Organic material is synthesized in the sunlit surface layer of the oceans and that is where most of it is decomposed back to carbon dioxide and other inorganic constituents. However, a small fraction escapes immediate degradation and makes its way into deeper waters, some as far as the bottom thousands of meters below the surface. This sinking organic material contributes to sequestering atmospheric carbon dioxide and to feeding the biota of the deep ocean. Degradation is carried out by a complex microbial community whose taxonomic composition and metabolic potential vary greatly along the kilometer-long trip to the ocean floor. The metabolic potential of microbial communities has been deduced from metagenomics, genomic information gained by sequencing the entire community. In PNAS, Bergauer et al. (1) use metagenomics and another omic approach to explore changes in microbial communities and metabolism from 100-m to over 4,000-m deep in the Atlantic Ocean. They find many changes, but more surprising were the similarities in the cellular proteins used by microbes to degrade organic material along this depth profile and to support microbial life in the deep ocean.
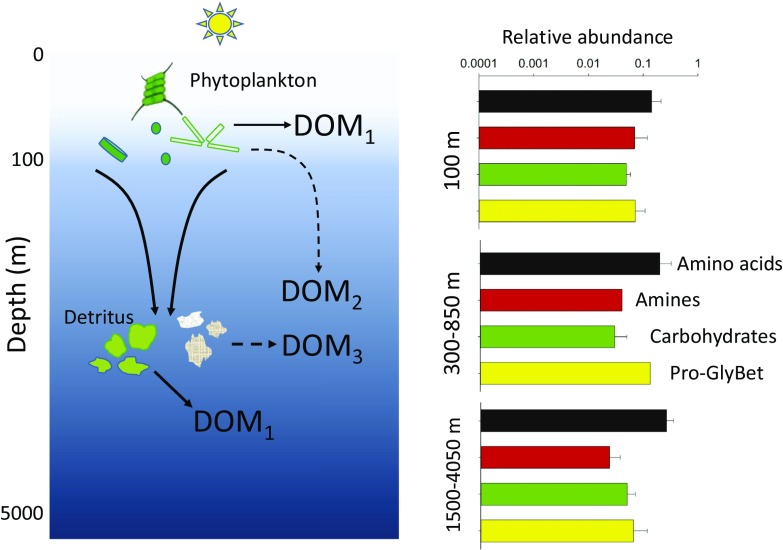
In the surface layer, bacteria use dissolved organic material (DOM) from phytoplankton, either produced directly or indirectly via release by grazers and viral lysis. The source for deep oceanic microbes is less obvious. One source is the organisms and particulate organic detritus produced in the surface layer that then sink to deeper waters. Because the biomass production of microbes on particles is much less than that by their free-living counterparts (2, 3), it is thought that sinking particulate organic material is broken up and solubilized to DOM (4) before being taken up by free-living heterotrophic bacteria and archaea (Fig. 1). Another potential source, albeit smaller than sinking particles, is the export of surface-layer DOM to deeper waters (5). The exported DOM differs from freshly produced DOM because only the compounds surviving immediate degradation are exported to deep waters. So the DOM components used by deep-sea bacteria and archaea could be similar to those in the surface layer if the main source is DOM from fresh detritus, or they could be quite different if the composition of the organic material has changed during its descent from the surface layer. Before the work by Bergauer et al. (1), most signs pointed to deep-sea DOM being quite different. Geochemical analyses indicate that the amount and chemical make-up of organic material change greatly with depth (6, 7), and large changes in microbial communities are also evident in metagenomic data and studies of 16S rRNA genes (8, 9).
Fig. 1.
Sources of organic material in the deep ocean and relative abundance of transport proteins. Heterotrophic microbes use dissolved organic material (DOM1) released by phytoplankton in the surface layer. Below the euphotic zone, microbes can use DOM exported to deep waters (DOM2) or DOM from the solubilization of sinking organisms and detritus. The composition of the organic material may be substantially modified during its descent or after solubilization (DOM3) or it may resemble DOM in the surface layer (DOM1). Dashed lines indicate that the several processes are likely needed to produce DOM. The bar graphs give the relative abundance (± SD of two to four samples) of ABC transporters for the indicated compounds based on data in figure 4 of Bergauer et al. (1). Amines, spermidine and putrescine; Pro-GlyBet, proline and glycine betaine.
