VIM in cell signaling is believed to be a consequence of its organization of signaling complexes inside the cell (19). The fact that antibody B1 induces phosphorylation of VIM is consistent with its role in signaling (29, 30). Since VIM is also on the cell surface, a similar organizing principle could account for our observation that soluble anti-VIM antibody induces cell signaling in stem cells. In this case, the antibody could either stabilize or destabilize the complex leading to signal transduction. We presently are studying the nature of the members of the VIM complex on the cell surface of bone marrow cells.
An issue raised by these studies concerns whether the bone marrow is a natural site for generation of microglia. Microglia can be derived from myeloid cells of the yolk sac during embryonic and fetal development before development of the BBB and bone marrow-derived monocytic cells (BMDM) in the adult (13, 31–33). However, the key question that remains is, under what conditions do bone marrow-derived microglia-like cells migrate to the brain in adult animals? While it is now generally accepted that they migrate to the brain after injury, it is more controversial as to whether BMDM can cross the BBB without specific induction of brain injury. Such injury includes the irradiation damage and attendant inflammation that accompanies the generation of chimeric mice for adoptive transfer, as was used here, or for infection (31). As a practical matter, the resolution of this controversy may be mute because brain injury might be exactly when one might want to induce microglia into the brain. In a certain sense it might be beneficial if microglia migrated only to injured brain regions because, when activated, these cells are not without their side effects. Therefore, selectivity based on injury might give a better therapeutic index.
As reported here, it appears that bone marrow cells can migrate to the brain and become microglia or microglia-like, but a driving force such as irradiation or inflammation is required. Autocrine migration-based selections may unearth the perhaps rare molecules involved in this driving force. As we have repeatedly shown, autocrine-based selections using membrane-bound antibodies to cytokine receptors have caused stem cells to respond and differentiate as if they had seen the natural ligand (receptor pleiotropism) (1–10). Perhaps the driving force in the present experiments is that the selected antibody provides an orthogonal activator of the brain-specific cytokine axis. Simply put, chemotactic factors from the brain may induce microglia to migrate to sites of injury in much the same way that macrophage migration is regulated in the periphery, and it is this process that is mimicked by B1 antibody (34).
Naturally, one might want to compare the therapeutic efficiency of induced microglia to antibodies against Aβ, particularly because of the mixed results seen in the extensive ongoing human trials with various Aβ antibodies. At present there is no basis for comparison, but scavenger cells might reasonably be expected to have effects that go beyond simple antibodies. Whether such effects, if any, are beneficial remains to be seen.
Materials and Methods
Mouse Strains and Cell Lines.
The following mouse strains were used: C57BL/6J, FVB/NJ, B6 (Cg)-Tyrc-2J Tg (UBC-mCherry) 1Phbs/J, 129S-Vimtm1Cba/MesDmarkJ, FVB-Tg (CAG-luc,-GFP) L2G85Chco/J, and B6.Cg-Tg (APPswe, PSEN1dE9)85Dbo/Mmjax (The Jackson laboratory). The HEK293T cell line was maintained in DMEM containing 10% FCS and penicillin and streptomycin (Gibco-Invitrogen). The Expi293F cell line was maintained in Expi293 Expression Media (Gibco-Invitrogen). Human CD34+ cells (All-Cells) and mouse bone marrow cells were cultured in StemSpan serum-free media with cytokine mixture 100 (STEMCELL Technologies), only serum-free media, or RPMI 1% FBS. Mice were housed and handled according to protocols approved by the Institutional Animal Care and Use Committee at The Scripps Research Institute. According to the Scripps Office for the Protection of Research Subjects Clinical Research Services, the study is not human subject research and does not require oversight by the Scripps Institutional Review Board.
Combinatorial Antibody Library and Transduction.
ScFv genes were obtained from a naive human combinatorial antibody library (1 × 1011 library diversity) and subcloned into a lentiviral vector. Lentivirus was produced in HEK293T cells by cotransfection of lentiviral vectors with the pCMVD8.91 and pVSVg viral packaging vectors at a ratio of 1:1:1. The mouse bone marrow cells were incubated with lentivirus for 3 d at 37 °C.
Bone Marrow Transplantation.
Bone marrow cells were transduced with the lentiviral antibody library at a multiplicity of infection of 2 and transplanted to lethally irradiated mice. The mice with transplanted bone marrow were maintained for 1 wk to 6 mo. The brains were perfused, fixed, harvested, and kept frozen at −80 °C. The antibody genes from the brain were amplified by PCR with primer pairs customized for our lentiviral vector, analyzed by electrophoresis, and recovered.
