Significance
How HIV reverse transcriptase (RT) engages its RNase H active site to degrade viral RNA genome has remained unknown. Here we show that only with a preferred sequence does an RNA/DNA hybrid adopt the specifically distorted structure to fit into RT for cleavage by RNase H. The conformation of RT for RNA hydrolysis is distinctly different from that for DNA synthesis and reveals a structural cavity, which can serve as a target for RT inhibition.
Keywords: RNase H, sequence specificity, polypurine tract, minor groove recognition, connection domain
Abstract
HIV-1 reverse transcriptase (RT) contains both DNA polymerase and RNase H activities to convert the viral genomic RNA to dsDNA in infected host cells. Here we report the 2.65-Å resolution structure of HIV-1 RT engaging in cleaving RNA in an RNA/DNA hybrid. A preferred substrate sequence is absolutely required to enable the RNA/DNA hybrid to adopt the distorted conformation needed to interact properly with the RNase H active site in RT. Substituting two nucleotides 4 bp upstream from the cleavage site results in scissile-phosphate displacement by 4 Å. We also have determined the structure of HIV-1 RT complexed with an RNase H-resistant polypurine tract sequence, which adopts a rigid structure and is accommodated outside of the nuclease active site. Based on this newly gained structural information and a virtual drug screen, we have identified an inhibitor specific for the viral RNase H but not for its cellular homologs.
HIV is the cause of the global AIDS pandemic. More than 70 million people have become infected, 35 million have died from AIDS-related illness, and 36.7 million people are currently living with HIV. HIV encodes four enzymatic activities: a protease, an integrase, a DNA polymerase, and RNase H. The latter two reside in a single polypeptide chain of the heterodimeric reverse transcriptase (RT) (1). Structures of HIV-1 RT engaging in DNA synthesis have been determined (2–4). How RT is inhibited by nonnucleoside (NNRTIs) and nucleotide-like RT inhibitors (NRTIs), and how drug resistance arises have been well established (5–7); however, how RNase H recognizes and cleaves viral genomic RNA in an RNA/DNA hybrid but spares polypurine tract RNA, which serves as the primer for DNA synthesis, has not emerged from previous structural studies. Because the molecular mechanisms underlying HIV-1 DNA polymerase, integrase, and protease activities have been established, their active sites have been successfully targeted using rational drug design to develop AIDS therapies (8). RNase H is the sole HIV-1–encoded enzyme for which no drug exists.
Hundreds of structures of HIV-1 RT, ranging from the apo form to complexes with inhibitors, nucleic acid substrates, or both, have been determined by X-ray crystallography. Its two subunits, p51 and p66, share the same coding sequence but have different tertiary structures (2, 5, 7). Both the polymerase and RNase H active sites reside in p66, and p51 plays a supportive role. The p51 subunit lacks the C-terminal RNase H domain, while the remaining fingers, thumb, palm, and connection domains undergo extensive domain rearrangement in comparison with p66 (Movie S1). The overall structure of RT is elongated, with the polymerase active site near one end and the RNase H 60 Å away at the other end.
In the HIV-1 RT structure captured in the DNA synthesis mode (2), the DNA duplex passes by the RNase H active site at a distance of at least 5.5 Å. The first structure of HIV-1 RT and an RNA/DNA hybrid complex, in which the RT was cocrystallized with “nucleic acid” without specification of RNA or DNA and then soaked in RNA/DNA hybrid, was reported in 2001 (9). The resulting structure of the purported RNA/DNA hybrid [Protein Data Bank (PDB) ID code 1HYS] is superimposable with the DNA duplex complexed with RT (PDB ID code 2HMI) reported by the same group (10) (SI Appendix, Fig. S1), and the RNA strand has severe steric clashes between adjacent O2′ and O4′ because of the overall B-form conformation of the hybrid (www.rcsb.org/pdb/explore.do?structureId=1hys; SI Appendix, Fig. S2). Whether an RNA/DNA hybrid or a DNA duplex, in these RT structures the closest approach between the nucleic acid backbone and the RNase H active site is 6.5 Å, and RNase H is not engaged in catalysis.
However, the difficulty in capturing the RNA strand in the RNase H active site of HIV RT stems not only from this distance, but also from the conformation of RNA/DNA hybrids. In contrast to the cellular RNases H, which recognize 4–10 bp of a hybrid and have little sequence preference (11, 12), HIV-1 RT binds a nucleic acid duplex of 20–22 bp from the DNA polymerase domain to beyond the RNase H active site (13). Structures of cellular RNase H complexed with an RNA/DNA hybrid in which the RNA strand is poised for cleavage have been determined (14, 15). When superimposing the “active” form of a cellular RNase H-RNA/DNA complex onto RT-substrate complexes (2), we find that the 20-bp duplex bound to RT must be unwound and bent in the middle to place the RNA strand in the viral RNase H active site for cleavage (15).
