Significance
Cell motility is a dynamic process that requires the directed application of force and continuous coordinated changes in cell adhesion and cytoskeletal architecture often in response to extracellular stimuli. Here we have defined a mechanism by which RSK2 can promote cell migration and invasion in response to promotility stimuli. We show that in response to these signals RSK2 directly binds the RhoGEF LARG and phosphorylates it, thereby promoting LARG activation of RhoA GTPases. Moreover, we find that RSK2 is important for epidermal growth factor activation of Rho GTPases. These results advance our understanding of cell motility, RSK kinase function, and LARG/RhoA activation by revealing that these pathways are integrated and the precise mechanism by which that is accomplished.
Keywords: RSK2, LARG, Rho GTPases, ARHGEF12, motility
Abstract
Directed migration is essential for cell motility in many processes, including development and cancer cell invasion. RSKs (p90 ribosomal S6 kinases) have emerged as central regulators of cell migration; however, the mechanisms mediating RSK-dependent motility remain incompletely understood. We have identified a unique signaling mechanism by which RSK2 promotes cell motility through leukemia-associated RhoGEF (LARG)-dependent Rho GTPase activation. RSK2 directly interacts with LARG and nucleotide-bound Rho isoforms, but not Rac1 or Cdc42. We further show that epidermal growth factor or FBS stimulation induces association of endogenous RSK2 with LARG and LARG with RhoA. In response to these stimuli, RSK2 phosphorylates LARG at Ser1288 and thereby activates RhoA. Phosphorylation of RSK2 at threonine 577 is essential for activation of LARG-RhoA. Moreover, RSK2-mediated motility signaling depends on RhoA and -B, but not RhoC. These results establish a unique RSK2-dependent LARG-RhoA signaling module as a central organizer of directed cell migration and invasion.
Cell migration and invasion are an essential part of developmental, physiological, and pathological processes including cancer invasion and metastasis. This is a dynamic process that requires the directed application of force and continuous coordinated changes in cell adhesion and cytoskeletal architecture (1, 2). While significant progress has been made in deciphering the multiple mechanisms that underlie spatiotemporal control of these processes, major gaps in our understanding remain. The p90 ribosomal kinases (RSKs) are involved in tumor cell growth and metastasis in several cancers, including lung, breast, melanoma, and glioblastoma multiforme (GBM) (3, 4). RSKs are serine/threonine kinases activated by extracellular signal-regulated kinases (ERK) MAP kinase and constitute four isoforms (RSK1–4) (5). RSKs act as downstream effectors of receptor tyrosine kinase (RTK)/Ras/ERK signaling (6, 7). All RSK isoforms contain two kinase domains: a central regulatory linker domain and a carboxyl terminal ERK docking site (8–10). The N-terminal kinase domain (NTKD) belongs to the AGC kinase family and is responsible for phosphorylation of all known substrates. The C-terminal kinase domain (CTKD) is a part of the CaM kinase family and regulates the NTKD. Full activation of RSK2 requires phosphorylation at multiple sites. Initially ERK phosphorylates the CTKD at Thr577 and a linker site at Ser369. The CTKD then phosphorylates Ser386 and thereby generates a docking site for PDK1. PDK1 binds and phosphorylates Ser227 in the NTKD, which fully activates it (11, 12). Phosphorylation at Tyr529 by either FGF receptor 3 (in myeloid cells) or Src and Fyn (in fibroblasts) enhances inactive ERK binding to RSK (13, 14). Integrin-mediated adhesion also activates RSK2 (15). Thus, RSKs are well positioned to be primary organizers of directed migration. RSKs coordinate migration by effecting changes in integrin-mediated cell adhesion through phosphorylation of filamin A (15) and through changes in transcription (16). RSKs also dramatically influence cytoskeleton dynamics although the mechanism is not fully understood.
Rho family GTPases are key regulators of cytoskeleton dynamics and control many cellular processes, including cell polarity and migration (17). While somatic mutations in Rho family GTPases are rare in human cancers, aberrant Rho signaling contributes to cancer cell proliferation, invasion, and metastasis (18). Importantly, activating mutations of various Rho guanine nucleotide exchange factors (GEFs) are found in GBM patients (18). Activity of Rho GTPases is well orchestrated during cell migration but significant gaps remain in our understanding of how the Rho GTPases are regulated in response to growth factors (19, 20). Given that RSKs and Rho GTPases regulate cell migration and influence the cytoskeleton in GBM, we examined the interplay between RSK2 and the Rho GTPases in migration and invasion of GBM cell lines. We report a unique RSK2 signaling pathway in which RSK2 forms a complex with LARG (ARHGEF12) and Rho GTPases and directly phosphorylates LARG, thereby activating Rho GTPases and driving directed cell migration and invasion.
Results
RSK Signaling Is Required for RhoA Activation by Diverse Stimuli.
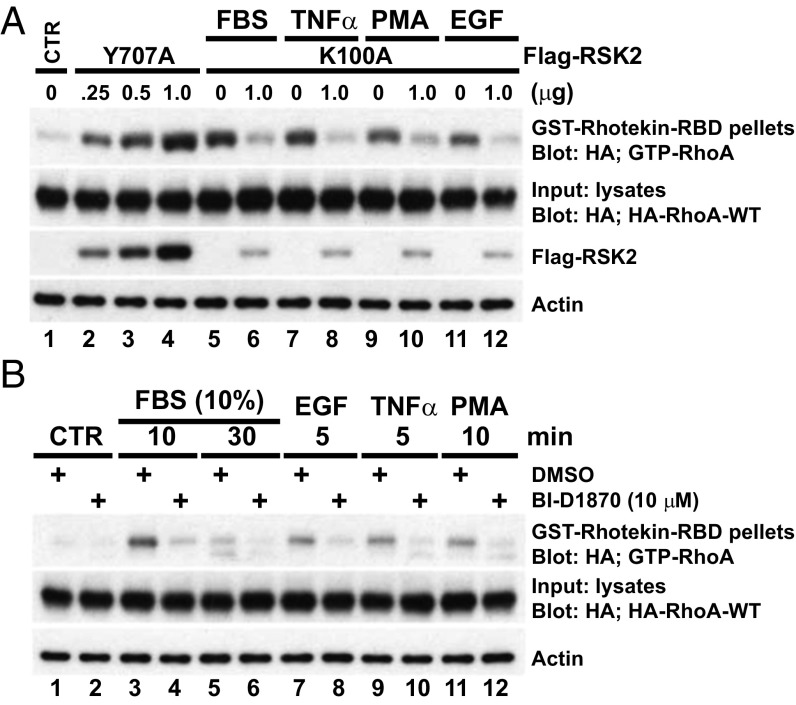
RSK2 drives migration and promotes metastasis of many tumor cell types, including GBM (3, 4). Previously we identified significant effects of RSK2 on the organization of the actin cytoskeleton and actin binding proteins such as filamin A in response to growth factors such as epidermal growth factor (EGF) (15). Rho GTPases regulate the actin cytoskeleton architecture and are well-established regulators of cell mobility and cancer invasion. We therefore determined the ability of RSK2 to activate Rho GTPases. GTP-bound RhoA was retrieved using GST-rhotekin-RBD as a probe. We used an active RSK2-Y707A mutant in which tyr707 in an inhibitory loop was mutated to alanine, which creates an active conformational change (21). Whereas empty vector expression had low levels of RhoA-GTP in serum-starved cells, expression of active RSK2-Y707A induced a dose-dependent increase of RhoA-GTP levels (Fig. 1A, lanes 2–4). In controls, RhoA expression levels were equal between samples. Importantly, expression of kinase dead RSK2-K100A blocked the increase of GTP-RhoA levels triggered by stimulation with FBS (10%, 10 min), tumor necrosis factor (TNFα, 50 ng/mL, 5 min), phorbol 12-myristate 13-acetate (PMA, 100 ng/mL, 10 min) or EGF (100 ng/mL, 5 min) (Fig. 1A, lanes 5–12), indicating that RSK2 activity is essential for these diverse extracellular signals to induce RhoA activation. Similarly, pretreatment with the RSK kinase-specific pharmacological inhibitor, BI-D1870 (10 μM, 30 min), prevented FBS, EGF, TNFα, and PMA-initiated GTP-RhoA activity (Fig. 1B). FBS and EGF activate RSK2 in these cells (Fig. S1). These data support an important, previously unknown role for RSK2 in mediating activation of RhoA in response to various promotility extracellular stimuli that activate the ERK MAP kinase pathway.
