Significance
The cholinergic system plays a critical role in neural modulation of the mammalian brain. Here, we generated a comprehensive atlas of the cholinergic system in the mouse brain via the whole-brain imaging and reconstruction system. In the whole-brain dataset, the cholinergic neurons were divided into three categories including cortical VIP neurons, long-range projection neurons, and brainstem motor neurons. After reconstructing the cholinergic neurons in a subregion of basal forebrain, we found that their projections to the forebrain and midbrain showed neuronal subgroups with distinct projection specificity. Our work presents three-dimensional information about the cholinergic system in the mouse brain, facilitating further studies of the cholinergic system.
Keywords: cholinergic neurons, tomography, whole-brain imaging, basal forebrain, single neuron reconstruction
Abstract
The cholinergic system in the brain plays crucial roles in regulating sensory and motor functions as well as cognitive behaviors by modulating neuronal activity. Understanding the organization of the cholinergic system requires a complete map of cholinergic neurons and their axon arborizations throughout the entire brain at the level of single neurons. Here, we report a comprehensive whole-brain atlas of the cholinergic system originating from various cortical and subcortical regions of the mouse brain. Using genetically labeled cholinergic neurons together with whole-brain reconstruction of optical images at 2-μm resolution, we obtained quantification of the number and soma volume of cholinergic neurons in 22 brain areas. Furthermore, by reconstructing the complete axonal arbors of fluorescently labeled single neurons from a subregion of the basal forebrain at 1-μm resolution, we found that their projections to the forebrain and midbrain showed neuronal subgroups with distinct projection specificity and diverse arbor distribution within the same projection area. These results suggest the existence of distinct subtypes of cholinergic neurons that serve different regulatory functions in the brain and illustrate the usefulness of complete reconstruction of neuronal distribution and axon projections at the mesoscopic level.
The cholinergic system regulates various aspects of brain function, including sensory processing (1), attention (2), sleep (3), and arousal (4), by modulating neural activity via acetylcholine receptors (5, 6). Most cholinergic neurons are located in subcortical regions and have axons that innervate many brain regions, including cortices and the hippocampus, as indicated by immunochemical staining (7). Previous studies identified cholinergic neurons using bacteria artificial chromosome (BAC)-mediated genetic labeling [choline acetyltransferase (ChAT):GFP] combined with immunohistochemistry, which does not fully reveal the distribution and projection patterns of cholinergic neurons over the entire brain (8, 9). Generating a mouse line expressing Cre recombinase specifically in cholinergic neurons greatly facilitated genetic labeling of the cholinergic system (10). In the present study, we obtained enhanced fluorescence labeling of cholinergic neurons in the mouse brain using a newly developed Cre-dependent fluorescence reporter mouse line, which was generated using a previously published strategy (10).
Three-dimensional images of neuronal distribution and axon projections are traditionally acquired by reconstructing brain slices over local brain regions through the visualization of fluorescent markers. Recent work has begun to reconstruct neural networks across the whole brain at the mesoscopic level, although cell-type specificity and the fine details of axonal arborization remain to be characterized (11, 12). To overcome these obstacles, tomography-based brain-wide precision imaging using image acquisition during continuous sectioning was recently developed to obtain whole-brain maps for neurons of interest (13–16). Using the latest tomography-based method (15), we constructed a comprehensive whole-brain map of genetically labeled cholinergic neurons in the mouse brain, which provides quantitative information on neuronal distribution and detail patterns of axonal projection. With reconstruction of single cholinergic neurons, highly diverse neuronal groups with distinct projection patterns for cholinergic neurons within a subregion of the basal forebrain were further revealed. This work highlights the potential contribution of single neuron projectomes in defining functional neuronal subtypes.
Results
Visualization of Genetically Labeled Cholinergic Neurons.