Purification of scFv-Fc Proteins.
The vector encoding the ScFv-Fc tag fusion protein was transfected into Expi293F cells for transient expression. Antibodies from the pooled supernatants were purified using HiTrap Protein G HP columns with an ÄKTAxpress purifier (GE). The buffer was exchanged to Dulbecco’s PBS (pH 7.4) and stored at 4 °C.
Immunoprecipitation and Mass Spectrometry.
For immunoprecipitation, mouse bone marrow cells were prepared and solubilized in lysis buffer. The lysates were incubated with B1 Ab for 2 h at 4 °C, followed by incubation with 50 μL of protein G-Sepharose beads (Pierce). The eluent was introduced into the linear trap quadrupole mass spectrometer from a nano-ion source with a 2-kV electrospray voltage. The analysis method consisted of a full MS scan with a range of 400–2,000 m/z followed by data-dependent MS/MS on the three most intense ions from the full MS scan. The raw data from the linear trap quadrupole were searched using the IPI human FASTA database with the MASCOT (https://www.matrixscience.com/) search engine.
Western Blot.
Cells were washed with PBS and then lysed in lysis buffer (50 mM Hepes, pH 7.2, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 10% glycerol, 1% Triton X-100). The lysates were then centrifuged at 12,000 × g for 15 min at 4 °C. The proteins were denatured in Laemmli sample buffer (5 min at 95 °C), separated by SDS/PAGE, and transferred to nitrocellulose membranes using the iBlot blotting system (Invitrogen). Membranes were blocked in PBS with Tween 20 (PBST) containing 5% BSA for 30 min before being incubated with antibodies for 3 h. VIM protein (Fitzgerald) and bone marrow lysates from C57BL/6J or VIM-deficient mice were used for identification. After washing the membranes several times with PBST, the blots were incubated with B1 Ab or horseradish peroxidase-conjugated anti-VIM or anti–β-actin antibody for 1 h. The membranes were then washed with PBST and developed by ECL. Phosphorylation was performed with phospho-AKT, ERK, p38, and VIM S38 (Cell Signaling Technology).
Flow Cytometry and Cell Sorting.
Cells were stained with anti-mouse CD45, CD14 (eBioscience), CD86 (BD Bioseciences), CD11b, CX3CR1, CD206, MHCII (Biolegend), and TMEM119 (Abcam). Stained cells were analyzed with a LSRII flow cytometer (Becton Dickinson).
Real-Time Quantitative PCR.
RNA from cells cultured with B1 Ab was extracted (Qiagen) for cDNA synthesis (Bio-Rad Laboratories). PCR was performed in triplicate using 400 ng of cDNA, the RT SYBR Green supermix, and a C1000 Thermal cycler (Bio-Rad Laboratories). Primer sets used were specific for human CX3CR1, IBA1, CD11b, CD68, TMEM119, HEXB, GFAP, SLC1A2, ALDH1LA, Olig1, Olig2, and MOG and for mouse ARG1, IL10, CD206, iNOS, TNFα, and IL1β. Primer sequences are in Dataset S1.
Immunohistochemistry and Immunofluorescent Confocal Microscopy.
Immunohistochemistry was performed on frozen whole-brain sections cut horizontally. Antibodies were diluted in 1× PBS containing 4% horse serum and 0.2% Triton-X100. Rat anti-mCherry (1:500; Invitrogen), goat anti-CX3CR1 (1:500; R&D system), rabbit anti-IBA1 (1:500; Wako), rabbit anti-GFAP (1:500; Wako), or rabbit anti-TMEM119 (kind gift from Ben Barres, Stanford University, Stanford, CA) were used as markers for microglia. Sections were incubated overnight with primary antibody, washed, and then incubated for 1 h with secondary antibody (goat anti-rabbit, goat anti-rat, or donkey anti-goat, 1:250; Invitrogen). Sections and coverslips were then mounted onto glass slides with anti-fade mounting medium with DAPI (ThermoFisher). Confocal microscopy was performed using a Zeiss LSM 710 laser-scanning confocal microscope.
Bioluminescence Imaging.
Bone marrow cells from luciferase-expressing transgenic mice [FVB-Tg (CAG-luc,-GFP) L2G85Chco/J] were transduced with the lentiviral B1 Ab and transplanted into lethally irradiated recipient mice (FVB/NJ). The mice were imaged 1 wk posttransplantation. CycLuc1 (END Millipore) was injected (100 μL of 5 mM solution in PBS) i.v. into recipient mice before acquiring images using the IVIS Lumina system (Perkin-Elmer). Images were acquired as a 60-s exposure/image. Regions of interest (ROI) were drawn around each brain, and the total number of counts within each ROI were recorded.