The omic approach used by Bergauer et al. (1) to address this question was metaproteomics, complemented by metagenomic data and single-cell genome sequences. Rather than genes and potential activities, metaproteomics examines all cellular proteins in a microbial community, yielding information about on-going metabolic reactions and the microbial taxa carrying them out. These proteins make up structures like flagella and adhesins, as well as enzymes and transport proteins that enable cells to take up compounds from the surrounding seawater. The transport proteins are our best clues about the organic and inorganic compounds actually being used by microbes. This question has been explored with metatranscriptomics (10), but transcripts are at least one step away from the biogeochemical function being carried out by the proteins sampled by metaproteomics. It is probably impossible to directly measure uptake of more than a few compounds by specific microbial taxa because marine organic material and the microbial community are both very diverse, each with thousands or more members. Metaproteomics has been used to explore organic material use in the surface layer (11) and a few other processes in a couple subsurface samples (12), but it has not been used extensively in the deep ocean where many questions about organic material degradation by bacteria and archaea remain unanswered.
Bergauer et al. (1) do find variation in the metaproteomes from the euphotic zone (100 m) down to the bathypelagic layer (1,000–4,000 m). Transport proteins increased from 23% of identified protein sequences in the euphotic zone to 39% in the bathypelagic layer. The relative abundance of ATP-binding cassette (ABC) transporters, the most abundant type of transporter at all depths, increased somewhat from 0.49 ± 0.13 (± SD) at 100 m to 0.66 ± 0.07 in waters >850 m [calculated with data from figure 2 in Bergauer et al. (1)]. These transporters are for compounds like amino acids, polyamines, and carbohydrates. Another class of membrane proteins, TonB-dependent (TBDT) transporters, were abundant in the shallow layers (100–500 m) but less so in deeper waters. These proteins include vitamin B12 receptors, iron transporters, and TonB-linked outer-membrane proteins thought to be crucial in using detrital peptides and carbohydrates.
The microbial taxa most important in degrading DOM also varied with depth, according to the origin of the transport proteins. Among proteobacterial classes and bacterial phyla, Alphaproteobacteria (40%), including the ubiquitous SAR11 clade, and Gammaproteobacteria (10%) accounted for the highest fractions of transport proteins in the euphotic and mesopelagic zones, whereas other Gammaproteobacteria (13%) and Deltaproteobacteria (11%) had the highest fractions in the bathypelagic layer. Most of the TBDT proteins were from Gammaproteobacteria, especially the SAR86 clade, and Bacteroidetes, indicating their important roles in biopolymer degradation. Few transport proteins were from archaea (2% vs. 69% for bacteria), suggesting that heterotrophic bacteria dominate DOM use throughout the water column in the Atlantic Ocean.
The archaeal proteins are consistent with our understanding of Thaumarchaeota ecophysiology, and provide new insights into the possible biogeochemical role of Euryarchaeota in oxic and suboxic oceans. Thaumarchaeota would not be expected to have many transporters for DOM because most members of this archaeal phylum are thought to be chemolithoautotrophs that gain energy from ammonia oxidation and carbon from carbon dioxide fixation (13). Thaumarchaeota, however, did account for nearly all proteins for ammonium transport (Amt), surprisingly even in the euphotic zone where heterotrophic bacteria and phytoplankton, both well known to use ammonium, are much more abundant. Much less is known about aerobic Euryarchaeota. Unlike methanogenic genera in this phylum, Euryarchaeota in Marine Groups II and III appear to be heterotrophic or photoheterotrophic (14) and in the Atlantic Ocean accounted for several proteins involved in amino acid and peptide transport, according to Bergauer et al. (1).
Although specific cellular proteins and their origins vary with depth, what is more striking are the similarities, especially in transport proteins, along the 4,000-m-depth profile. All clusters of orthologous groups of proteins, better known as COGs, in the metaproteomic data were present throughout the entire water column sampled by Bergauer et al. (1). Transport proteins for some compounds, like amino acids and carbohydrates, varied with depth (Fig. 1), but the differences are small compared with the huge variation in environmental conditions
In PNAS, Bergauer et al. use metagenomics and another omic approach to explore changes in microbial communities and metabolism from 100-m to over 4,000-m deep in the Atlantic Ocean.
and microbial community composition from the euphotic zone down to the bathypelagic layer. One concern is that the databases essential for deciphering protein function and origin may be inadequate for examining the use of unknown organic compounds by poorly characterized microbes, both potentially abundant in the deep ocean. We also have to assume that the relative abundance of a transport protein for an organic compound faithfully mirrors the relative uptake of that compound. Nevertheless, the metaproteomic data suggest that the DOM used by microbes was roughly similar throughout the depth profile.