Fig. 1.
RSK2 activity is required for RhoA activation in response to diverse stimuli. (A) U87MG cells expressing HA-RhoA-WT were transfected with Flag-RSK2-Y707A vector or dominant negative (DN) Flag-RSK2-K100A. Flag-EV transfection was used as control. Cells were serum starved for 24 h before stimulation with FBS (10%, 10 min), TNFα (50 ng/mL, 5 min), PMA (100 ng/mL, 10 min), or EGF (100 ng/mL, 5 min). Five hundred micrograms of total cell lysates was subjected to GST-rhotekin-RBD precipitation. The levels of GTP-RhoA were determined by immunoblotting with anti-HA antibody. Equal loading of HA-RhoA proteins and expression levels of transfected Flag-RSK2 proteins are shown. Results are representative of seven independent experiments. (B) U87MG cells expressing HA-RhoA-WT were serum starved and then stimulated as indicated after pretreatment with BI-D1870 (10 μM). GTP-RhoA was detected by rhotekin assay as described above. Results are representative of four independent experiments.
RSK2 Associates with Rho Isoforms, but Not Rac1 or Cdc42.
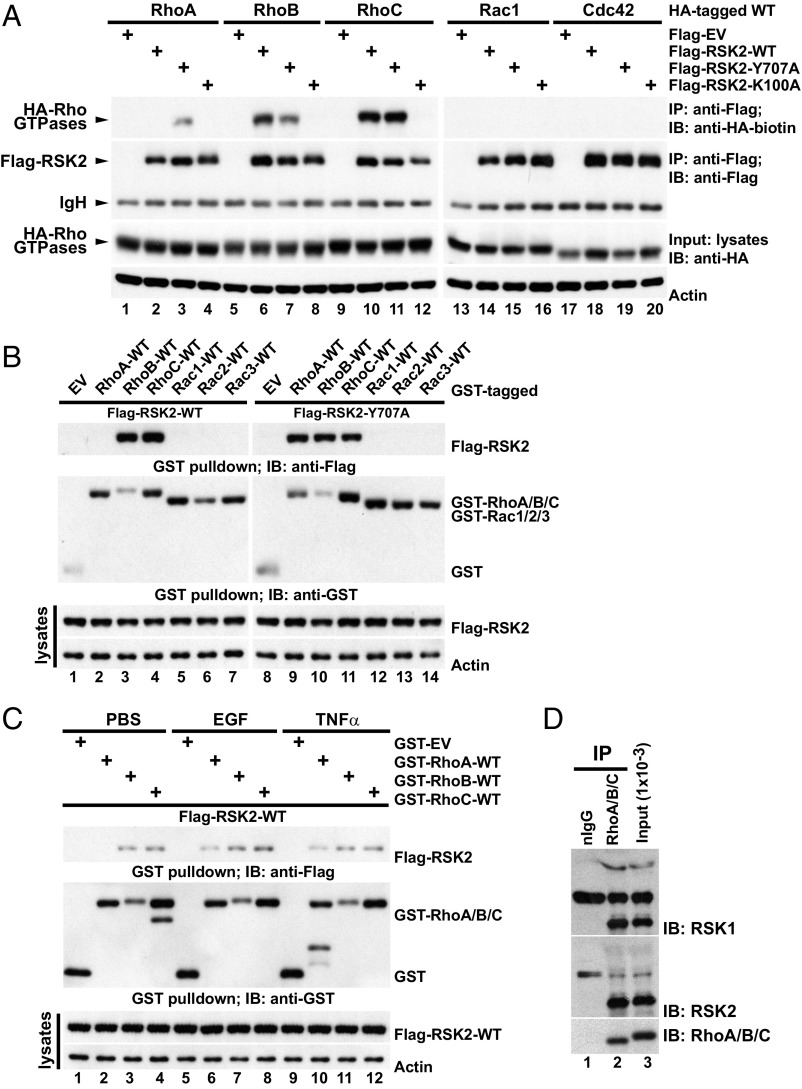
To define how RSK2 influences RhoA activity, we determined whether RSK2 associates in a complex with Rho GTPases in a GBM cell line, U87MG, in which RSK2 mediates migration (4). In serum-starved cells, active RSK2-Y707A immunoprecipitated all three Rho isoforms (Fig. 2A), whereas WT-RSK2 bound to both RhoB (lane 6) and RhoC (lane 10), but not RhoA (lane 2). Moreover, a dominant negative (DN) RSK2 bearing a K100A mutation that creates an inhibitory kinase domain conformation failed to bind to these Rho isoforms (Fig. 2A, lanes 5, 9, and 12). The interaction was further validated by a complementary directed GST pulldown (Fig. 2B) in which GST-WT-RhoA, -B, or -C successfully coprecipitated active RSK2-Y707A, whereas RSK2-WT was precipitated only by GST-RhoB and -C.
Fig. 2.
RSK2 forms a complex with RhoA GTPases. (A) U87MG cells transfected with the indicated Flag-RSK2 and wild-type (WT) HA-Rho GTPase constructs and serum starved. Lysates were subjected to anti–Flag-RSK2 immunoprecipitation (IP) and the coprecipitated HA-tagged Rho GTPases were detected by immunoblotting with biotinylated anti-HA antibody. The IP efficiency of Flag-RSK2 proteins and the equal loading of HA-Rho GTPases were determined. A representative of seven independent experiments is shown. (B) U87MG cells expressing GST-tagged WT-Rho GTPases together with Flag-RSK2-WT or Flag-RSK2-Y707A and serum starved. GST-Rho GTPases were recovered by GST pulldown, and coprecipitated Flag-RSK2 proteins were detected by immunoblotting with anti-Flag antibody. The precipitated GST-fused protein and equal loading of the Flag-RSK2 proteins are shown. The results shown are representative of three independent experiments. (C) U87MG cells were transfected with Flag-RSK2-WT and the indicated GST-Rho constructs and serum starved before stimulation with EGF (100 ng/mL, 5 min) or TNFα (50 ng/mL, 15 min). Lysates were subjected to GST pulldown. Coprecipitated Flag-RSK2 proteins were detected by immunoblotting with anti-Flag monoclonal antibody, while recovered GST-Rho proteins were determined by anti-GST immunoblotting. A representative of four independent experiments is shown. (D) U87MG cell lysate was subjected to immunoprecipitation with anti-RhoA/B/C antibody and bound proteins were recovered by incubation with protein-G Sepharose. The presence of coprecipitated RSK1/2 was determined by immunoblotting with RSK1 or -2 antibodies. The input levels of RSK1/2 and the immunoprecipitated Rho proteins are shown. nIgG, normal rabbit control IgG. A representative of four independent experiments is shown.
