Whole-brain atlas of the mouse cholinergic system
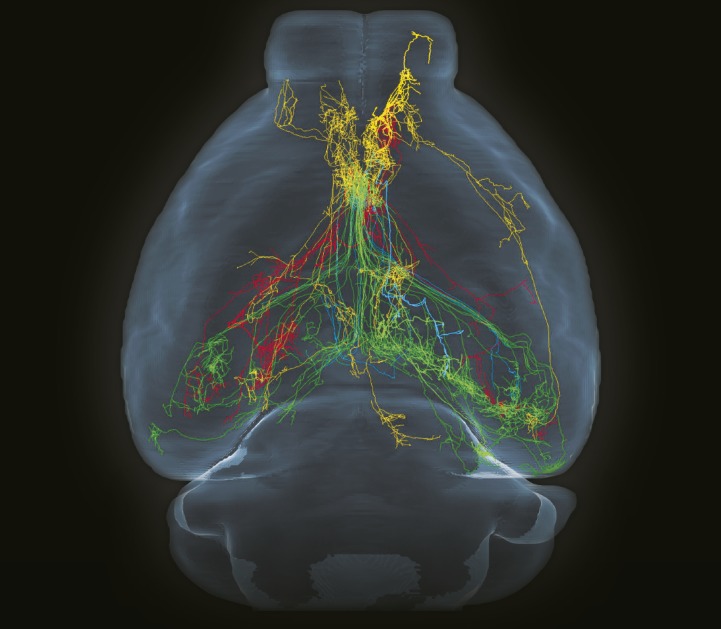
Horizontal view of 50 cholinergic neurons in basal forebrain reconstructed from whole-brain database. Image courtesy of Yefei Li (artist).
The cholinergic system modulates neuronal activity tied to critical functions, including memory, learning, behavior, sensory processing, and sleep–wake cycles. Researchers have begun to pinpoint the location and distribution of cholinergic neurons to tease out detailed network maps of this neural pathway. Xiangning Li et al. (pp. 415–420) present a comprehensive atlas of the murine cholinergic system that was constructed using an enhanced method of genetic labeling to tag cholinergic neurons and tomography-based, 1-µm resolution whole-brain imaging. Using the brain-wide map, the authors reconstructed complete axonal arbors from single neurons, traced patterns of axonal projections connecting cortical and subcortical regions, and quantified the number and volume of cholinergic neurons for 22 areas of the mouse brain. In addition, the authors analyzed a subregion of the forebrain and identified distinct categories of cholinergic neurons with projection-specific subgroups that connect to the forebrain and midbrain. The findings offer an approach to map the cholinergic system that could be applied to the human brain for unraveling disorders such as Alzheimer’s disease and chronic insomnia, according to the authors. — T.J.
Assessing the contribution of rare variants to type 2 diabetes
Much of the variance in the risk of type 2 diabetes (T2D) is thought to be due to rare DNA sequence variations with large effects, but assessing the contributions of such rare variants has been challenging. To examine the role of rare variants, Goo Jun et al. (pp. 379–384) conducted a deep whole-genome analysis of 1,034 individuals from 20 large Mexican-American pedigrees with a high prevalence of T2D. The authors failed to identify significant evidence of association between any individual rare variants and T2D or related traits, such as fasting glucose or insulin levels, suggesting that large-effect rare variants explain only a small fraction of the genetic risk of these traits in this sample. The authors also used gene expression data on 21,677 transcripts for 643 individuals in 17 of the 20 pedigrees and identified evidence of large-effect rare-variant cis-expression quantitative trait loci (eQTLs) that could not be detected in population studies. The results suggest that large numbers of rare eQTLs have larger biological effects than common variants but only make a minor contribution to overall expression heritability. Large-effect rare variants are unlikely to have a major contribution to the risk of diabetes and related traits in the sample of families studied, according to the authors. — S.R.