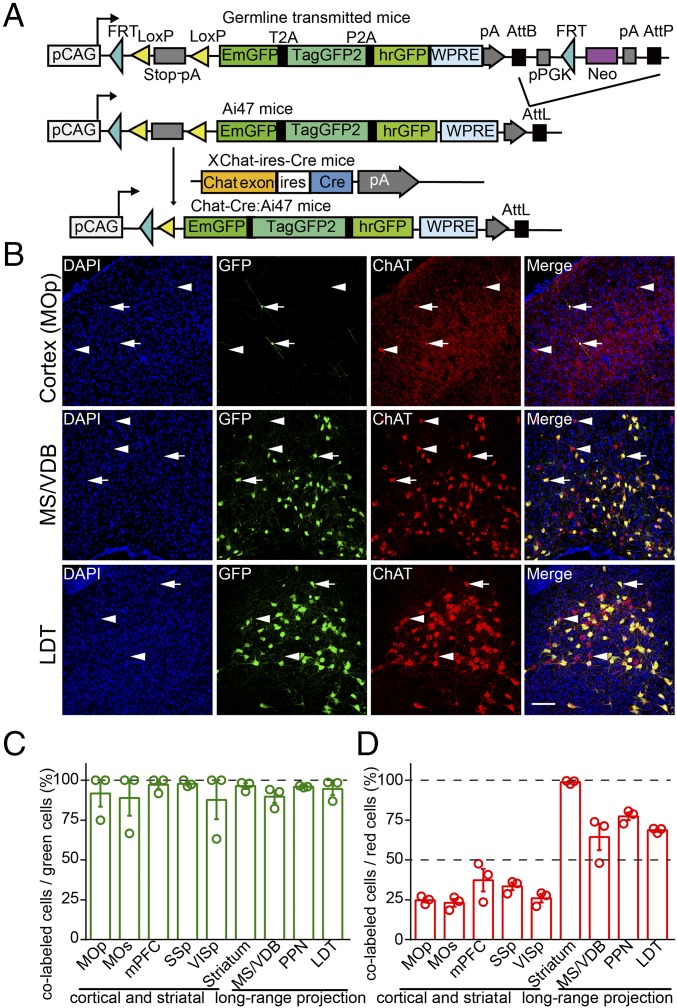
To genetically label cholinergic neurons, we crossed Chat-ires-Cre mice with Ai47 reporter mice (Chat-ires-Cre:Ai47 mice), in which the cassette containing Emerald-GFP (EmGFP), TagGFP2, and humanized Renilla-GFP (hrGFP) was expressed in a Cre-dependent manner (Fig. 1A). To ensure the accuracy of genetic labeling of cholinergic neurons, we performed immunostaining against ChAT on brain slices collected from various brain regions of Chat-ires-Cre:Ai47 mice. We found that most GFP-positive neurons in Chat-ires-Cre:Ai47 mice (89–98% in cortical and subcortical brain regions) also expressed ChAT, indicating that the genetic labeling used in this study faithfully marked cholinergic neurons in the mouse brain (Fig. 1 B–D and SI Appendix, Fig. S1 and Table S1). The Ai47 reporter containing three GFP genes yielded much higher fluorescence intensity than the single GFP cassette used in the previous Ai3 line (10) and was used to quantify neuronal number and soma volume. To trace the projection patterns of single basal forebrain cholinergic neurons, we used another approach involving injection of Cre-dependent AAV-CAG-flex-GFP virus into the basal forebrain of Chat-ires-Cre mice (SI Appendix, Fig. S2A).
Fig. 1.
Generation of Chat-ires-Cre:Ai47 mice, experimental workflow, and analysis of expression specificity of Chat-ires-Cre:Ai47 mice. (A) Schematic diagram illustrating the generation of Chat-ires-Cre:Ai47 transgenic mice. Breeding germline-transmitted mice with the Rosa26-PhiC31 deletor line induced in vivo recombination between the AttB and AttP sites, with concomitant deletion of the intervening pPGK-neomycin selectable marker cassette to produce Ai47 mice. Breeding Ai47 mice with Chat-ires-Cre mice yielded Chat-ires-Cre:Ai47 mice, in which GFP is expressed specifically in ChAT-positive neurons. (B) Immunohistochemical analysis of expression specificity of Chat-ires-Cre:Ai47 mice. GFP and ChAT double-positive neurons, GFP-negative, and ChAT-positive neurons are indicated by arrows and arrowheads. (Scale bar: 100 μm.) (C) Colocalization of GFP and ChAT. Percentage of GFP-positive neurons expressing ChAT in different brain regions. (D) Labeling ratios of GFP-positive neurons to ChAT-positive neurons. Percentage of ChAT-positive neurons expressing GFP in different brain regions. Data in C and D are presented as the mean ± SEM; n = 30 slices from three mice for every brain region.
The location of genetically labeled cholinergic neurons showing GFP fluorescence within the whole brain was defined using the newly developed brain-wide positioning system (SI Appendix, Fig. S2B) (15), in which simultaneous two-channel fluorescence imaging of GFP and propidium iodide (PI) staining is performed to provide precise position information on labeled neurons within the brain tissue (Fig. 2A). The sectioning procedure and the obtained sections are illustrated in SI Appendix, Fig. S2 C and D and Movie S1. The continuous 3D dataset allowed us to locate the brain region and each labeled cholinergic neuron in coronal, sagittal, and horizontal sections, as illustrated for several regions containing cholinergic neurons (SI Appendix, Fig. S2 E–H). The overall location of cholinergic neurons identified by this method is largely consistent with previous findings using Chat-ires-Cre mice (17). Furthermore, the PI-staining map provided cytoarchitectonic landmarks for the precise location of GFP-positive neurons (SI Appendix, Fig. S3 A and B). To align our map with the existing atlas, we provided datasets for 100-μm coronal sections with a horizontal resolution of 10 μm (down-sampled from our original 0.32-μm dataset; SI Appendix, Fig. S3 C–H) and overlaid our PI-staining map with the Paxinos brain atlas (18) and the Allen Reference Atlas (19, 20) by aligning the tissue borders and landmarks (SI Appendix, Fig. S3 C and D) (21, 22). The locations of most nuclei identified in the present work matched well with those described in the existing brain atlases, as illustrated by the map at the brainstem where cholinergic neurons are highly concentrated (SI Appendix, Fig. S3 E–H). As shown by the dataset from the same mouse brain, the alignment with Allen Reference Atlas was precisely matched for all brain regions (SI Appendix, Fig. S3I). For simplicity, we use the Allen Reference Atlas nomenclature to annotate brain regions in this study.