In 2013, we reported a 3.3-Å cocrystal structure of HIV-1 RT and an RNA/DNA hybrid in the presence of the NNRTI efavirenz (EFV), which enhances RNase H activity and inhibits DNA polymerase by keeping RT in an open inactive form (16). The RNA/DNA hybrid is significantly underwound just upstream of the RNase H active site. The curvature and orientation of the RNA strand are compatible for cleavage, but the scissile phosphate remains >4 Å away from the HIV-1 RNase H active site. After screening various lengths and sequences of DNA/RNA hybrids, we have obtained a cocrystal structure of an HIV-1 RT–RNA/DNA hybrid at 2.65-Å resolution, in which just two base pairs of the hybrid upstream from the scissile phosphate are changed and the RNA strand is poised for cleavage by the RNase H. Furthermore, we have obtained three additional cocrystal structures of HIV-1 RT with different RNA/DNA hybrids at resolutions ranging from 2.44 to 3.25 Å that reveal the importance of substrate sequence for viral RNase H cleavage.
Results
Finding a Sequence Efficiently Cleaved by HIV-1 RNase H.
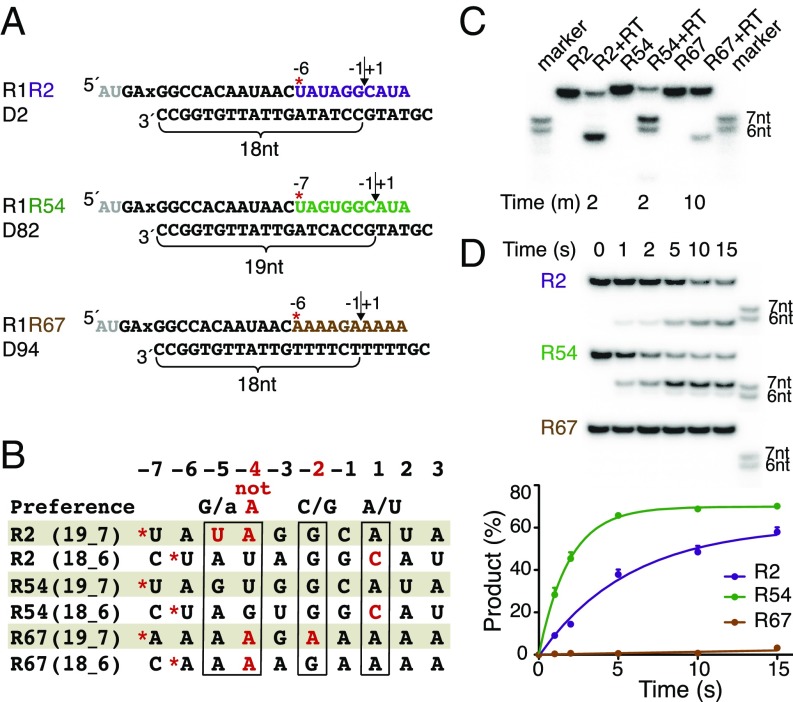
Our initial structure of an RT-substrate complex contains a 22-bp RNA/DNA hybrid with a 5′-RNA overhang of 5 nt, which includes an abasic site at the templating position to inhibit DNA synthesis (Fig. 1A; R1R2/D2). The RNA strand is composed of two oligos (10 and 17 nt) for the purpose of efficient crystallization screening (16), and the resulting RNA/DNA hybrid contains a designed nick (Fig. 1A). The phosphate nearest to the RNase H active site is 19 bp from the 3′ end of the DNA strand and 7 bp from the nick (Fig. 1 A and B) (16). This potential scissile phosphate is well oriented but outside of the RNase H cleavage site. Initially, we suspected that this displacement from the cleavage site was due to crystal lattice contacts made by the RNA/DNA hybrid, and we tried to alter lattice contacts by varying the ends of the RNA/DNA hybrid. After many failures, we realized that the RNA/DNA substrate has an unsuitable sequence (Fig. 1B).
Fig. 1.
Sequence preference of HIV-1 RT for RNase H cleavage. (A) Three sequence variations of RNA/DNA hybrids for cleavage and cocrystallization with RT. R60 is similar to R1 but 2 nt shorter (gray) on the 5′-end. * indicates the 5′-end of the second RNA strand after the prenick. The preferred RNase H cleavage site of each substrate is indicated by a vertical arrowhead. (B) Sequence comparison of two alternative cleavage sites of the three RNA/DNA variations shown in A with the known favorable sequence (17). The forbidden adenine at position −4 is highlighted in red. (C and D) Extended (C) and single-turnover (D) assays of cleaving three substrate variations by HIV-1 RT. The size markers correspond to the 6- and 7-nt cleavage products of R54.
Compared with RNA sequences efficiently cleaved by HIV-1 RNase H (17), uracil and adenine (UA) at the −5 and −4 positions from the cleavage site in our initial RNA/DNA substrate are disfavored by RNase H (Fig. 1B). When we change the sequence of the second half of the hybrid from the nonpreferred UA (R2) to the preferred guanine-uracil sequence (R54, the 54th RNA tried in the RT project), the cleavage sites and efficiency of these hybrids change dramatically in solution (Fig. 1). The R2 hybrid is not cleaved at the 19th base pair as crystallized, which would generate a 7-nt product. Instead, a 6-nt product (Fig. 1 A and C) cleaved 18 bp from the 3′ end of DNA is generated because by shifting 1 bp, the −2, −4, and −5 positions have the preferred sequence (Fig. 1 B and C). In contrast, the R54 hybrid is cleaved efficiently at 19 bp from the DNA 3′ end and less efficiently at 18 bp, generating 7- and 6-nt products, respectively (Fig. 1C). In single turnover reactions, the R54 hybrid is more efficiently cleaved than the R2 hybrid (Fig. 1D), and the 7-nt cleavage product of the R54 hybrid is more abundant than the 6-nt product. The combined cleavage efficiency of R54 hybrid (6 and 7 nt) is at least threefold higher than that of the R2 hybrid (Fig. 1D), and the cleavage efficiency of the R54 hybrid at 19 bp (7-nt product) is infinitely greater than the undetectable cleavage at the corresponding site in the R2 hybrid. For a reputedly non–sequence-specific nuclease, the difference made by a 2-bp change was surprising.