These data suggested that RSK2 may constitutively associate with RhoB and -C, whereas the association of RSK2 and RhoA appears to be activation dependent. Indeed, stimulation of the starved U87MG cells with EGF (100 ng/mL, 5 min) or TNFα (50 ng/mL, 15 min) was required for the formation of a RSK2-RhoA complex (Fig. 2C, lanes 6 and 10 vs. lane 2). Interestingly, different forms of RSK2 all failed to precipitate Rac1 and Cdc42 (Fig. 2A). Conversely, RSK2 proteins were not detectable in Rac1, -2, or -3 pulldowns (Fig. 2B). We further found that in serum-stimulated cells, endogenous RSK1 and RSK2 proteins were retrieved by immunoprecipitation using a RhoA/B/C-specific antibody (Fig. 2D), further verifying an interaction between RSK2 and Rho under physiological conditions.
Rho Is Required for RSK2-Mediated Cell Migration and Invasion.
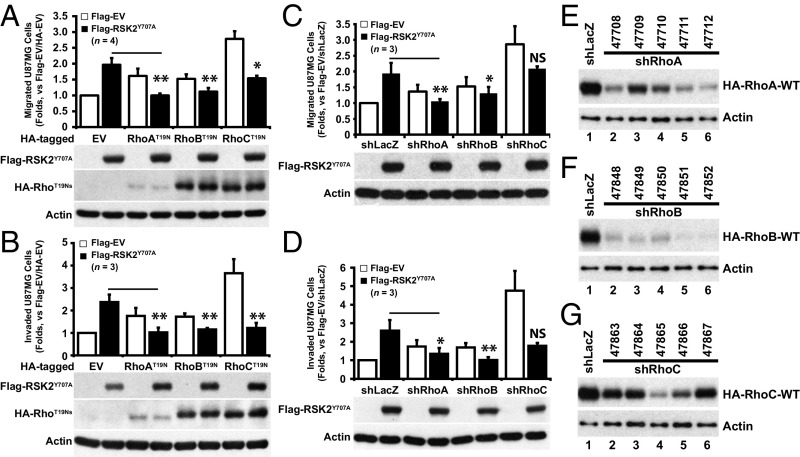
To examine whether Rho signaling is required for RSK2-mediated cell migration and invasion, Rho signaling was inhibited in U87MG cells by expression of DN-Rho-T19Ns or Rho silencing by shRNA. Expression of active RSK2-Y707A promoted cell migration (Fig. 3A) and invasion (Fig. 3B). This was also true to a lesser extent of expression of DN-RhoA-T19N and DN-RhoB-T19N. Interestingly DN-RhoC-T19N alone promoted extensive migration and invasion (Fig. 3 A and B). RSK2-Y707A–induced migration and invasion was significantly inhibited by DN-RhoA and DN-RhoB but not to the same extent by DN-RhoC (Fig. 3 A and B). Active RSK2-Y707A combined with the blockade of Rho signaling disrupted RhoC-mediated cell migration and invasion under the same experimental conditions. The modest increase in motility due to expression of the DN-RhoA/B constructs is unexpected and may be due to a number of effects, including titration of upstream activators from parallel pathways (such as Rac and Cdc42). Moreover, RhoC appears to have a distinct role.
Fig. 3.
RSK2-stimulated cell migration requires Rho GTPase activity. (A and B) U87MG cells were transfected with Flag-EV or Flag-RSK2-Y707A together with HA-tagged DN-Rho isoforms or HA-EV and subjected to G418 selection and serum starved (24 h) before Transwell migration (A) or Matrigel invasion (B) assays. Results are presented as mean ± SD of four independent experiments each performed in triplicate. (C and D) U87MG cells infected with indicated rLenti-shRNA against different Rho isoforms were transfected with Flag-RSK2-Y707A. shLacZ infection and Flag-EV transfection were used as controls. Cells were subjected to G418 and puromycin selection and serum starved before Transwell assays. The results are presented as mean ± SD of three independent experiments performed in triplicate. (E–G) Lentivirus shRNA-mediated Rho silencing. U87MG cells expressing human WT-HA-Rho isoforms were transduced with the indicated lenti-shRNA against human RhoA (E), RhoB (F), or RhoC (G). Cells were subjected to puromycin selection before lysate preparation, and the levels of HA-Rho isoforms were detected by anti-HA immunoblotting. Actin was used as a loading control. For all figures, error bars represent SD. Statistical significance between the indicated sample versus EV+RSK2Y707A is *P < 0.05, **P < 0.01, or NS (not significant, P ≥ 0.05). Refer to Dataset S1 A and B for all P values.
To eliminate potential off-target effects of DN-Rho expression, individual Rho isoforms were knocked down using recombinant lentivirus containing shRNAs directed against human RhoA (Fig. 3E), RhoB (Fig. 3F), or RhoC (Fig. 3G). Knockdown of RhoA and RhoB was effective, while knockdown of RhoC was modest (Fig. 3 E–G). Loss of RhoA, -B, or -C promoted increased cell migration and invasion with RhoC knockdown showing the biggest increase (Fig. 3 C and D). Silencing of RhoA and RhoB resulted in a significant inhibition of active RSK2-mediated cell migration and invasion, whereas the limited RhoC silencing did not (Fig. 3 C and D). We further found that RSK2 colocalized with each Rho GTPase as determined by immunofluorescence of FBS-treated cells (Fig. S2). Taken together, these results establish a key yet unsuspected role of RSK2-RhoA/B signaling in cell migration and invasion. They also reveal that RSK2 activity may have differing effects on RhoA, -B, and -C.
Mapping the RSK2 Sequences Required for Complex Formation with Rho GTPases.
The RSK2-RhoA association is dependent on the activation status of RSK2 (Fig. 2). The full activation of RSK2 requires sequential phosphorylation at several serine and threonine sites by the coordinated activity of a number of protein kinases (7). To map RSK2 sequences involved in the formation of the RSK2-Rho complex, we generated RSK2-NTKD and -CTKD truncations (Fig. S3A) and a series of phosphorylation mimetic (to glutamic acid, E) or deficit (to alanine, A) mutants at the residues critical for full RSK2 activation (Fig. S3B). RSK2-WT and -Y707 but not -K100A mutants were detected in the GST-RhoA pulldown, while the NTKD also associated with RhoA, whereas CTKD and CTKD-Y707A did not (Fig. S3 C and D). Phosphomimetic mutants including RSK2-T577E, -S386E, and -S227E associated with RhoA but the phospho-deficit mutants RSK2-T577A, -S386A, and -S227A did not (Fig. S3 C and D). Moreover, dual T365A/S369A mutations eliminated the interaction (Fig. S3C, lane 6). However, the single alanine mutation at threonine 365 (T365A) did not disrupt the association (Fig. S3C, lane 4), whereas the S369A resulted only in a reduction of the association (Fig. S3C, lane 5). These findings support the hypothesis that fully activated RSK2 associates with RhoA, potentially at a site in the linker region of the NTKD truncation.
RSK2-RhoA Signaling in Cell Migration and Invasion Is Dependent on RSK2-T577.
We next determined the ability of the phosphomimetic mutants to induce RhoA activation. Expression of RSK2-S386E (Fig. S3E, lanes 5–7) and RSK2-T577E (lanes 8–10) significantly activated a dose-dependent increase of GTP-RhoA in a manner similar to RSK2-Y707A (lanes 11 and 12). While S227 phosphorylation is critical for full RSK2 activation, the RSK2-S227E failed to activate RhoA (lanes 2–4), suggesting additional phosphorylation events may be necessary for RSK2 to activate RhoA. Control cells (Flag-EV) maintained low levels of GTP-RhoA and RhoA protein loading was equal (Fig. S3E), indicating that the change in GTP-RhoA activity was not a consequence of changes in RhoA expression.