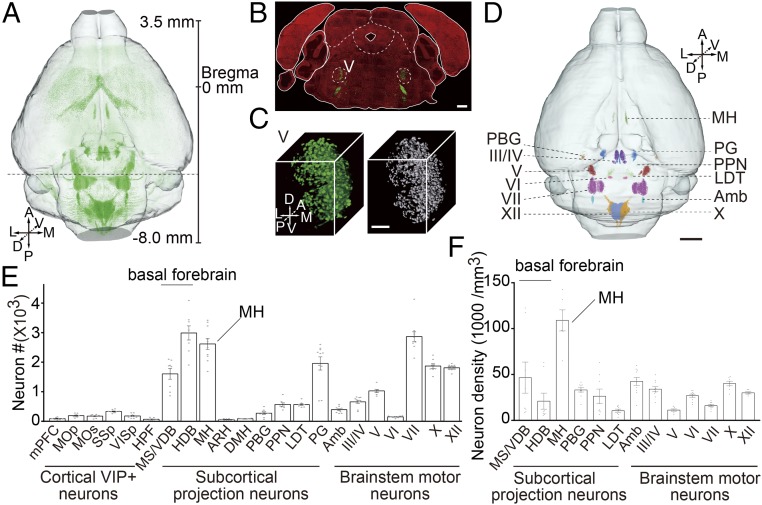
Fig. 2.
Quantitation of the distribution of cholinergic neurons in whole mouse brain. (A) Horizontal view of genetically labeled cholinergic neurons in the whole brain. Maximum-intensity projections were reconstructed at a resolution of 2 × 2 × 2 μm. Green points indicate GFP-positive cholinergic neurons. Scale bar represents distance from Bregma. A, anterior; D, dorsal; L, lateral; M, medial; P, posterior; V, ventral. (B) Merged image of coronal sections (GFP and PI signals) at the position indicated in A shows cholinergic neurons in the motor nucleus of trigeminal (V). (Scale bar: 500 μm.) (C) Overview of the 3D distribution of cholinergic neurons in V (Left). The neuron soma is distinguished using Imaris software and reconstructed in 3D space (Right). (Scale bar: 100 μm.) (D) Visualization of the anatomical localization and neuronal distribution of 3D-reconstructed subdivisions in midbrain and hindbrain. (E) Numbers of cholinergic neurons in brain regions of a single hemisphere [from five brains, one-way ANOVA, P < 0.0001, F (12, 104) = 59.99]. (F) Density of cholinergic neurons in midbrain and hindbrain of the single hemisphere shown in C [from five brains, one-way ANOVA, P < 0.0001, F (11, 88) = 12.79].
Quantitation of Cholinergic Neuronal Number and Soma Volume.
To quantify the exact number of cholinergic neurons in different regions of the mouse brain, we acquired the dataset of labeled cholinergic neurons at a resolution of 0.32 × 0.32 × 2 μm. Based on PI-stained cytoarchitectonic structure and the resolution of single soma in regions densely packed with cholinergic neurons, such as the motor nucleus of the trigeminal (V) (Fig. 2 B and C), we manually reconstructed the 3D neuronal distribution of representative subcortical regions. These regions included the nucleus of the diagonal band (NDB); medial habenula (MH); pedunculopontine nucleus (PPN); laterodorsal tegmental nucleus (LDT); and the pontine gray (PG); as well as brainstem motor nuclei, including the oculomotor nucleus (III), trochlear nucleus (IV), V, facial motor nucleus (VII), nucleus ambiguus (Amb), dorsal motor nucleus of vagus (X), and hypoglossal nucleus (XII) (Fig. 2D). The total number of cholinergic neurons in these subcortical regions represents 71.7 ± 2.0% (n = 5 mice) of all cholinergic neurons (average 5.67 × 105) in the entire mouse brain. We further determined the number of cholinergic neurons in 22 brain areas, including six cortical regions, nine subcortical regions known to provide cholinergic modulatory neurons, and seven brainstem motor nuclei (Fig. 2E and SI Appendix, Table S2). Notably, cholinergic neurons were distributed in an uneven and clustered manner in many brain regions, and neuronal density was highly variable (by a more than 10-fold difference) among various brain regions (Fig. 2F). Such a comprehensive comparison of neuronal distributions over all major cholinergic brain areas within the same mouse has not been performed previously.
Projection Patterns of the Cholinergic Neurons in Motor Nuclei in the Brainstem.
The 2-μm z-axis resolution allowed for quantitative analysis of the soma volume of cholinergic neurons. Interestingly, we found that the soma volume varied significantly among different brain regions, with up to an eightfold difference (SI Appendix, Fig. S4 and Table S3). Cholinergic neurons in the hypoglossal nucleus (XII) had the largest soma volume (2.73 ± 0.08 × 103 μm3). Neurons in brainstem motor nuclei are generally larger than those in the forebrain, suggesting the existence of region-specific subtypes of cholinergic neurons that serve different functions.