We further tested the RNase H cleavage of a polypurine tract sequence (R67) by changing the second half of the hybrid while retaining the sequence of the first half that contacts the DNA polymerase domain (R1) (Fig. 1A). Unsurprisingly, cleavage of the R67 hybrid is barely detectible in a single-turnover assay (Fig. 1D). With prolonged incubation in a steady-state reaction, the sole detectable product is 6 nt, and not 7 nt (Fig. 1C), likely because Ade at the −2 position is unfavorable (Fig. 1B). The RNA cleavage results of these hybrids by HIV-1 RT agree with the preferred sequence identified by Schultz et al. (17), and support the importance of the −2 (favoring G/C) and −4 positions (disfavoring A) for determining substrate specificity.
Crystal Structure of the HIV-1 RNase H Engaged in RNA Cleavage.
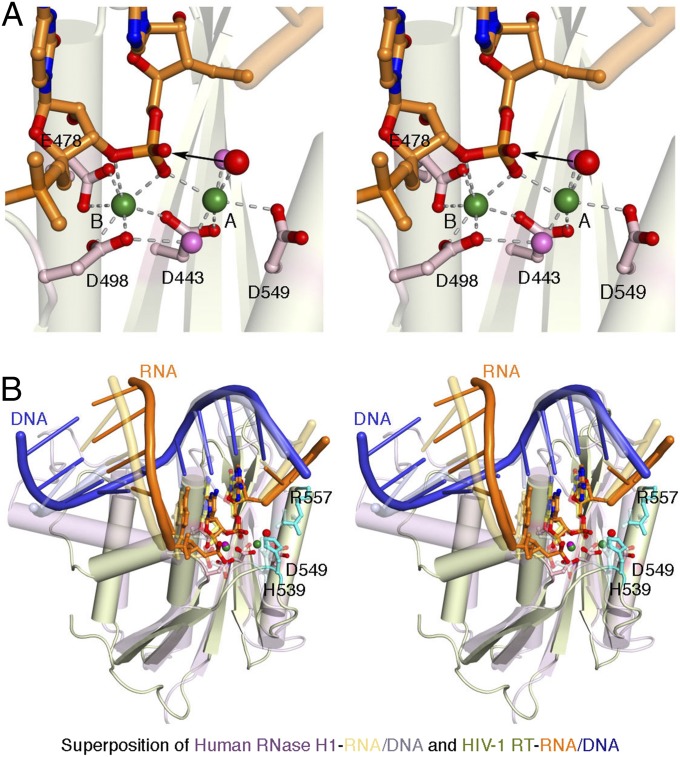
We obtained RT cocrystals with all three RNA/DNA hybrids shown in Fig. 1A in the presence of Ca2+ and EFV. Wild-type RT was used, and the catalytic cofactor Mg2+ was replaced with Ca2+, which supports enzyme-substrate complex formation but not catalysis (15, 18, 19). The diffraction quality of these crystals was improved by trimming the 5′-overhang on the RNA strand from 5 nt to 3 nt (R60), and by coexpression and copurification of RT p51 and p66 subunits rather than mixing of the cell lysates of individually expressed subunits before purification (16). Two different types of cocrystals with the RNase H-favored sequence (R54) were obtained, one with a single dG overhang at the 5′-end of the DNA strand (crystal 1) and the other with a dCG dinucleotide overhang (crystal 2), with X-ray diffraction to 2.65 and 2.44 Å, respectively (SI Appendix, Table S1). With the 2-nt trimmed RNA template and the modified protein preparation procedure, the RNase H-disfavored sequence (R2) produced a 2.89-Å structure (crystal 3; SI Appendix, Table S1), which is an improvement from the original 3.3 Å (PDB ID code 4B3O) (16). Despite the resolution improvement and the small but discernible structural changes in the finger and thumb domains of RT and the second half of the RNA/DNA hybrid (SI Appendix, Fig. S3), the scissile phosphate of the RNA remained outside of the RNase H active site in crystal 3. Finally, we produced cocrystals of RT with the R67 polypurine tract hybrid (crystal 4; SI Appendix, Table S1), which resulted in a 3.25-Å structure.
All of these crystals are in the P3121 space group and isomorphous to the previously published structure, 4B3O (16). In the four newly refined structures, the potential scissile phosphate on the RNA strand (closest to the RNase H active site) is always 19 bp from the 3′-end of the DNA strand and 7 bp from the nick site (Fig. 2 and SI Appendix, Fig. S4). Consequently, only R54 RNA presents a favorable sequence for RNase H cleavage within the crystal lattice. In crystal 1 (R54), the scissile phosphate is ideally situated in the RNase H1 active site, and the active site is completed with two Ca2+ ions (Fig. 2A). The A site Ca2+ ion, which has six ligands including nucleophilic water, and the B site Ca2+ ion, are coordinated by the four conserved active site carboxylates and the scissile phosphate, similar to the two divalent cations bound in the active site of cellular RNase H1 (14, 15) (Fig. 2B).