Phagocytosis Assay.
The phagocytosis assay was conducted with DAPI-labeled FluoSpheres Fluorescent Microspheres (Invitrogen). Human CD34+ cells were differentiated into microglia by the B1 Ab in a six-well plate in vitro. Microbeads were sonicated and diluted (1:80) with RPMI medium (Invitrogen) without FBS. The diluted solution was then mixed with culture medium and incubated for 2 h. To determine the phagocytic event, microglial engulfment was analyzed by an IN Cell Analyzer 6,000 (GE) during incubation at 37 °C.
Aβ Peptide Aggregation in Vitro.
The Aβ peptide aggregation assay was conducted with Beta-Amyloid (1–42) HiLyte Fluor 488-labeled (Anaspec). Human CD34+ cells were differentiated into microglia by the B1 Ab in a six-well plate in vitro. Aβ peptide (20 µM) was mixed with culture medium and incubated for 12 h. The Aβ peptide uptake experiment was analyzed by fluorescence microscopy (Zeiss).
Plaque Deposition Analysis.
Mouse brains were perfused, fixed in 4% paraformaldehyde (PFA) for 24 h (4 °C), cryoprotected with 30% sucrose in PBS (4 °C), and frozen in dry ice. Serial coronal sections (50 μm thick) were collected from the genu of the corpus callosum to the caudal hippocampus. Sections (each separated by 300 μm) were stained with biotinylated HJ3.4 (Aβ 1–16) antibody (gift from Jason Ulrich, Washington University in St. Louis, St. Louis) to visualize Aβ-immunopositive plaques. Immunostained sections were imaged using a Leica scanner. Quantitative analysis of the percentage of area covered by HJ3.4 was performed using the ImagePro program.
Supplementary Material
Acknowledgments
We thank Professor Ben Barres (Stanford University) for the gift of the TMEM119 antibody and Michal Bajo [The Scripps Research Institute (TSRI)], Anita San Soucie (TSRI), William Kiosses (TSRI), Shuli Kang (TSRI), Young Jun Kang (TSRI), F. Chris Bennett (Stanford University), Jason Ulrich (Washington University), Ian Wilson (TSRI), Tamas Bartfai (TSRI), and Paul Greengard (Rockefeller University) for helpful discussions and advice. This is manuscript #29364 from The Scripps Research Institute. This work was supported by the JPB Foundation and Zebra Biologics.
Footnotes
The authors declare no conflict of interest.
Data deposition: Raw data from RNA-seq analyses have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107006 (accession no. GSE107006).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719259115/-/DCSupplemental.
References
- 1.Han KH, et al. An agonist antibody that blocks autoimmunity by inducing anti-inflammatory macrophages. FASEB J. 2016;30:738–747. doi: 10.1096/fj.15-281329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yea K, Xie J, Zhang H, Zhang W, Lerner RA. Selection of multiple agonist antibodies from intracellular combinatorial libraries reveals that cellular receptors are functionally pleiotropic. Curr Opin Chem Biol. 2015;26:1–7. doi: 10.1016/j.cbpa.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Lerner RA, et al. Antibodies from combinatorial libraries use functional receptor pleiotropism to regulate cell fates. Q Rev Biophys. 2015;48:389–394. doi: 10.1017/S0033583515000049. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Xie J, Lerner RA. A proximity based general method for identification of ligand and receptor interactions in living cells. Biochem Biophys Res Commun. 2014;454:251–255. doi: 10.1016/j.bbrc.2014.10.085. [DOI] [PubMed] [Google Scholar]
- 5.Xie J, et al. Prevention of cell death by antibodies selected from intracellular combinatorial libraries. Chem Biol. 2014;21:274–283. doi: 10.1016/j.chembiol.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, et al. Selecting agonists from single cells infected with combinatorial antibody libraries. Chem Biol. 2013;20:734–741. doi: 10.1016/j.chembiol.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Yea K, et al. Converting stem cells to dendritic cells by agonist antibodies from unbiased morphogenic selections. Proc Natl Acad Sci USA. 2013;110:14966–14971. doi: 10.1073/pnas.1313671110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie J, Zhang H, Yea K, Lerner RA. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc Natl Acad Sci USA. 2013;110:8099–8104. doi: 10.1073/pnas.1306263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Wilson IA, Lerner RA. Selection of antibodies that regulate phenotype from intracellular combinatorial antibody libraries. Proc Natl Acad Sci USA. 2012;109:15728–15733. doi: 10.1073/pnas.1214275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yea K, et al. Agonist antibody that induces human malignant cells to kill one another. Proc Natl Acad Sci USA. 2015;112:E6158–E6165. doi: 10.1073/pnas.1519079112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mildner A, et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 12.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 13.Ginhoux F, Lim S, Hoeffel G, Low D, Huber T. Origin and differentiation of microglia. Front Cell Neurosci. 2013;7:45. doi: 10.3389/fncel.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muffat J, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosselin D, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chernoivanenko IS, Minin AA, Minin AA. Role of vimentin in cell migration. Ontogenez. 2013;44:186–202. doi: 10.7868/s0475145013030026. [DOI] [PubMed] [Google Scholar]
- 19.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Helfand BT, Chou YH, Shumaker DK, Goldman RD. Intermediate filament proteins participate in signal transduction. Trends Cell Biol. 2005;15:568–570. doi: 10.1016/j.tcb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Mitra A, et al. Cell-surface vimentin: A mislocalized protein for isolating csVimentin(+) CD133(−) novel stem-like hepatocellular carcinoma cells expressing EMT markers. Int J Cancer. 2015;137:491–496. doi: 10.1002/ijc.29382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du N, et al. Cell surface vimentin is an attachment receptor for enterovirus 71. J Virol. 2014;88:5816–5833. doi: 10.1128/JVI.03826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mor-Vaknin N, Punturieri A, Sitwala K, Markovitz DM. Vimentin is secreted by activated macrophages. Nat Cell Biol. 2003;5:59–63. doi: 10.1038/ncb898. [DOI] [PubMed] [Google Scholar]
- 24.Jiang SX, Slinn J, Aylsworth A, Hou ST. Vimentin participates in microglia activation and neurotoxicity in cerebral ischemia. J Neurochem. 2012;122:764–774. doi: 10.1111/j.1471-4159.2012.07823.x. [DOI] [PubMed] [Google Scholar]
- 25.Wohl SG, Schmeer CW, Friese T, Witte OW, Isenmann S. In situ dividing and phagocytosing retinal microglia express nestin, vimentin, and NG2 in vivo. PLoS One. 2011;6:e22408. doi: 10.1371/journal.pone.0022408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamphuis W, et al. GFAP and vimentin deficiency alters gene expression in astrocytes and microglia in wild-type mice and changes the transcriptional response of reactive glia in mouse model for Alzheimer’s disease. Glia. 2015;63:1036–1056. doi: 10.1002/glia.22800. [DOI] [PubMed] [Google Scholar]
- 27.Kumar N, et al. Requirement of vimentin filament assembly for beta3-adrenergic receptor activation of ERK MAP kinase and lipolysis. J Biol Chem. 2007;282:9244–9250. doi: 10.1074/jbc.M605571200. [DOI] [PubMed] [Google Scholar]
- 28.Virtakoivu R, et al. Vimentin-ERK signaling uncouples slug gene regulatory function. Cancer Res. 2015;75:2349–2362. doi: 10.1158/0008-5472.CAN-14-2842. [DOI] [PubMed] [Google Scholar]
- 29.Barberis L, et al. Leukocyte transmigration is modulated by chemokine-mediated PI3Kgamma-dependent phosphorylation of vimentin. Eur J Immunol. 2009;39:1136–1146. doi: 10.1002/eji.200838884. [DOI] [PubMed] [Google Scholar]
- 30.Li QF, et al. Critical role of vimentin phosphorylation at Ser-56 by p21-activated kinase in vimentin cytoskeleton signaling. J Biol Chem. 2006;281:34716–34724. doi: 10.1074/jbc.M607715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okonogi N, et al. Cranial irradiation induces bone marrow-derived microglia in adult mouse brain tissue. J Radiat Res (Tokyo) 2014;55:713–719. doi: 10.1093/jrr/rru015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asheuer M, et al. Human CD34+ cells differentiate into microglia and express recombinant therapeutic protein. Proc Natl Acad Sci USA. 2004;101:3557–3562. doi: 10.1073/pnas.0306431101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinze A, Stolzing A. Differentiation of mouse bone marrow derived stem cells toward microglia-like cells. BMC Cell Biol. 2011;12:35. doi: 10.1186/1471-2121-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLarnon JG. Microglial chemotactic signaling factors in Alzheimer’s disease. Am J Neurodegener Dis. 2012;1:199–204. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.