With 3D reconstruction of the cholinergic fibers, we identified several axonal projection patterns of motor nuclei in the brainstem, including III, IV, V, VII, X, and XII (SI Appendix, Fig. S5 A–F). The nerve fibers from IV have a branch before they penetrate into superior medullary velum, where the contralateral fibers converge together. The cholinergic neurons in III and X project in a posterolateral downward direction with multiple fiber bundles. The neurons in XII have a main bundle of fibers in a downward direction but no obvious branches. The cholinergic nerve from VII projects through genu of the facial nerve and branch into two routes, while some fibers pass through the medial longitudinal fascicle to ipsilateral genu of the facial nerve. The main branch is VIIn, and the other one passes through the vestibular nerve to go outside of the brain and has branches in the ventral cochlear nucleus (schematically illustrated in SI Appendix, Fig. S5G), indicating that the fibers in the facial nerve may play roles in cochlear modulation.
Long-Range Axonal Projections of Cholinergic Neurons in the Basal Forebrain.
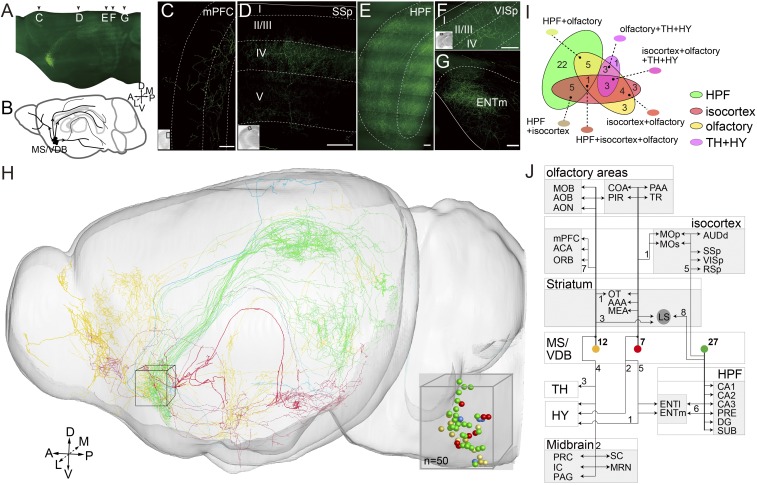
Previous studies have shown that cortical projections of cholinergic neurons in the basal forebrain exhibit an anteroposterior preference (23). To examine the detailed patterns of axonal projections of cholinergic neurons, we injected Cre-dependent AAV-CAG-flex-GFP virus into a subregion of the basal forebrain of adult Chat-ires-Cre mice, consisting of the medial septal nucleus (MS) and the vertical limb of the diagonal band nucleus (VDB) (abbreviated hereafter as MS/VDB), and performed whole-brain imaging of individual neurons at 1-μm mesoscopic resolution. Despite being only one subregion of the basal forebrain, the MS/VDB was found to send cholinergic projections to many cortical and subcortical regions, as well as the hippocampus (Fig. 3 A and B), consistent with a previous report (24). Briefly, we found that axonal fibers were widely spread out in the medial prefrontal cortex (mPFC) (Fig. 3C). Few cholinergic terminals were present in sensory and motor cortices, which were interestingly distributed in a layer-specific pattern (Fig. 3D). In the primary somatosensory area (SSp), most terminals targeted layer 4, whereas in the primary visual area (VISp), terminals were distributed across all layers, with higher density in layer 2/3 (Fig. 3 D and F). These data suggest that cholinergic modulation may act at different stages of sensory processing in a modality-specific manner. Furthermore, we found that axons from many MS/VDB cholinergic neurons sent projections to the hippocampus (HPF) and the medial entorhinal cortex (ENTm) (Fig. 3 E and G).
Fig. 3.
Mesoscopic projectome of cholinergic neurons in the basal forebrain. (A) The 3D view of cholinergic neurons in the MS/VDB labeled with AAV-CAG-FLEX-GFP virus. The resolution is 2 × 2 × 2 μm. (B) Diagram of the cholinergic projection from MS/VDB showing different routes to the olfactory bulb, frontal cortex, somatosensory cortex, hippocampal formation, and subcortical regions. Maximum-intensity projection of the coronal section showing the cholinergic terminals in mPFC (C), SSp (D), HPF (E), VISp (F), and ENTm (G). The length of the z stack is 100 μm. (H) A total of 50 cholinergic neurons were reconstructed from the whole-brain database. The soma of these reconstructed neurons focused in MS/VDB are shown in 3D. (I) Distribution of the 50 reconstructed neurons with their target areas. The numbers of neurons are labeled. (J) Schematic diagram illustrating the major projection patterns of the cholinergic neurons in VDB. The numbers of neurons are labeled. (Scale bar: 100 μm.)