Fig. 2.
The RT-RNA/DNA structure in crystal 1. (A) Closeup stereoview of the RNase H active site. The arrowhead marks the nucleophilic attack of the scissile phosphate. Coordination of the two Ca2+ ions is indicated by the dashed lines. Oxygen atoms are shown in red, nitrogen in blue, and water molecules in pink or red (nucleophile). (B) Stereoview of the superposition of human RNase H1 and HIV-1 RT. The cellular RNase H is shown in pink, with the RNA and DNA in light orange and blue, respectively. The retroviral RNase H is shown in light green, and the RNA/DNA hybrid is in orange and blue. In addition to the catalytic carboxylates, H539 and R557 of RT are shown as sticks.
The proper positioning of the RNA strand and scissile phosphate for RNase H cleavage by RT can be easily perturbed by slight changes of crystal lattice contacts, as demonstrated in crystal 2. With just one additional nucleotide at the 5′-end of the DNA primer, 4 bp of the hybrid, including the scissile phosphate, pivots away by 2 Å from the RNase H active site and by 3.5 Å near the DNA 5′-end (SI Appendix, Fig. S4A). The pivoted RNA strand is no longer poised for cleavage. If higher resolution and lower B factors are indicators of increased structural stability, the RNase H active configuration in crystal 1 is likely a less-stable and higher-energy state than the RNase H inactive configuration in crystal 2.
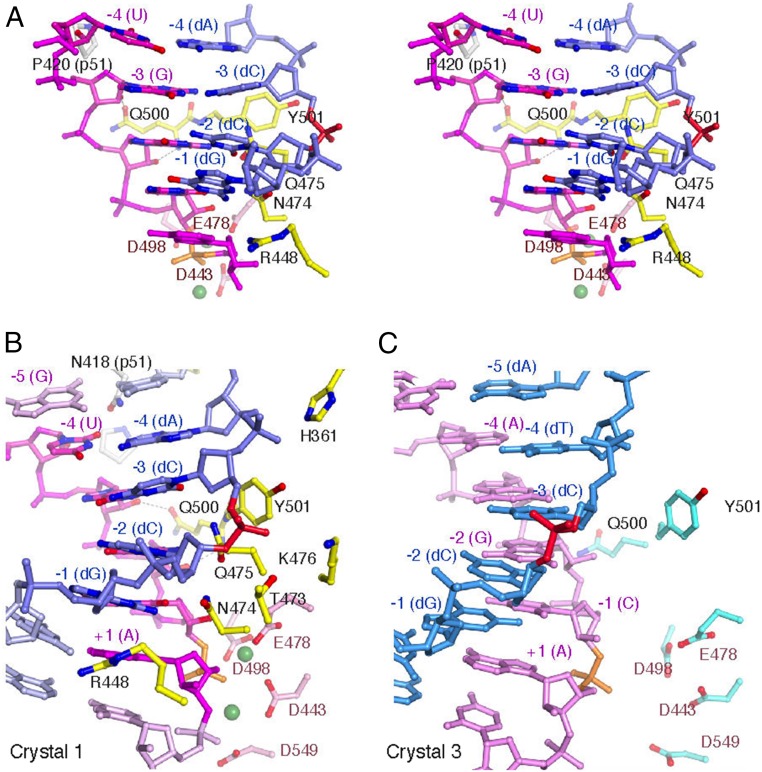
Reasons for the strong sequence bias of retroviral RNase H are revealed by the structures of crystals 3 and 4. The difference resulting from the 2-nt substitution between R54 (crystals 1 and 2) and R2 substrate (crystal 3) is dramatic (SI Appendix, Fig. S4B). Of the two halves of the RNA/DNA hybrid (before and after nick), the first half near the polymerase domain changes little, but the duplex curvature of the second half, which is bound to the RNase H domain, deviates significantly. Together with a clamshell-like slight opening of the p66 subunit (resulting in an increased distance between the polymerase and RNase H active site), the noncleavable hybrid (R2 in crystal 3) is less contracted than the preferred sequence (R54 in crystal 1), and the scissile phosphate is ∼2.5 Å further from the RNase H active site in crystal 3 (Fig. 3; SI Appendix, Fig. S4B; and Movie S2).
Fig. 3.
Interactions of RNase H with RNA/DNA hybrids. (A) Stereoview of the sharp turn at the −3 position in crystal 1. The RNA/DNA hybrid is shown as blue (DNA) and pink (RNA) sticks. The scissile phosphate is shown in orange, and the −3 phosphate of the DNA strand is highlighted in red. Protein side chains contacting the RNA/DNA hybrid (yellow) and the active site (pink) are shown in ball-and-stick representation. The Ca2+ ions are shown as green spheres. (B) Bending of the RNA/DNA hybrid at the −3 position in Crystal 1. Seven base pairs (−5 to +2) are shown, with −1 to −4 in darker colors. (C) The lack of corresponding RNase H–RNA/DNA interactions and absence of the deformation of the RNA/DNA hybrid in crystal 3.