The ability of these mutants to drive cell migration and invasion was also examined. Surprisingly, the RSK2-NTKD fragment significantly induced tumor cell migration and invasion similar to the mutant RSK2-Y707A, whereas RSK2-WT, RSK2-S227E, and RSK2-CTKD did not (Fig. S3 F and G). The RSK2-T577E mutant promoted increased cell migration and invasion, in a fashion significantly exceeding the activity of either RSK2-Y707A or -NTKD mutants. Unlike RSK2-Y707A, this unexpected invasive behavior of T577E mutation was impaired by DN-RhoA and DN-RhoB, but not DN-RhoC (Fig. S4 A and B). The differences between these two RSK2 mutants may be attributed to their ability to promote selective signaling or due to differences in the magnitude of endogenous Rho activation. Indeed, RSK2-T577E predominantly interacts with RhoA, while RSK2-Y707A interacts with both RhoA and -C (Fig. S4C). Taken together, these results suggest that phosphorylation of RSK2 T577 plays a dominant role in inducing invasion by RSK2 and reveal the RSK2-T577E mutant as a potent activated mutant of RSK2 that mimics endogenous RSK2 activity.
RSK2 Binds Directly to Active Nucleotide-Bound Rho GTPases.
Initial in vitro interaction analysis indicated that RSK2 did not directly interact with naïve nucleotide-free Rho GTPases (Fig. S5). Rho GTPases naturally exist in both nucleotide-free (inactive) and nucleotide-bound (active) states and our results indicated the activation-dependent association of RSK2 and RhoA (Fig. 2). To test the possibility that RSK2 preferentially interacts with nucleotide-bound Rho GTPases, recombinant MBP-His6-Rho GTPases were preloaded with either GDP or nonhydrolyzable GTPγS and subsequently subjected to recombinant GST-RSK2 pulldown (Fig. S6). RSK2 captured both GTPγS- and GDP-bound forms of RhoB or RhoC with a similar affinity (Fig. S6, Middle two panels), whereas RSK2 preferentially interacted with GTPγS-loaded RhoA (Fig. S6, Upper). In contrast, activated Rac1 failed to interact with RSK2 (Fig. S6, Lower). These interactions were specific, since GST alone did not precipitate any of these Rho proteins. Surprisingly, RSK2-K100A also retained similar affinity in the interaction (Fig. S6). It is possible that the recombinant RSK2-K100A may not fold correctly in vitro to form the inhibitory conformation that forms in vivo.
The phosphorylation of Rho GTPases allows for various regulatory roles, including the disassociation from RhoGDI (22, 23), and contributes to their cellular functions, including tumor cell migration and invasion (22, 23). Both RhoA and RhoC possess potential Ser73 phosphorylation sites within evolutionarily conserved RSK consensus sequences (RxRxxS/T): RhoA, RLRPL73SYPDTD and RhoC, RLRPL73SYPQTD. In our hands, RSK2 failed to directly phosphorylate either RhoA or RhoC (Fig. S7).
RSK2 Phosphorylates LARG and Induces LARG-Dependent RhoA Activation.
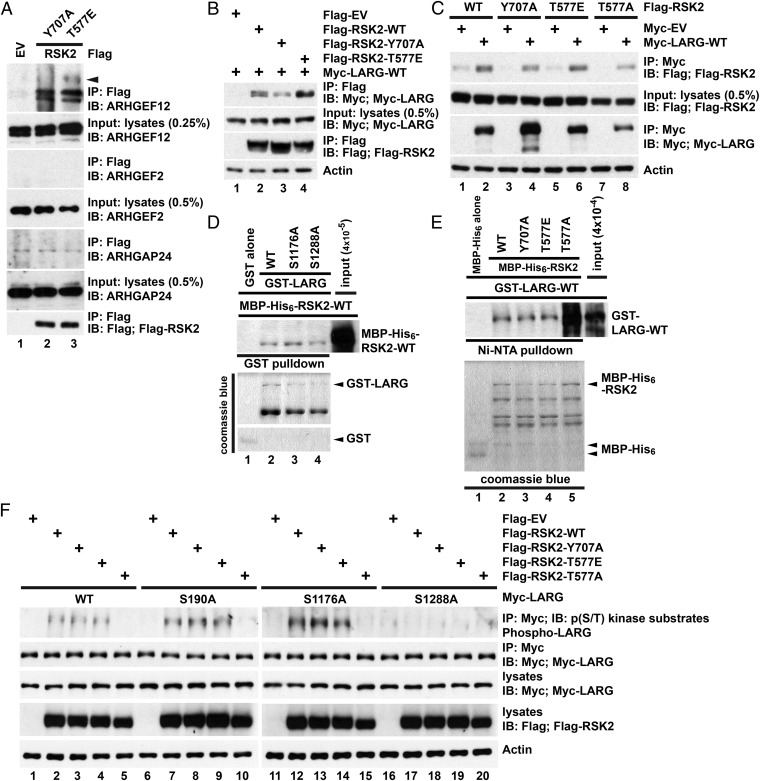
We further determined whether RhoGEFs and/or RhoGAPs were in the RSK2-RhoA complex. An array of potential RSK substrates were identified via data mining and included ARHGEF12 (LARG), ARHGEF2, and ARHGAP24. We therefore investigated if any of these Rho regulators associated with the RSK2-Rho complexes. We found that both active RSK2-Y707A and RSK2-T577E precipitated endogenous LARG, but not ARHGEF2 or ARHGAP24 (Fig. 4A). This interaction was confirmed by coimmunoprecipitation using expressed Flag-RSK2 and Myc-LARG proteins (Fig. 4 B and C). Interestingly, the phospho-deficient T577A mutation retained the ability to bind LARG (Fig. 4C, lane 8), although it completely abolished the RSK2 association with RhoA (Fig. S3C). Furthermore, the interaction appears to be direct since in vitro interaction analysis indicated a similar pattern in the interaction between different forms of purified RSK2 and LARG (Fig. 4 D and E). Interestingly, we also found a robust interaction between pure LARG-WT and RSK2-T577A (Fig. 4E, lane 5).
Fig. 4.
RSK2 forms a signaling complex composed of Rho GTPases and the RhoGEF, LARG. (A) Active RSK2 precipitates endogenous LARG. A total of 10 mg of U87MG cell lysates with overexpressed Flag-RSK2-Y707A or Flag-RSK2-T577E was incubated with anti-Flag antibody at 4 °C overnight and bound proteins were detected by immunoblotting with indicated antibodies. The arrow indicates possible phosphorylated LARG. (B and C) Association of RSK2 and LARG. U87MG cells were transfected with indicated Flag- or Myc-tagged constructs and serum starved for 24 h before lysate preparation. Flag (C)- or Myc (D)-tagged proteins were immunoprecipitated by anti-Flag or -Myc antibodies, respectively. Bound proteins were detected by immunoblotting with anti-Flag (RSK2) or -Myc (LARG) antibodies. Results are representative of three independent experiments. (D and E) RSK2 interacts directly with LARG. GST-LARG proteins and MBP-His6-RSK2-WT were incubated with either glutathione Sepharose (D) or Ni-NTA His-Bind resin (E). Bound MBP-His6-RSK2-WT (D) or GST-LARG (E) was determined by immunoblotting. (F) RSK2 phosphorylates LARG on Ser1288 residue. U87MG cells were cotransfected with Flag-RSK2 constructs and the indicated Myc-LARG constructs. The Myc-LARG proteins were immunoprecipitated after serum starvation for 24 h. LARG phosphorylation was determined by immunoblotting anti-Myc immunoprecipitates with a phospho-AKT substrate-specific antibody. Results are representative of three independent experiments.