To further define the projection pattern of cholinergic neurons, we traced the arborization of 50 well-separated cholinergic neurons in the MS/VDB throughout the brain using the 3D database and reconstruction protocol (Fig. 3H and SI Appendix, Fig. S6A and Movie S2). The axon-targeted areas were determined by alignment with the Allen Reference Atlas (SI Appendix, Fig. S6B). These neurons were classified into different categories according to differences in the target brain areas. Projection targets for all 50 neurons are illustrated in Fig. 3 H and I and SI Appendix, Fig. S6A. Although axons from most neurons exhibited multiple targets, the specific target sets differed. As shown in Fig. 3I, different subsets of cholinergic neurons with overlapping targets could be defined, with olfactory areas, hippocampus, and isocortex as the three major targets. The complete target areas of all 50 neurons are shown in SI Appendix, Fig. S7, which reveals the complexity of the projection patterns among these neurons.
We further defined three major categories of projection patterns for 46 neurons based on their projections to the three major target areas, with four remaining neurons belonging to other possible categories (Fig. 3J). The first category comprised 27 neurons with projections to the hippocampal formation, with subsets projecting only to different hippocampal regions and the entorhinal cortex (n = 14), as well as to the striatum (n = 8) or isocortex (n = 5), in addition to the hippocampal formation. The second category comprised 12 neurons projecting to the olfactory cortex, with subsets projecting to other olfactory areas (n = 3), the isocortex (n = 7), thalamus (n = 3), or midbrain (n = 2), in addition to the olfactory cortex. The third category comprised 7 neurons projecting to noncortical olfactory areas, with two projecting to the hypothalamus in addition to the olfactory areas (Fig. 3J). The remaining 4 uncategorized neurons (due to the low number of neurons traced) consisted of neurons that projected only to the isocortex cortex (n = 3) and palidium (n = 1) (blue colored dots in SI Appendix, Fig. S6A). The three major categories of projections appeared to be biased to three preferential routes after exiting from the basal forebrain, as shown in Fig. 3H.
Furthermore, individual cholinergic neurons can project long distances to distinct brain regions through different projection routes. For example, neuron 1 had axons that simultaneously terminated in olfactory areas, the isocortex [including secondary motor areas (MOs) and mPFC], thalamus (TH), and midbrain [periaqueductal gray (PAG)] (Fig. 3J and SI Appendix, Fig. S6C and Movie S3). We found that adjacent neurons in the basal forebrain could have significantly different projection patterns. For instance, neuron 12 (located adjacent to neuron 1) projected to hippocampal CA1 and the entorhinal cortex, whereas neuron 1 projected to olfactory areas, the isocortex, TH, and PAG (SI Appendix, Fig. S6 C and D). Two others (neurons 5 and 13), located only 6 μm away, participated in different pathways (neuron 5 projected to olfactory-related areas, while neuron 13 projected to HPF) (SI Appendix, Fig. S6E), suggesting that adjacent neurons may participate in different functions. Moreover, the targeting specificity of these cholinergic neurons was also indicated by the fact that projection routes often cross-talk with the terminal arbors of other categories, as illustrated in SI Appendix, Fig. S6F, in which an olfactory area-targeting neuron (neuron 22) that targeted both the piriform area (PIR) and the lateral part of the entorhinal area (ENTl), whereas another basal forebrain neuron (neuron 42) only target ENTl.
Layer-Specific Distribution of Cortical Cholinergic Neurons.
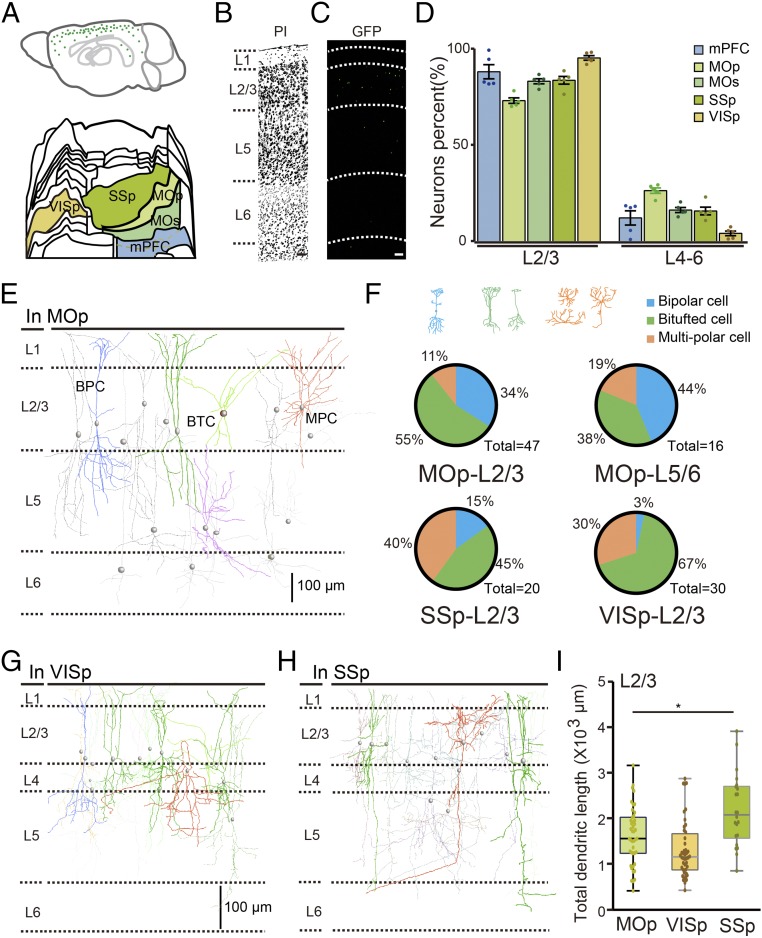