The structure of the polypurine tract in crystal 4 shows a straighter hybrid (R67) due to the strong base stacking of polypurines and a more extended RT to accommodate the stiffer and more extended hybrid than that in crystal 3 (R2). The scissile phosphate of R67 is displaced by >4 Å from the cleavage position. Surprisingly, dT at the −5 position from the scissile phosphate is flipped out of the duplex and into a cavity between the p51 and p66 subunits. This cavity exists only when RNase H is complexed with an RNA/DNA hybrid (R2, R54, or R67). The significance and implication of this cavity is discussed below.
Key Interactions Dictate the Sequence Context for RNA Cleavage.
The distortion of the RNA/DNA hybrid that leads to position the scissile phosphate in the RNase H active site occurs at 3 bp upstream (−3 position) from the scissile phosphate. The DNA strand is pushed away from the protein by Y501, Q475, and N474, and the RNA strand is pulled in by Q500 (Figs. 3A and 4). The adjacent deoxyribonucleotides (−1 and −2) are surrounded by R448, T473, N474, Q475, and K496. The tight interactions of R488, Q475, and Y501 with the phosphosugar backbone exclude 2′-OH groups at the −1 to −3 positions (Fig. 3A) and are thus specific for DNA in the hybrid. Specificity for the RNA strand is achieved chiefly by the active site residues and Q500, which form hydrogen bonds with the −2 and −3 riboses (Fig. 3 A and B). Such “reading” of RNA/DNA hybrids is conserved in cellular RNase H1 (Fig. 2B) (14, 15).
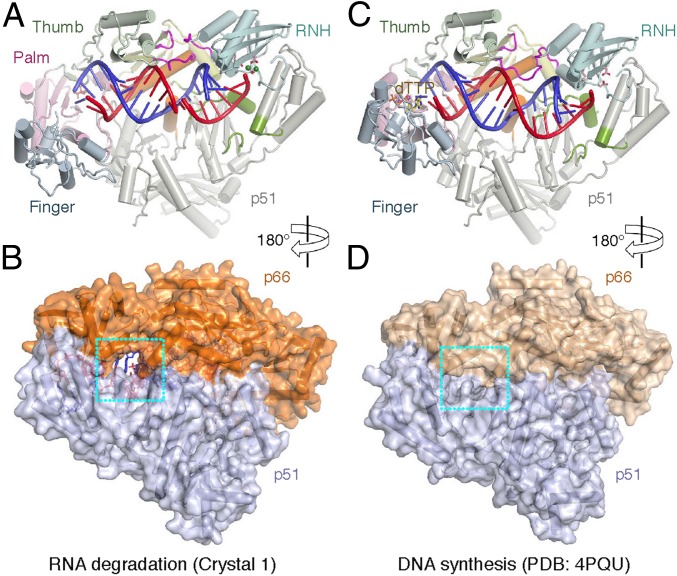
Fig. 4.
The cavity between p66 and p51 in the RNA cleavage mode. (A and B) Front (A, cartoon) and back (B, space-filling) views of crystal 1, representing the RNA degradation mode. (C and D) Front (C) and back (D) views of RT engaging in the RNA-templated DNA synthesis (PDB ID code 4PQU). The thumb domains (p66) of the two structures are superimposed. The three loop regions that undergo extensive conformational changes between the RNA degradation and DNA synthesis modes are highlighted in magenta. The cavity at the p51-p66 interface in the RNA degradation mode (B) and the corresponding region in the DNA polymerization mode (D) are boxed in cyan.
The sharp bend at the −3 position coupled with a severe tilt and shift of the DNA base to the major groove leads to the sequence preference observed for HIV-1 RNase H. If dT were at the −4 position, then the methyl group on its C5 would clash severely with both the base and deoxyribose at the −3 position (<2.4 Å) (Fig. 3B). Adenine is disfavored in the RNA strand at the −4 position, most likely to avoid the dT opposite it. When the RNA at −4 is an adenine as in the R2 hybrid, there is no bend at the −3 position, and its phosphate backbone is >4 Å further from Y501 than that of the R54 hybrid (Fig. 3 B and C). In the absence of close contacts with RT, the R2 hybrid is more relaxed than R54, and its scissile phosphate is 2.5 Å away from the RNase H active site. Owing to the sharp turn in the RNase H active form, the base pair at the −2 position assumes a strong propeller twist (Fig. 3 A and B), which may explain why a −2 G/C base pair with three hydrogen bonds is favored over A/T for RNase H cleavage (Fig. 1B).
Two Competing Modes of HIV1-1 RT for DNA Synthesis and RNA Degradation.
To convert from the DNA synthesis to the RNA hydrolysis mode, RT undergoes a seesaw motion. If we superimpose the thumb domains (p66) of the two functional modes and thus leave the 3′-end of the primer anchored in the polymerase active site, the finger domain moves away from the thumb (opening) and the RNase H domain moves toward the thumb, thus closing the gap between them, while the RNA/DNA hybrid after the midpoint (from the 3′-primer end) becomes underwound and tilted toward the RNase H active site (Fig. 4 and Movie S3). The closing of the thumb and RNase H domains requires significant repacking of three loops (G333-G335, A355-N363, and Q509-S513), as described previously (16). Furthermore, the p51-p66 subunit interface is altered. In the RNA degradation-compatible mode, the two RT subunits are further apart at the fingers-fingers and connection-connection interfaces.