The activation of LARG is regulated by phosphorylation (24). There is a series of potential phosphorylation sites within LARG, including the Cdk1 substrates of Ser190 and Ser1176 (24), and a RSK2 consensus site at Ser1288: FPRYRTA1288SQGPQTDS. We found that expression of active RSK2-Y707A or -T577E mutants was sufficient to induce ligand-independent LARG phosphorylation (Fig. 4F, lanes 2–4), while expression of phospho-deficient RSK2-T577A mutant was not (Fig. 4F, lane 5). We next examined whether RSK2 phosphorylated one of the candidate sequences in LARG. LARG phosphorylation was retained in LARG protein bearing S190A (Fig. 4F, lanes 7–9) or S1176A (Fig. 4F, lanes 12–14) mutations, whereas the S1288A mutation completely abrogated LARG phosphorylation by RSK2 (Fig. 4F, lanes 17–19). Note that the S1288A mutation had only a modest effect on binding of active RSK2 to LARG complexes (Fig. S8). The S190A and S1176A mutations also maintained interaction with RSK2 (Fig. S8). These data define LARG as a functional substrate of RSK2 with RSK2-mediated phosphorylation occurring at S1288.
FBS and EGF Stimulation Promote Endogenous RSK2 Binding to LARG and RSK2-Dependent Binding of LARG to RhoA.
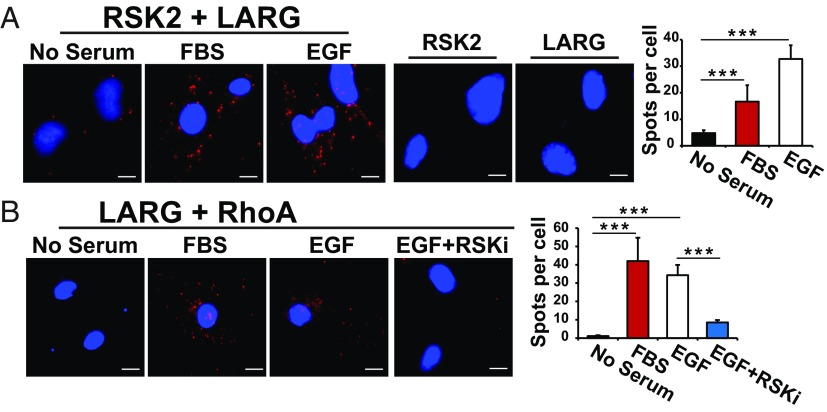
To determine whether FBS and EGF stimulation of cells influences the interaction of endogenous RSK2, LARG, and RhoA proteins, we utilized the proximity ligation assay. This is a method by which you can detect, visualize, and quantify endogenous protein–protein interactions within the cell. Using this approach, we found that both FBS and EGF stimulation of U373MG cells induced the formation of an RSK2-LARG complex in the cytoplasm (Fig. 5A). Minimal association of RSK2 and LARG was seen in unstimulated serum-starved cells. Similarly, EGF and FBS both induced formation of an endogenous LARG-RhoA complex in the cytoplasm. Importantly, preincubation of the cells with the RSK inhibitor BI-D1870 blocked LARG-RhoA interaction in the cell (Fig. 5B). These data support a model in which EGF or FBS stimulation leads to formation of an endogenous RSK2-LARG-RhoA complex in the cell cytoplasm.
Fig. 5.
Activation of RSK2 by FBS or EGF stimulates endogenous RSK2 binding to LARG and LARG to RhoA. (A) U373MG cells were starved for 24 h and then stimulated with FBS (10%, 10 min) or EGF (100 ng/mL, 5 min). Endogenous RSK2 and LARG protein complex formation was determined by proximity ligation assay as described in Materials and Methods. RSK2 or LARG antibody alone was used as negative controls. Complex formation is indicated where there are distinct red spots. Complex formation in cells from each slide was quantified by ImageJ software and is presented as bar graph. (B) Starved overnight U373MG cells were treated with RSK inhibitor (RSKi, BI-D1870) for 30 min before stimulation. Complex formation is indicated where there are distinct red spots. Complex formation in cells from each slide was quantified by ImageJ software and is presented as bar graph. n = 5. (Scale bar, 5 μM.) ***P < 0.001.
RSK2 Activates RhoA GTPase, Migration, and Invasion Through Effects on LARG.
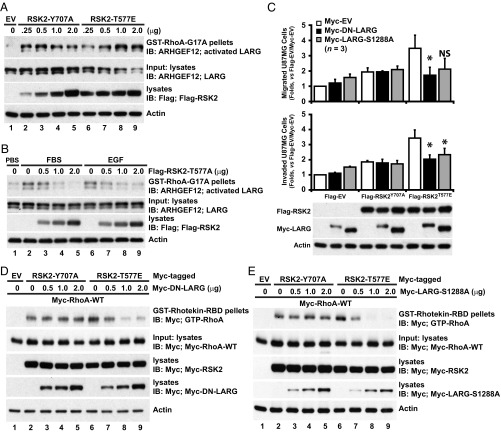
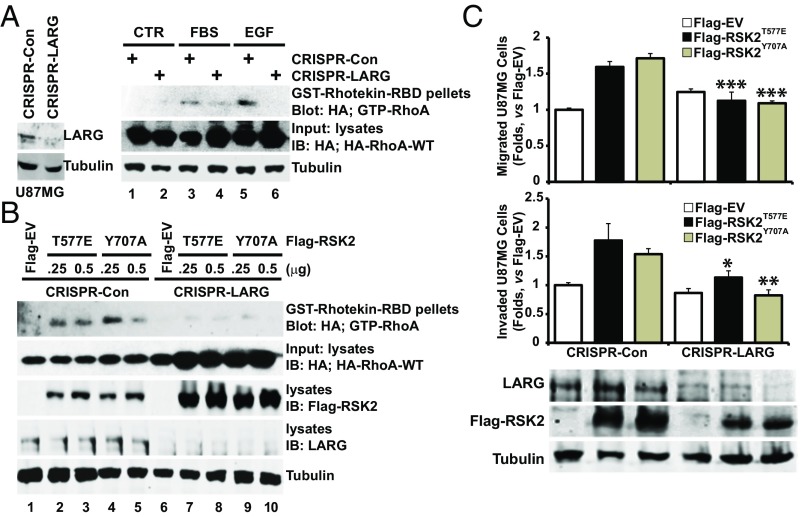
The previous results suggest a mechanism by which RSK2 promotes cellular invasion in response to exogenous signals by phosphorylating and activating LARG, leading to RhoA activation. Therefore, RSK2-T577A and LARG-S1288A are anticipated to act as dominant negative forms that interfere with activation of this signaling cascade. Indeed, we found expression of activated RSK2-Y707A or RSK2-T577E resulted in increased levels of activated LARG (Fig. 6A, lanes 2–9), while FBS or EGF-triggered LARG activation was inhibited by RSK2-T577A in a dose-dependent manner (Fig. 6B). Therefore, we next tested whether LARG activity is required for invasive RSK2-T577E signaling. Expression of either DN-LARG or S1288A-LARG significantly inhibited RSK2-T577E-mediated cell migration and invasion (Fig. 6C). These results indicate LARG can act as a RSK2 RhoGEF for Rho GTPases. Indeed, expression of DN-LARG (Fig. 6D) or LARG-S1288A (Fig. 6E) inhibited RSK2-T577E activation of RhoA. In these assays, the dominant-negative LARG constructs did not alter RSK2-Y707A induced migration and invasion (Fig. 6C) or RhoA activation (Fig. 6 D and E). Because dominant negatives can have nonspecific or incomplete effects, we further investigated whether RSK2 activation of RhoA GTPase requires LARG by using LARG-deleted cell lines developed using CRISPR/Cas9 targeting. We found that both FBS and EGF activation of RhoA was significantly impaired in LARG-deleted cells (Fig. 7A). Furthermore, deletion of LARG also significantly impaired activation of RhoA by both RSK2-Y707A and RSK2-T577E activated forms of RSK2 (Fig. 7B). Moreover, deletion of LARG equivalently impaired induction of migration and invasion by both activated RSK2 mutants (Fig. 7C). Taken together, these results point to a RSK2-LARG-RhoA signaling axis that regulates the invasive behavior of cells in response to environmental signals.