We observed GFP-labeled cholinergic neurons distributed throughout the entire cerebral cortex (Fig. 4A and Movie S1). Although ChAT-positive neurons have been found in the cerebral cortices of rodents (8, 25), the distribution and morphology of cholinergic neurons in the cortex are not fully understood. First, we performed in situ hybridization against vasoactive intestinal polypeptide (VIP) and immunofluorescence staining against GFP, parvalbumin, and somatostatin to examine subtypes of cortical cholinergic neurons. Over 90% of the cholinergic neurons in mouse cortex contained VIP, consistent with previous findings based on single-cell sequencing (SI Appendix, Fig. S8 and Table S4) (26). Next, we quantitated the cholinergic neurons in mPFC, primary motor area (MOp), MOs, SSp, and VISp (Fig. 4A). The number of cholinergic neurons was 73 ± 13 and 330 ± 18 in the medial prefrontal cortex and SSp, respectively (SI Appendix, Table S2). Using PI staining, we assessed the layer distribution pattern of cholinergic neurons in the cortex (Fig. 4 B and C). As shown in Fig. 4D, more than half of the cholinergic neurons are distributed in layer 2/3 of the prefrontal, motor, somatosensory, and visual cortices. The exact numbers of cholinergic neurons in layer 2/3 or layer 4–6 across various cortical regions are shown in SI Appendix, Table S5.
Fig. 4.
Illustration of cortical cholinergic neurons at single-cell resolution. (A) Cholinergic neurons distributed in the cortex, with the green points indicating the neurons. The flattened view of the mouse cortex in the reference atlas (18) showed the anatomical locations of the mPFC, MOp, MOs, SSp, and VISp. (B) Inverted image of the PI-staining figure showed the layer of the motor cortex. (C) The cholinergic neurons distributed in different layers of the motor cortex. (D) Percentage of cholinergic neurons in layer 2/3 and layer 4–6 of the mPFC, MOp, MOs, VISp, and SSp from five mice. (E) Reconstructed cholinergic neurons in the MOp exhibited bipolar cell (BPC), bitufted cell (BTC), and multipolar cell (MPC) patterns. (F) Statistical results showing various percentages of three morphological neurons distributed in the motor cortex, sensory cortex, and visual cortex (n = 63, 20, and 30, respectively). Reconstruction of cholinergic neurons in the SSp cortex (G) and VISp cortex (H). (I) The dendritic length of cholinergic neurons in layer 2/3 of the SSp cortex showed differences compared with those in the MOp and VISp cortices (unpaired t test, P = 0.0031, t = 3.066).
Taking advantage of 3D imaging, we examined the morphological features of these cortical cholinergic neurons in fine detail. When we acquired the dataset of the cholinergic system at a resolution of 0.32 × 0.32 × 1 μm (x, y, and z axes), we found that the dendritic arbors of cortical cholinergic neurons in S1 strikingly extended over several hundreds of micrometers, strongly suggesting that cortical cholinergic neurons may have critical functions, given their wide inputs (Fig. 4 E, G, and H). We reconstructed 150 individual cholinergic neurons at 1-μm resolution from all layers of MOp, VISp, and SSp. Based on the somatodendritic configurations, cholinergic neurons showed multiple types, including bipolar, bitufted, and multipolar morphology (Fig. 4F). Clearly, layer 2/3 cholinergic neurons are distinct from neurons located in layer 5/6, leading to the highly diverse 3D morphologies of cholinergic neurons in the MOp, VISp, and SSp cortices (Fig. 4 E, G, and H). For cholinergic neurons in layer 2/3 of MOp, more than half of all dendritic branches were distributed in layer 1 and layer 5, suggesting that these neurons mainly receive inputs from layer 1 and layer 5 (Fig. 4E). The dendritic branches of cholinergic neurons in the SSp cortex appeared to be significantly more complex than those in the MOp cortex, suggesting that cholinergic neurons in the SSp may receive more inputs. Although the morphology of these neurons was not complete due to a brightness issue during genetic labeling, the currently reconstructed single neuron still extended over a 500 × 400 × 300 μm range, and the total dendritic length of a single neuron in the SSp was ∼3 mm (Fig. 4I).