Looking from the back side of the RT (opposite from where nucleic acid substrate binds), an oval-shaped cavity of 8 × 12 Å appears between the p51 and p66 subunits when RT is in the RNA-degradation–compatible mode, and the deoxyribonucleotides at the −4 and −5 positions from the RNase H cleavage site become solvent-exposed on both major and minor groove sides (Fig. 4 A and B). The cavity, which persists in crystals 1–4 and 4B3O (16), is the largest when dT at the −5 position is flipped into it in crystal 4. This cavity disappears in the DNA synthesis mode, and the minor groove side of nucleic acid substrate is shielded from the bulk solvent whether the template is DNA or RNA (PDB ID codes 1RTD, 3KK2, and 4PQU) (2–4) (Fig. 4 C and D).
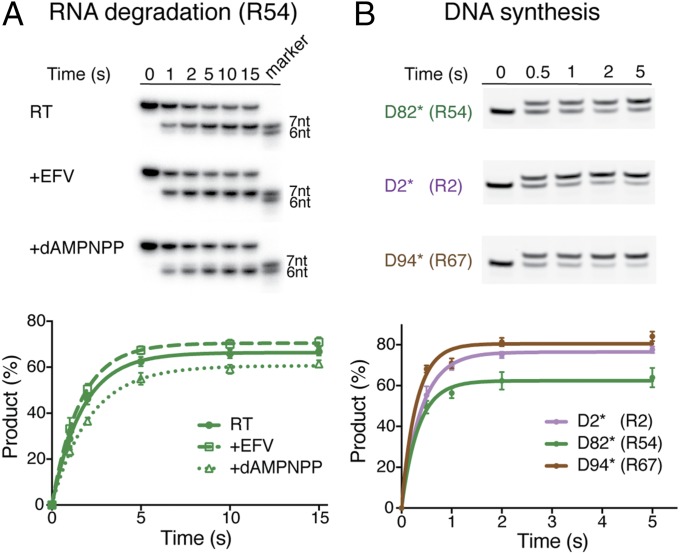
Whether DNA synthesis and RNA degradation can occur simultaneously has been a topic of debate (20, 21). It is well established that DNA synthesis occurs more frequently and at a much faster rate than RNA hydrolysis during reverse transcription (22). As detailed above, the two functional states of RT are structurally nonequivalent. It has been reported that engaging the DNA 3′-end in the synthesis mode inhibits RNA degradation (23), while inhibition of DNA synthesis by NNRTIs promotes 3′-DNA–directed RNA degradation (24, 25). Using the RNA/DNA hybrid preferred by RNase H hydrolysis (R54) in single-turnover assays, we confirmed that inhibition of DNA synthesis by the NNRTI EFV enhances RNA hydrolysis (Fig. 5A). The presence of EFV in cocrystals of RT and RNA/DNA likely stabilizes the RNA degradation mode. In contrast, prolonged engagement of DNA polymerization by a nonreactive nucleotide analog (dAMPNPP) reduces RNA hydrolysis (Fig. 5A).
Fig. 5.
The competing DNA synthesis and RNA hydrolysis modes of RT. (A) Single-turnover assay of RNase H cleavage (of R54) by HIV-1 RT alone and in the presence of 100 µM EFV or 500 µM dAMPNPP. The 32P-labeled R54 is preferably cleaved 19 bp from the 3′-end of the DNA to produce 7 nt. In the presence of dAMPNPP, the cleavage site is shifted to 18 bp from the 3′-DNA, and cleavage efficiency is reduced. (B) Single-turnover assays of dATP incorporation into R54, R2, and R67 hybrids by the RNase H-inactivated HIV-1 RT (D498N). DNA primers are 5′-labeled with fluorescein amidite.
We then asked whether engaging in RNA degradation reduces the efficiency of DNA synthesis. To do so, we conducted single-turnover assays of the RNase H-defective RT (D498N) using preferred (R54), nonpreferred (R2), or polypurine tract (R67) RNA/DNA hybrids. We found that incorporation of dATP is most efficient with the RNase H-resistant polypurine tract and least efficient with the RNase H-preferred substrate (Fig. 5B). To eliminate the possibility that such differences are due to different substrate affinities, we determined the overall binding constants of these hybrids to RT and found them to be similar, ranging from 6.7 nM to 10.4 nM (SI Appendix, Fig. S5). These data indicate that local conformation of RNA/DNA hybrids around the RNase H active site influences the rate of DNA synthesis. The better the hybrid binds to the RNase H active site, the less DNA synthesis occurs. The incompatible structures and the competing nature of RNA degradation and DNA synthesis suggest that RT cannot perform the two functions simultaneously.
An Inhibitor of RT Targeting the p51-p66 Interface.