Planets with similarities to hot Jupiters
Hot Jupiters are a class of Jupiter-size gas planets with an orbital period that is less than 10 days, indicating that the planets travel close to their host stars. The origin of hot Jupiters remains unclear. Subo Dong et al. (pp. 266–271) used data from NASA’s Kepler satellite in conjunction with China’s Large Sky Area Multi-Object Fiber Spectroscopic Telescope to study the relationship between Kepler planets with short orbital periods, such as hot Jupiters, and the host stars’ metallicity, which is defined as the portion of a star that is not hydrogen or helium. The authors discovered a population of Neptune-size planets, referred to as “Hoptunes,” with short orbital periods. Approximately 1% of solar-type stars contain Hoptunes. Like hot Jupiters, Hoptunes often orbit stars that have higher abundances of metal than the Sun, and the frequency of Hoptunes and hot Jupiters increases with the amount of iron in their host stars. Furthermore, both hot Jupiters and Hoptunes are often associated with single-planet systems, despite nearly half of the Kepler planets being discovered in multiplanet systems. The authors found few Saturn-size planets similar to hot Jupiters and Hoptunes. The similarities among hot Jupiters and Hoptunes suggest that both types of exoplanets may have similar origins, according to the authors. — L.C.
Transmission potential of dengue virus

Ae. aegypti. Image courtesy of the CDC/James Gathany.
Dengue, a self-limiting arboviral disease, is transmitted by Aedes aegypti mosquitoes. The disease has no treatment, and efforts to suppress Ae. aegypti populations have failed to control the spread of dengue in most endemic countries, raising a need for improved vaccines and disease management approaches. Lauren Carrington et al. (pp. 361–366) explored differences in dengue virus (DENV) transmission between wild-type (WT) Ae. aegypti and Ae. aegypti infected with the wMel strain of the bacterium Wolbachia, which has been associated with a reduced susceptibility to dengue in Ae. aegypti mosquitoes. In direct human–mosquito feeding experiments involving dengue patients from Vietnam, the authors found that wMel-infected mosquitoes exhibited reduced susceptibility to DENV and had an extended incubation period for the virus compared with WT mosquitoes. Next, the authors compared DENV transmission potential in mosquitoes reared under field and laboratory conditions using indirect blood feeding and found that field-reared WT mosquitoes were more likely than lab-reared WT mosquitoes to harbor infectious virus. Moreover, the relative difference between WT and wMel mosquitoes with infectious virus was greater for field-reared mosquitoes than lab-reared mosquitoes, suggesting that laboratory conditions may underestimate the magnitude of the anti-DENV blocking effect in wMel mosquitoes. According to the authors, the findings carry implications for the biocontrol of DENV and other arboviruses. — C.S.
How genes and experience influence birdsong

Bengalese finch.
Previous studies have shown that differences in birdsong within a species reflect variation in learning, but some song heterogeneity could result from genetic diversity. David Mets and Michael Brainard (pp. 421–426) tutored genetically diverse juvenile Bengalese finches (Lonchura striata domestica) on synthetic songs that varied in tempo, which is a learned and quantifiable feature of birdsong that is controlled by central neural circuitry. Young birds tutored on a medium-tempo synthetic song sang learned songs with a broad range of tempos and with learned tempos that strongly correlated with paternal birds’ tempos, despite never having heard the paternal birds sing. Under the synthetic tutoring paradigm, the paternal birds’ genetic makeup and the tempo of the synthetic song explained approximately 55% and 21% of the variability in the young birds’ tempos, respectively. In a further analysis, living, unrelated, adult males tutored another group of hatchlings. Under live tutoring, the paternal birds’ genetic makeup and the tempo of the live tutors’ songs explained approximately 16% and 53% of variability in the young birds’ tempos, respectively—a reversal of the balance between the genetic and experiential factors observed under synthetic tutoring. According to the authors, heritable genetic contributions influence individual variation in song tempo, but the degree to which genes drive song tempo varies with the quality of instruction. — L.C.