That observation and other metaproteomic data have implications for understanding organic material degradation and carbon cycling in the deep ocean. That the type of DOM used by microbes varied little with depth implies that deep ocean bacteria rely mainly on DOM released from fresh sinking particles produced in the surface layer. These results are consistent with Follett et al. (15), who found that 30% of deep-sea DOM was recent and likely from fresh particulate detritus. There is no evidence that bacteria use exported DOM or DOM from modified particulate detritus whose composition differs substantially from surface-layer DOM. The metaproteomic data also highlight the redundancy of microbial communities in organic matter degradation. The taxonomic composition of the communities varied greatly along the depth profile as expected, but without equally large variation in transport proteins and apparent DOM utilization. As one microbial taxon declined with depth, another increased and took over its role in DOM degradation.
Bergauer et al. (1) mined their metaproteomic data for other nuggets about the ecology of oceanic microbes and the biogeochemical processes behind the cycling of carbon and other elements along the 4,000-m-depth profile. One intriguing question Bergauer et al. mention but do not pursue is the importance of chemolithotrophy versus heterotrophy in the deep ocean and whether organic carbon produced by chemolithoautotrophs is high relative to the organic carbon supplied by sinking particles. Of course, this question and others need to be explored by more work using omic technologies and other approaches. In the meantime, Bergauer et al. provide fascinating data to address some fundamental questions about organic carbon degradation and the carbon cycle in the deep ocean.
Supplementary Material
Footnotes
The author declares no conflict of interest.
See companion article on page E400.
References
- 1.Bergauer K, et al. Organic matter processing by microbial communities throughout the Atlantic water column as revealed by metaproteomics. Proc Natl Acad Sci USA. 2018;115:E400–E408. doi: 10.1073/pnas.1708779115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho BC, Azam F. Major role of bacteria in biogeochemical fluxes in the ocean’s interior. Nature. 1988;332:441–443. [Google Scholar]
- 3.Karl DM, Knauer GA, Martin JH. Downward flux of particulate organic matter in the ocean: A particle decomposition paradox. Nature. 1988;332:438–441. [Google Scholar]
- 4.Smith DC, Simon M, Alldredge AL, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–142. [Google Scholar]
- 5.Hansell DA. Recalcitrant dissolved organic carbon fractions. Annu Rev Mar Sci. 2013;5:421–445. doi: 10.1146/annurev-marine-120710-100757. [DOI] [PubMed] [Google Scholar]
- 6.Kaiser K, Benner R. Biochemical composition and size distribution of organic matter at the Pacific and Atlantic time-series stations. Mar Chem. 2009;113:63–77. [Google Scholar]
- 7.Wakeham SG, Lee C, Hedges JI, Hernes PJ, Peterson MJ. Molecular indicators of diagenetic status in marine organic matter. Geochim Cosmochim Acta. 1997;61:5363–5369. [Google Scholar]
- 8.Sunagawa S, et al. Tara Oceans coordinators Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 9.Salazar G, et al. Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 2016;10:596–608. doi: 10.1038/ismej.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poretsky RS, Sun S, Mou X, Moran MA. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol. 2010;12:616–627. doi: 10.1111/j.1462-2920.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sowell SM, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 12.Mattes TE, et al. Sulfur oxidizers dominate carbon fixation at a biogeochemical hot spot in the dark ocean. ISME J. 2013;7:2349–2360. doi: 10.1038/ismej.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahl DA, de la Torre JR. Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol. 2012;66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- 14.Haro-Moreno JM, Rodriguez-Valera F, López-García P, Moreira D, Martin-Cuadrado A-B. New insights into marine group III Euryarchaeota, from dark to light. ISME J. 2017;11:1102–1117. doi: 10.1038/ismej.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Follett CL, Repeta DJ, Rothman DH, Xu L, Santinelli C. Hidden cycle of dissolved organic carbon in the deep ocean. Proc Natl Acad Sci USA. 2014;111:16706–16711. doi: 10.1073/pnas.1407445111. [DOI] [PMC free article] [PubMed] [Google Scholar]