Discussion
In this study, we provide a comprehensive atlas of the cholinergic system using genetic labeling and tomography-based whole-brain imaging technology. Various methods have been developed to measure the number and projections of cholinergic neurons in many brain regions. Data from 3D measurements of the whole brain are essential for developing a comprehensive understanding of the cholinergic system (27). For the whole-brain imaging system, we can image all of the GFP+ neurons and count them through the whole brain. The possible reasons for the underestimated ratio of GFP+ neurons may be for valid labeling by the genetic mouse. Because the Chat-ires-cre may only express in special developmental stages, only part of the Chat+ neurons express GFP in Chat-ires-cre: Ai47 mice at 3–6 mo old (when we imaged). Even so, in most of the subcortical regions, almost 98% of the ChAT+ neurons are labeled by GFP in the crossed mice. As indicated in Fig. 2, the density of cholinergic neurons is not uniform across different brain regions, even within the same brain nucleus. Because classic studies using 2D methods only analyzed a few sections, the distribution pattern of cholinergic neurons should be viewed as an oversimplified model.
Reconstructing complete neuronal morphology from discontinuous sections in consecutive tissue sections is difficult, due to tissue sectioning and alignment across different sections (28, 29). Moreover, the limited 3D data cannot be used to trace the long-range projection fibers of cholinergic neurons. As shown in SI Appendix Fig. S5, the branches of motor nerves that enter into different nerve bundles or routes can be distinguished from the continuous 3D dataset. Previous studies comparing different parallel experiments or using multiple anatomical tracers suggested that cholinergic neurons in adjacent areas of the basal forebrain may project to interconnected cortical areas (28–30). The present study showed that single cholinergic neurons can project to multiple areas. Most target areas of individual cholinergic neurons are interconnected, such as the olfactory bulb connecting to the piriform cortex and entorhinal cortex (31), while the subregions of the hippocampal complex have connections with the striatum (lateral septal nucleus), isocortex (retrosplenial area), and hypothalamus (32). The mediodorsal thalamus shares reciprocal connectivity with the prefrontal cortex and modulates the prefrontal activity (33). The projection patterns of individual neurons can be diversified, while adjacent neurons have different projection patterns, such that no specific relationship between soma location within MS/VDB and axonal target areas exists (Fig. 3H and SI Appendix, Fig. S6A). These results revealed a possible model for the cholinergic projectome in the basal forebrain, suggesting that a single cholinergic neuron projects to associated functional areas, while neighboring neurons can project disparately across the cortical and subcortical areas, in contrast to a previous hypothesis (34).
The morphology and location of these neurons agree with previous descriptions of cholinergic neurons in the cortex (8). We found that over 90% of the cholinergic neurons in the mouse cortex contained VIP (SI Appendix, Fig. S6 and Table S4). More than half of the cholinergic neurons were distributed in layer 2/3, and for those neurons in layer 2/3 of MOp, more than half of all dendritic branches were distributed in layer 1 and layer 5. Moreover, we found that the dendritic branches of cholinergic neurons in the SSp cortex appeared to be significantly more complex than those in the MOp cortex. Intrinsic cholinergic cortical interneurons in the SSp are ideally suited to exert a restricted modulatory effect on small cortical units.
The whole-brain atlas of the cholinergic system established in this study may serve as a framework for further functional studies of cholinergic nuclei in the brain. Thus, subsequent circuit-level investigations may identify the structure and function of cholinergic innervation. Furthermore, this 3D atlas may shed light on brain disorders, such as Alzheimer’s disease and sleep disorders, involving dysfunction of the cholinergic system in the brain (3, 35).
Materials and Methods
Chat-ires-Cre mice (018957) were obtained from The Jackson Laboratory. The use and care of animals complied with the guidelines of the Animal Advisory Committee at the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. A complete description of the materials and methods is provided in SI Appendix, SI Materials and Methods. Abbreviations of brain regions are summarized in SI Appendix, Table S6, according to the Allen Reference Atlas.
Supplementary Material
Acknowledgments
We thank Dr. Mu-ming Poo for his critical comments on this work; Tonghui Xu, Yuxin Li, Can Zhou, Zhuonan Duan, Qiuyuan Zhong, and Hong Ni for help with experiments and data analysis; the Optical Bioimaging Core Facility of Huazhong University of Science and Technology (HUST) for support with data acquisition; and the Analytical and Testing Center of HUST for spectral measurements. This work was supported by the National Key Scientific Instrument & Equipment Development Program of China (2012YQ030260); the Science Fund for Creative Research Group of China (61421064); National Natural Science Foundation of China Grants 91432105, 91432111, 81527901, and 31625013; and the Director Fund of Wuhan National Laboratory for Optoelectronics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Illustrations of the images used in this work can be found at the Visible Brain-Wide Networks site: vbn.org.cn/2D/neuron.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703601115/-/DCSupplemental.