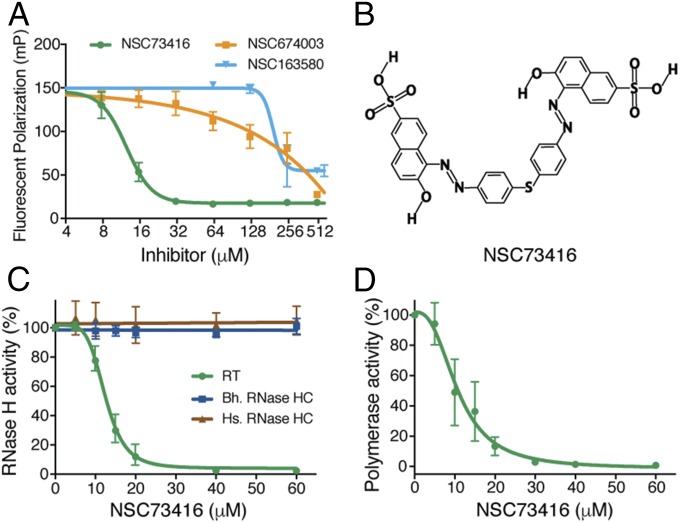
The cavity between the p51 and p66 subunits 4–5 bp upstream from the RNase H cleavage site provides a site for small molecules to bind to RT. Using this cavity as a target and an in-house developed virtual screening pipeline (LvsPipe) (26), we screened 260,000 compounds in the NCI DTP (Developmental Therapeutics Program) library and identified potential inhibitors. We obtained 46 compounds from DTP and tested them in fluorescence-based RNA/DNA substrate binding, RNase H, and DNA polymerase activity assays. Three compounds inhibited nucleic acid binding and both enzymatic activities of HIV-1 RT, and the best one inhibited all three activities at 10–15 µM and had no effect on bacterial and human RNase H1 (Fig. 6). The mechanism of action of these inhibitors differs from that of the NRTIs and NNRTIs currently in clinical use, which do not inhibit either substrate binding or RNase H activity, but is consistent with the expected effects of interfering with the p51-p66 interface.
Fig. 6.
Characterization of an RT inhibitor targeting the p51-p66 interface. (A) Three compounds from the NCI DTP library inhibit the fluorescence-labeled RNA/DNA hybrid binding to RT at various concentrations in the presence of Ca2+. (B) Structure diagram of NSC73416. Diagrams of the other two compounds are provided in SI Appendix, Fig. S8. (C) NSC73416 inhibits the HIV-1 RNase H with IC50 = 12 µM, but has no effects on human (Hs) or bacterial (Bh) RNase H1. (D) NSC73416 inhibits the HIV-1 polymerase activity with IC50 = 10 µM.
Discussion
Our determination of an RT structure with RNA substrate in the RNase H active site reveals the factors that prevented crystallization of HIV-1 RT in the RNA degradation mode in previous studies. Although the RNase H activity of RT is reported to be sequence-dependent (17, 27), the mechanism has remained unclear until now. We find that the strong sequence preference is due to a distorted substrate structure necessary for RNase H cleavage rather than base-specific interactions with the protein. An rA/dT pair at the −4 position prevents the necessary structure distortion for RNA degradation and thus is incompatible with RNase H cleavage. In solution, HIV-1 RT is able to find preferred sequences in most substrates. In crystals, because the substrate cannot slide relative to RT as in solution, the sequence effect is binary: cut or no cut (Figs. 1 and 3). The lack of proper crystal lattice contacts and a favorable sequence for RNase H cleavage explains the 2-decade absence of an HIV-1 RT structure in the RNA degradation mode. Despite the report of sequence preference by retroviral RNases H in 2009 (27), unfavorable sequences are continuously used in kinetic and structural studies of HIV-1 RNase H (4, 20).
The severely distorted substrate conformation for RNase H cleavage appears to be favored by the nick that we introduced for crystallization, and crystals obtained with the nicked RNA diffract X-rays to higher resolutions compared with those without the nick in the RNA (16). Nevertheless, in solution, RNA cleavage by RT occurs at the same sites and with similar efficiencies regardless of whether or not the RNA strand is prenicked (SI Appendix, Fig. S6).
Lattice contacts formed by the polymerase domains, which have been suggested to explain the inactive RNase H (28), have no effect, as we had foreseen (29), and are confirmed to be inconsequential here. Between the DNA synthesis and RNA degradation modes, the thumb domain, which forms the primer grip adjacent to the polymerase active site, interacts similarly with RNA/DNA and DNA/DNA duplexes (Fig. 4). This may explain why RNA is still hydrolyzed by RNase H when the DNA end is cross-linked to the thumb or polymerase active site (21).
Between the two functional modes of RT, the p51-p66 interface changes substantially from the fingers domain to the RNase H domain (Movie S3). Conversion of RT from DNA synthesis mode to RNA degradation mode by soaking crystals in an NNRTI solution (nevirapine) leads to significant changes at the fingers interface of p51-p66 but not at the connection interface or the RNase H domain, so the result is an incomplete switch and an unengaged RNase H active site (PDB ID code 4PUO) (4) (SI Appendix, Fig. S7). Interconversion of the two RT functional states might not be kinetically detectable (20) if it occurs faster than hydrolysis by RNase H; however, the two modes have evidently different structures, compete for a shared substrate, and may not occur simultaneously.
Competition for substrate by RNase H naturally decreases the dwelling time of an RNA/DNA hybrid in the polymerase active site, and thus may reduce the chance of incorporating mismatched dNTPs, which bind RT less well than correct dNTPs. Therefore, RNase H activity may indirectly increase the fidelity of DNA synthesis. Nature’s ploy of assigning two competing enzymatic activities in one molecule is often found in DNA polymerases that have both the DNA synthesis and endonuclease proofreading activities, and the result is enhanced fidelity (30, 31). We may also take advantage of such duality to target alternative sites for inhibition. In addition to the inhibitor found by targeting the p51-p66 interface, RNA oligonucleotides with sequences preferred by RNase H may be modified with 2′-methyl, -fluoro, or -amine groups to resist degradation and used to indirectly inhibit the DNA polymerase activity of HIV-1 RT.