Fig. 6.
RSK2 activation of RhoA signaling in cell migration and invasion requires LARG. (A) RSK2-T577E activates LARG. U87MG cells were transfected with varying amounts of Flag-RSK2-T577E. Flag-EV transfection was used as a negative control. Cells were serum starved before cell lysate preparation. Activated LARG (ARHGEF12) was captured by GST-RhoA-G17A pulldown and detected by immunoblotting with anti-LARG antibody. The levels of LARG and Flag-RSK2 are shown. Results are representative of four independent experiments. (B) U87MG cells transfected with Flag-RSK2-T577A and serum starved were stimulated by FBS (10%, 10 min) or EGF (100 ng/mL, 5 min). Activated LARG was retrieved by GST-RhoA-G17A pulldown and detected by immunoblotting with anti-LARG antibody. Expression of LARG and Flag-RSK2-T577A levels is shown. (C) RSK2-T577E–induced U87MG cell migration and invasion requires LARG activity. U87MG cells were cotransfected with Flag-RSK2-T577E and Myc-LARG-S1288A or -DN-LARG. Flag-EV and Myc-EV transfection was used as controls. Transfected cells were serum starved before migration and invasion assays. Cell migration (Upper) and invasion (Middle) were assessed by Transwell assays. Expression levels of exogenous RSK2 and LARG proteins are shown (Lower). Refer to Dataset S1C for all P values. *P < 0.05. (D and E) Inhibition of LARG activity disrupts RSK2-T577E–triggered RhoA activation. U87MG cells expressing Myc-RhoA-WT were transfected with Myc-RSK2-T577E (1 μg) in the presence or absence of Myc-tagged DN-LARG (D) or LARG-S1288A (E). Cells were starved before lysate preparation and GST-rhotekin-RBD pulldown analysis. GTP-RhoA levels were determined by immunoblotting pulldown precipitate with anti-Myc antibody. Expression levels of Myc-RhoA-WT, Myc-RSK2, and Myc-LARG are determined by immunoblot and shown. Results are representative of three independent experiments.
Fig. 7.
Deletion of LARG impairs RSK2 activation of RhoA and cell migration and invasion. (A and B) LARG was deleted in U87MG cells by CRISPR/Cas9-targeted deletion as described in Materials and Methods. LARG-deleted cells and nonspecific CRISPR/Cas9-treated control cells expressing HA-RhoA-WT were stimulated with FBS (10%, 10 min), EGF (100 ng/mL, 5 min) (A), or transfected with Flag-EV, Flag-RSK2-T577E, or Y707A (B). GTP-RhoA was retrieved using GST-rhotekin-RBD, and GTP-RhoA levels were determined by immunoblotting with anti-HA antibody as described above. Expression levels of HA-RhoA, LARG, and Flag-RSK2 were determined by immunoblotting. (C) RSK2-induced migration and invasion was impaired in LARG-deleted U87MG cells. CRISPR/Cas9-mediated nonspecific control or LARG-deleted cells were transfected with Flag-EV, RSK2-T577E, and Y707A. Transfected cells were enriched by G418 selection and serum starved for 24 h before migration and invasion assays. Cell migration (Upper) and invasion (Middle) were assessed by Transwell assays. Refer to Dataset S1D for all P values. Expression levels of RSK2 and LARG proteins are shown (Lower). *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
RSKs have emerged as central regulators of migration and invasion, however the mechanisms mediating invasive RSK dependent signaling remain incompletely understood. We previously reported a key role for RSK2 in GBM invasion (4) and RSK2 promotes metastasis of various tumor types (3, 25). Here, we present evidence for a signaling axis in which RSK2 activates a LARG-dependent RhoA signaling cascade in cell migration and invasion. The data support a model in which RSK2 directly binds to the RhoGEF LARG (ARHGEF12) in response to EGF or FBS stimulation and phosphorylates it at Ser1288. LARG then binds and activates RhoA GTPase in response to EGF or FBS stimulation in a RSK2-dependent manner. RSK2-mediated phosphorylation of LARG and subsequent activation of RhoA GTPase promote cellular migration and invasion. We have further identified an active phosphomimetic mutation at residue Thr577 of RSK that induces LARG and RhoA GTPase activation and subsequent cell migration and invasion. Thr577 phosphorylation is the initial event leading to the phosphorylation and full activation of RSK2. In addition, neither S386E (required for PDK1 docking) or S227E (critical for NTKD activation) exhibited activity similar to RSK2-T577E in RhoA activation or cell motility. Thr577E phosphorylation and the phosphomimetic may therefore be useful tools to help define the pathophysiological significance of RSK2 in human disease.
RSK2 does not interact with inactive nucleotide-free naïve Rho isoforms (Fig. S5), whereas it directly interacts with active nucleotide-bound Rho isoforms (Fig. S6). The conformational changes upon nucleotide loading to Rho GTPases appear to be necessary for this direct interaction. RSK2 does not possess a functional GEF or GAP domain (7). Therefore, it is likely that RSK2 activates RhoA GTPase via phosphorylation of the Rho-specific RhoGEF LARG, which in turn, facilitates GTP-loading of RhoA, creating a conformation necessary for the formation of the RSK2-LARG-RhoA complex. LARG belongs to a regulator of G protein signaling (RGS) domain-containing RhoGEF family and acts exclusively as a RhoGEF, without activity toward either Rac1 or Cdc42 (26), which is in agreement with our finding that RSK2 directly interacts with Rho GTPases but not Rac1 or Cdc42. Sequences in the RSK2 linker domain including S369 and S386 appear to be essential for RSK2 binding to RhoA GTPases. However, the minimum sequences necessary for the direct interaction between RSK2 and LARG remain unclear. In addition to a Dbl homology (DH) domain (GEF domain) and a Pleckstrin homology domain (PH, RhoA binding), LARG has a N-terminal PDZ domain and a middle RGS domain necessary for coupling to different effectors and/or anchoring to the plasma membrane (27, 28). Interestingly, both the phospho-defective LARG-S1288A mutant and the DH/PH-deletion DN-LARG mutant retained the ability to bind RSK2 (Fig. S8). Currently the exact interaction sequence between RSK2 and LARG remains undefined.
LARG can act as GEF for all three Rho isoforms (26). Whether LARG relays active RSK2 signaling to RhoB or RhoC remains to be investigated. In addition, RSK2 interacts with all three Rho isoforms, however, only RhoA and RhoB are required for the RSK2-mediated cell migration and invasion. Therefore, RSK2 effects on RhoB and RhoC appear to be different from RhoA and thorough investigation of the spatiotemporal effects of RSK2 on each Rho family member is necessary to clarify these differences. RhoA/B/C are highly homologous with only a few amino acid sequence differences (29). Nevertheless, different Rho isoforms have the ability not only to regulate distinct signaling pathways but also to crosstalk among themselves in cell remodeling and migration (17, 20, 30, 31). Several studies have demonstrated spatiotemporal signaling modules that precisely regulate the signaling of distinct Rho GTPases at different subcellular locations where they regulate specific cellular functions by coupling to different effectors (20). Moreover, RhoA activity is also required for cell migration, and extracellular cues may induce a distinct pattern of RhoA activation during this cellular process (20, 32). Thus, it is possible that RSK2 forms a unique Rho signaling node to integrate extracellular signals to selected invasive signaling cascades. A recent report described a phosphorylation-mediated molecular switch required for the spatiotemporal activation of Rac1 and Rho GTPases in EGF- or PDGF-driven cell migration (22). RSK2 may play a similar role through its phosphorylation of LARG.