References
- 1.Minces V, Pinto L, Dan Y, Chiba AA. Cholinergic shaping of neural correlations. Proc Natl Acad Sci USA. 2017;114:5725–5730. doi: 10.1073/pnas.1621493114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarter M, Hasselmo ME, Bruno JP, Givens B. Unraveling the attentional functions of cortical cholinergic inputs: Interactions between signal-driven and cognitive modulation of signal detection. Brain Res Brain Res Rev. 2005;48:98–111. doi: 10.1016/j.brainresrev.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Xu M, et al. Basal forebrain circuit for sleep-wake control. Nat Neurosci. 2015;18:1641–1647. doi: 10.1038/nn.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szymusiak R. Magnocellular nuclei of the basal forebrain: Substrates of sleep and arousal regulation. Sleep. 1995;18:478–500. doi: 10.1093/sleep/18.6.478. [DOI] [PubMed] [Google Scholar]
- 5.Everitt BJ, Robbins TW. Central cholinergic systems and cognition. Annu Rev Psychol. 1997;48:649–684. doi: 10.1146/annurev.psych.48.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Sarter M, Parikh V. Choline transporters, cholinergic transmission and cognition. Nat Rev Neurosci. 2005;6:48–56. doi: 10.1038/nrn1588. [DOI] [PubMed] [Google Scholar]
- 7.Houser CR, Crawford GD, Salvaterra PM, Vaughn JE. Immunocytochemical localization of choline acetyltransferase in rat cerebral cortex: A study of cholinergic neurons and synapses. J Comp Neurol. 1985;234:17–34. doi: 10.1002/cne.902340103. [DOI] [PubMed] [Google Scholar]
- 8.von Engelhardt J, Eliava M, Meyer AH, Rozov A, Monyer H. Functional characterization of intrinsic cholinergic interneurons in the cortex. J Neurosci. 2007;27:5633–5642. doi: 10.1523/JNEUROSCI.4647-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 10.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zingg B, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A, et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science. 2010;330:1404–1408. doi: 10.1126/science.1191776. [DOI] [PubMed] [Google Scholar]
- 14.Ragan T, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong H, et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat Commun. 2016;7:12142. doi: 10.1038/ncomms12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seiriki K, et al. High-speed and scalable whole-brain imaging in rodents and primates. Neuron. 2017;94:1085–1100 e6. doi: 10.1016/j.neuron.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 17.Harris JA, et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8:76. doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paxinos G, Franklin KBJ, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2nd Ed Academic; San Diego: 2001. [Google Scholar]
- 19.Dong HW. 2008. Allen Reference Atlas: A Digital Color Brain Atlas of the C57Black/6J Male Mouse (Wiley, Hoboken, NJ)
- 20.Kuan L, et al. Neuroinformatics of the Allen mouse brain connectivity atlas. Methods. 2015;73:4–17. doi: 10.1016/j.ymeth.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, et al. TDat: An efficient platform for processing petabyte-scale whole-brain volumetric images. Front Neural Circuits. 2017;11:51. doi: 10.3389/fncir.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng J, et al. A quantitative analysis of the distribution of CRH neurons in whole mouse brain. Front Neuroanat. 2017;11:63. doi: 10.3389/fnana.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloem B, et al. Topographic mapping between basal forebrain cholinergic neurons and the medial prefrontal cortex in mice. J Neurosci. 2014;34:16234–16246. doi: 10.1523/JNEUROSCI.3011-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Williams J, Nathans J. Complete morphologies of basal forebrain cholinergic neurons in the mouse. Elife. 2014;3:e02444. doi: 10.7554/eLife.02444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong DM, Saper CB, Levey AI, Wainer BH, Terry RD. Distribution of cholinergic neurons in rat brain: Demonstrated by the immunocytochemical localization of choline acetyltransferase. J Comp Neurol. 1983;216:53–68. doi: 10.1002/cne.902160106. [DOI] [PubMed] [Google Scholar]
- 26.Seung HS. Reading the book of memory: Sparse sampling versus dense mapping of connectomes. Neuron. 2009;62:17–29. doi: 10.1016/j.neuron.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Benes FM, Lange N. Two-dimensional versus three-dimensional cell counting: A practical perspective. Trends Neurosci. 2001;24:11–17. doi: 10.1016/s0166-2236(00)01660-x. [DOI] [PubMed] [Google Scholar]
- 28.Zaborszky L, et al. Neurons in the basal forebrain project to the cortex in a complex topographic organization that reflects corticocortical connectivity patterns: An experimental study based on retrograde tracing and 3D reconstruction. Cereb Cortex. 2015;25:118–137. doi: 10.1093/cercor/bht210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gielow MR, Zaborszky L. The input-output relationship of the cholinergic basal forebrain. Cell Rep. 2017;18:1817–1830. doi: 10.1016/j.celrep.2017.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaborszky L, Buhl DL, Pobalashingham S, Bjaalie JG, Nadasdy Z. Three-dimensional chemoarchitecture of the basal forebrain: Spatially specific association of cholinergic and calcium binding protein-containing neurons. Neuroscience. 2005;136:697–713. doi: 10.1016/j.neuroscience.2005.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamichi K, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fanselow MS, Dong HW. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolkan SS, et al. Thalamic projections sustain prefrontal activity during working memory maintenance. Nat Neurosci. 2017;20:987–996. doi: 10.1038/nn.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz W, Rudy B. Spatiotemporal specificity in cholinergic control of neocortical function. Curr Opin Neurobiol. 2014;26:149–160. doi: 10.1016/j.conb.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craig LA, Hong NS, McDonald RJ. Revisiting the cholinergic hypothesis in the development of Alzheimer’s disease. Neurosci Biobehav Rev. 2011;35:1397–1409. doi: 10.1016/j.neubiorev.2011.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.