Materials and Methods
The materials and methods used in this study, including protein and nucleic acid preparations; structure determination; assays for RNA/DNA hybrid binding, RNA cleavage, and DNA synthesis; and virtual and experimental drug screening, are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank B. Shen for the suggestion of virtual screening, and R. Craigie and D. Leahy for a critical reading of the manuscript. This research was supported by the National Institutes of Health Intramural AIDS-Targeted Antiviral Program (IATAP) and the National Institute of Diabetes and Digestive and Kidney Diseases (Grant DK036144-11, to W.Y.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6BSG, 6BSH, 6BSI, and 6BSJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719746115/-/DCSupplemental.
References
- 1.Freed EO. HIV-1 replication. Somat Cell Mol Genet. 2001;26:13–33. doi: 10.1023/a:1021070512287. [DOI] [PubMed] [Google Scholar]
- 2.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 3.Lansdon EB, et al. Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex. J Mol Biol. 2010;397:967–978. doi: 10.1016/j.jmb.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Das K, Martinez SE, Bandwar RP, Arnold E. Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: Insights into requirements for RNase H cleavage. Nucleic Acids Res. 2014;42:8125–8137. doi: 10.1093/nar/gku487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlain PP, et al. Crystal structures of zidovudine- or lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215. J Virol. 2002;76:10015–10019. doi: 10.1128/JVI.76.19.10015-10019.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarafianos SG, et al. Structure and function of HIV-1 reverse transcriptase: Molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pau AK, George JM. Antiretroviral therapy: Current drugs. Infect Dis Clin North Am. 2014;28:371–402. doi: 10.1016/j.idc.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarafianos SG, et al. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J, et al. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 11.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tadokoro T, Kanaya S. Ribonuclease H: Molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 13.Champoux JJ, Schultz SJ. Ribonuclease H: Properties, substrate specificity and roles in retroviral reverse transcription. FEBS J. 2009;276:1506–1516. doi: 10.1111/j.1742-4658.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Nowotny M, et al. Structure of human RNase H1 complexed with an RNA/DNA hybrid: Insight into HIV reverse transcription. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Lapkouski M, Tian L, Miller JT, Le Grice SFJ, Yang W. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat Struct Mol Biol. 2013;20:230–236. doi: 10.1038/nsmb.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz SJ, Zhang M, Champoux JJ. Multiple nucleotide preferences determine cleavage-site recognition by the HIV-1 and M-MuLV RNases H. J Mol Biol. 2010;397:161–178. doi: 10.1016/j.jmb.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Rosta E, Yang W, Hummer G. Calcium inhibition of ribonuclease H1 two-metal ion catalysis. J Am Chem Soc. 2014;136:3137–3144. doi: 10.1021/ja411408x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carafoli E, Krebs J. Why calcium? How calcium became the best communicator. J Biol Chem. 2016;291:20849–20857. doi: 10.1074/jbc.R116.735894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figiel M, et al. Coordination between the polymerase and RNase H activity of HIV-1 reverse transcriptase. Nucleic Acids Res. 2017;45:3341–3352. doi: 10.1093/nar/gkx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suo Z, Johnson KA. Effect of RNA secondary structure on RNA cleavage catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1997;36:12468–12476. doi: 10.1021/bi971218+. [DOI] [PubMed] [Google Scholar]
- 23.Purohit V, Roques BP, Kim B, Bambara RA. Mechanisms that prevent template inactivation by HIV-1 reverse transcriptase RNase H cleavages. J Biol Chem. 2007;282:12598–12609. doi: 10.1074/jbc.M700043200. [DOI] [PubMed] [Google Scholar]
- 24.Shaw-Reid CA, et al. Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry. 2005;44:1595–1606. doi: 10.1021/bi0486740. [DOI] [PubMed] [Google Scholar]
- 25.Radzio J, Sluis-Cremer N. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage, leading to diminished zidovudine excision. Mol Pharmacol. 2008;73:601–606. doi: 10.1124/mol.107.038596. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, et al. A selective small molecule DNA2 inhibitor for sensitization of human cancer cells to chemotherapy. EBioMedicine. 2016;6:73–86. doi: 10.1016/j.ebiom.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz SJ, Zhang M, Champoux JJ. Preferred sequences within a defined cleavage window specify DNA 3′ end-directed cleavages by retroviral RNases H. J Biol Chem. 2009;284:32225–32238. doi: 10.1074/jbc.M109.043158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das K, Sarafianos SG, Arnold E. Structural requirements for RNA degradation by HIV-1 reverse transcriptase. Nat Struct Mol Biol. 2013;20:1341–1342. doi: 10.1038/nsmb.2725. [DOI] [PubMed] [Google Scholar]
- 29.Lapkouski M, Tian L, Miller JT, Le Grice SF, Yang W. Reply to “Structural requirements for RNA degradation by HIV-1 reverse transcriptase”. Nat Struct Mol Biol. 2013;20:1342–1343. doi: 10.1038/nsmb.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunkel TA. Exonucleolytic proofreading. Cell. 1988;53:837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- 31.Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.