Aberrant RSK2 signaling has been implicated in the pathogenesis of diverse human diseases including Coffin-Lowry syndrome (33, 34) and cancer (3). Despite the fact that RSK2 somatic mutations are rare in cancers, hyperactive RSK2 signaling is common in many types of cancer (35). Indeed, GBM patients who have high RSK2 signaling have a poor prognosis with lower survival rate and shorter life span after diagnosis (4). RTK/Ras signaling-mediated activation of both Raf/MEK/ERK and PI3K/AKT contribute to the invasiveness of GBM (36, 37). RSK2 can act as a downstream effector that integrates these two signaling cascades (3). In addition, hyperactivaton of RhoGEFs and RhoGTPases can promote invasion of various tumors (38). Recently, strategies designed to interfere with the interaction of LARG or RhoA have shown effects in inhibiting tumor transformation (39, 40). Our findings establish a specific role for RSK2 in controlling LARG-RhoA signaling in response to diverse invasive signals. Thus, targeting this unique RSK2-dependent signaling may provide additional benefits to those GBM patients who have aberrant RSK2 signaling. Inhibition of RSK signaling has shown promising effects in several preclinical models (3). Future studies are necessary to determine the therapeutic value of targeting RSKs alone or in combination with targeting LARG/Rho or chemo/radiotherapies in GBM. In conclusion, our results identify an unanticipated RSK2-LARG-RhoA signal transduction pathway that regulates cancer cell motility and may provide therapeutic possibilities.
Materials and Methods
Cell Culture and Transfections.
Human glioblastoma U87MG is a commonly studied grade IV glioma cell line (41). Previously, we have successfully established the U87MG cell line as an reliable cellular model for the elucidation of RSK2-mediated function in cell motility. U87MG cells were obtained from the National Cancer Institute’s Tumor Repository. Human embryonic kidney HEK293T cells were obtained from American Type Culture Collection. All cells were maintained in DMEM supplemented with 10% (vol/vol) FBS (Life Technologies), 100 units/mL penicillin, and 100 µg/mL streptomycin. Except indicated otherwise, U87MG cells were seeded at 5 × 104/cm2, while HEK293T cells were seeded at 1 × 105/cm2 in six-well plates for all experiments involving DNA transfection. U87MG cells were transfected by either Lipofectamine 2000 DNA transfection reagent (Life Technologies) or Effectene DNA transfection reagent (Qiagen) following the manufacturer’s instructions. HEK293T cells were transfected with calcium phosphate following standard protocol described elsewhere. The cells were incubated with DNA complex overnight (24 h) before being cultured in complete medium. All cells were tested for mycoplasma (LT07-188, Lonza).
Generation of LARG Knockout Cells Using CRISPR/Cas9 Targeting.
U87MG cells were seeded in six-well plates and transfected with 1 μg of LARG (ARHGEF12) CRISPR/Cas9 knockout plasmid (sc-418063, Santa Cruz) or control CRISPR/Cas9 knockout plasmid with GeneJuice (70967, Millipore). At 48 h posttransfection, individual GFP positive cells were isolated by the use of a FACS-Aria cell sorter (Becton Dickinson) and seeded into a 96-well plate. The cell clones were expanded and clones in which LARG was deleted were confirmed by immunoblot (IB) analysis.
In Vivo Protein–Protein Association Analysis.
The intracellular protein–protein association was analyzed using either GST protein pulldown or immunoprecipitation (IP). U87MG cells were transfected with different epitope-tagged RSK2, LARG, and Rho family of GTPases. The cells were further incubated for 48 h to allow maximum protein expression before being applied to different treatments as indicated in the text. Detergent-soluble cell lysates were prepared using kinase lysis buffer and 1–2 mg of total cell lysates were subjected to either GST pulldown using 20 μL of glutathione-coupled Sepharose (GE Healthcare), or immunoprecipitation using 2 μg of antibody together with 20 μL of protein-G Sepharose (GE Healthcare) in a final volume of 1 mL. The reaction was incubated at 4 °C for 1 or 2 h by end-to-end rotation and the resin was recovered by brief centrifugation at 4 °C, 500 × g for 5 min. The resin was then extensively washed with kinase lysis buffer once, kinase lysis buffer supplemented with 0.5 M NaCl twice, and kinase lysis buffer twice. The bound proteins were released by boiling in 1× Laemmli loading buffer for 5 min and then fractionated by SDS/PAGE. The presence of individual proteins was determined by immunoblotting with antibody specific to the epitope tag that was fused to the protein of interest, while the expression levels of protein of interest were also determined.
In Situ Proximity Ligation Assay.
Duolink proximity ligation assays were performed according to the manufacturer’s instructions (MilliporeSigma). Briefly, U373MG cells were starved overnight and treated with either 10% FBS or 100 ng/mL EGF for 10 min. Cells were then fixed in 3.7% formaldehyde for 15 min at room temperature (RT) and washed with PBS. Cells were permeabilized with 0.05% Triton X-100 for 10 min at RT. Cells were blocked by Duolink in situ blocking solution for 1 h at 37 °C and then incubated overnight with LARG (22441-1-AP, Proteintech) and RSK2 (sc-9986, Santa Cruz Biotechnology) or LARG and RhoA (sc-418, Santa Cruz Biotechnology) antibodies. Detection was performed according to the manufacturer’s protocol. Briefly, probes for anti-mouse and anti-rabbit were incubated for 1 h at 37 °C and then washed twice with Duolink wash buffer A. Ligation solution was added to each sample for 30 min at 37 °C and washed twice with Duolink wash buffer A. Amplification solution was added overnight at 37 °C in a dark container. Samples were washed with Duolink wash buffer B two times and mounted with DAPI. The red dot images were taken under ZEISS Axiovert 200 M microscope. Quantification was performed using ImageJ software.
Determination of RhoA and LARG Activation.
A GST-fused RhoA-binding domain (RBD) of Rhotekin protein in pGEX-2T vector (pGEX-2T-RBD) was purchased from Addgene as probe for the pulldown of GTP-RhoA, while a GST-RhoA-G17A in pGEX-4T1 vector was generated by mutagenesis as probe to retrieve activated LARG (42). The recombinant GST-fused proteins were expressed in Escherichia coli strain BL21-DE3 (lysE) as described in the “recombinant protein expression section,” and then extracted and coupled to glutathione-agarose (Sigma) resin following the protocol described for the preparation of GST-Raf-RBD–coupled glutathione resin (43). U87MG cells were manipulated using indicated cotransfection, pretreatment with kinase inhibitor, and/or extracellular stimuli before the activation analysis. Cells were serum starved in serum-free DMEM for 24 h before treatment and/or cell lysates preparation. Cell monolayers were washed with ice-cold PBS once and cell lysates prepared using GST-pulldown lysis buffer (GPLB) [Hepes (pH 7.4), 20 mM; KCl, 100 mM; glycerol, 10%; DTT, 1 mM; PMSF, 1 mM; Triton X-100, 0.5%]. Five hundred micrograms of total cell lysates was then incubated with glutathione agarose beads containing approximately 10 μg of GST-fused probes in a total volume of 1 mL with end-to-end rotation at 4 °C for 1 h. The resin was recovered by centrifugation (500 × g, 5 min) at 4 °C and extensively washed sequentially with GPLB once, GPLB supplemented with 1 M NaCl twice, and finally with GPLB twice. Bound activated RhoA or LARG was released by boiling in 1× Laemmli sample loading buffer for 5 min and detected by immunoblotting with anti-HA, -Myc (RhoA), or -ARHGEF12 (LARG) antibodies. The equal loading of overexpressed HA- or Myc-tagged RhoA-WT, as well as endogenous LARG proteins, was determined by immunoblotting total cell lysates. To determine the ability of RSK2 to induce RhoA activation, U87MG cells expressing HA-RhoA-WT were transfected with different amounts of plasmid constructs containing Flag-tagged RSK2-Y707A, -S227E, -S386E, or -T577E mutant. To define the requirement of RSK2 signaling for extracellular stimulus-induced RhoA activation, U87MG cells were either transfected with Flag-tagged DN-RSK2 (Flag-RSK2-K100A, 1.0 μg) or pretreated with RSK inhibitor, BI-D1870 (10 μM, 30 min). The cells were serum starved for 24 h in the presence of BI-D1870 (10 µM) or not and stimulated with FBS (10%), TNFα (50 ng/mL), PMA (100 ng/mL), or EGF (100 ng/mL) for the indicated duration. DMSO vehicle pretreatment or Flag-EV transfection was used as controls. To determine the ability of RSK2 to activate LARG, U87MG cells were transfected with different amounts of Flag-RSK2-Y707A or -T577E constructs. To evaluate the requirement of RSK2-T577 phosphorylation for ligand-induced LARG activation, U87MG cells were transfected with increased amounts of Flag-RSK2-T577A as indicated and serum starved for 16 h before being stimulated with FBS (10%, 10 min) or EGF (100 ng/mL, 5 min). To determine the contribution of LARG to RSK2-induced RhoA activation, U87MG cells expressing Myc-RhoA-WT were transfected with Myc-EV, Myc-LARG-DN, or Myc-LARG-S1288A in the presence of either Myc-RSK2-Y707A or Myc-RSK2-T577E as indicated, and serum starved for 16 h before lysate preparation.
Cell Migration and Invasion Assays.
Analysis of cell migration and invasion was done using Transwell permeable supports or Matrigel matrix-coated supports with 8 μm pore polyester membrane inserts (BD-Costar) according to the manufacturer’s instructions. Briefly, U87MG cells, manipulated as described in the text, were serum starved for 24 h before being dislodged by pipetting. Single cell suspensions were then prepared and adjusted to 1 × 106 cells per milliliter using serum-free DMEM. A total of 0.2 mL of cell suspension was added into the top Transwell inserts. Triplicated inserts were used for each group and three separate experiments were performed to gain significant statistical difference. For the migration assays, inserts were placed into a well in 24-well plates with the low chamber containing 0.5 mL of serum-free DMEM and further incubated for 6 h in a 37 °C incubator with 5% CO2 humidified air. For the invasion assays, the inserts were inserted into a well in 24-well plates with the low chamber containing 0.5 mL of DMEM supplemented with 10% of FBS and further incubated for 24 h in a 37 °C incubator with 5% CO2. Migrated/invaded cells were collected and analyzed using a BD-Beckman cell counter with each sample being counted twice, while total cell number in the cell suspension was also counted and calculated. The percent of migrated/invaded cells was calculated by dividing the numbers of migrated/invaded cells by the total cell number being loaded. The fold induction was then calculated by dividing the percent of migrated/invaded cells in testing samples with that in the negative control sample. To determine the effects of DN-Rho isoforms on RSK2-Y707A or -T577E mutant-induced cell migration and invasion, U87MG cells were transiently transfected with 2 μg of Flag-RSK2-Y707A or Flag-RSK2-T577E in presence or absence of HA-RhoA-T19N, HA-RhoB-T19N, or HA-RhoC-T19N (1 μg) and the transfected cells were enriched by G418 selection (400 μg/mL, VWR) for 24 h before starvation. Flag-EV and HA-EV transfection were used as controls. To examine the requirement of Rho signaling in RSK2-mediated cell migration and invasion, U87MG cells transiently transfected with Flag-RSK2-Y707A (2 μg) were transduced with rLenti-shRNA against human RhoA, -B, or -C. Flag-EV transfection and shLacZ lentivirus transduction were used as controls. The cells were then subjected to G418 (400 μg/mL) and puromycin (2 μg/mL) selection for 24 h before starvation. To determine the ability of different RSK2 mutations or truncations to induce cell migration and invasion, U87MG cells were transiently transfected with 2 μg of Flag-RSK2 mutants and selected by G418. To determine the contribution of LARG activity to RSK2-induced cell migration and invasion, U87MG cells expressing Flag-EV, Flag-RSK2-Y707A, or -T577E (2 μg) were transfected with Myc-EV, Myc-LARG-DN, or Myc-LARG-S1288A (1 µg) and the transfected cells were selected by incubation with G418 (400 μg/mL).
Statistical Analysis.
To ensure the reproducibility of results, all experiments were performed independently three to seven times as indicated in the figure legends. These are biological replicates. For migration and invasion analysis, three separate Transwell inserts were used for each testing sample in individual experiments. To reduce potential error, the cell number from each well was counted three times to acquire the average of cell counts. Three to four technical replicates were performed per analysis. The statistical significance of differences between biological replicates was calculated using an unpaired Student’s t test and all P values are shown in Dataset S1.
Additional information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Junfang Ji for helpful discussions. This work was supported by grants from the National Institute of General Medicine (R01GM088266 to J.W.R. and R01GM104984 to M.L.M.) and the Victoria S. and Bradley Geist Foundation (J.W.R.). The content of this article does not necessarily reflect the position or policy of the US government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708584115/-/DCSupplemental.
References
- 1.Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: Integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sulzmaier FJ, Ramos JW. RSK isoforms in cancer cell invasion and metastasis. Cancer Res. 2013;73:6099–6105. doi: 10.1158/0008-5472.CAN-13-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sulzmaier FJ, et al. RSK2 activity mediates glioblastoma invasiveness and is a potential target for new therapeutics. Oncotarget. 2016;7:79869–79884. doi: 10.18632/oncotarget.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauge C, Frödin M. RSK and MSK in MAP kinase signalling. J Cell Sci. 2006;119:3021–3023. doi: 10.1242/jcs.02950. [DOI] [PubMed] [Google Scholar]
- 6.Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J. 2012;441:553–569. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- 7.Anjum R, Blenis J. The RSK family of kinases: Emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 8.Roux PP, Richards SA, Blenis J. Phosphorylation of p90 ribosomal S6 kinase (RSK) regulates extracellular signal-regulated kinase docking and RSK activity. Mol Cell Biol. 2003;23:4796–4804. doi: 10.1128/MCB.23.14.4796-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frödin M, Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol. 1999;151:65–77. doi: 10.1016/s0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 10.Smith JA, Poteet-Smith CE, Malarkey K, Sturgill TW. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J Biol Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 11.Frödin M, Jensen CJ, Merienne K, Gammeltoft S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen CJ, et al. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J Biol Chem. 1999;274:27168–27176. doi: 10.1074/jbc.274.38.27168. [DOI] [PubMed] [Google Scholar]
- 13.Kang S, et al. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang S, et al. Epidermal growth factor stimulates RSK2 activation through activation of the MEK/ERK pathway and src-dependent tyrosine phosphorylation of RSK2 at Tyr-529. J Biol Chem. 2008;283:4652–4657. doi: 10.1074/jbc.M709673200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gawecka JE, et al. RSK2 protein suppresses integrin activation and fibronectin matrix assembly and promotes cell migration. J Biol Chem. 2012;287:43424–43437. doi: 10.1074/jbc.M112.423046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doehn U, et al. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol Cell. 2009;35:511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 18.Fortin Ensign SP, Mathews IT, Symons MH, Berens ME, Tran NL. Implications of Rho GTPase signaling in glioma cell invasion and tumor progression. Front Oncol. 2013;3:241. doi: 10.3389/fonc.2013.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertz O. Spatio-temporal Rho GTPase signaling–Where are we now? J Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 21.Poteet-Smith CE, Smith JA, Lannigan DA, Freed TA, Sturgill TW. Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family. J Biol Chem. 1999;274:22135–22138. doi: 10.1074/jbc.274.32.22135. [DOI] [PubMed] [Google Scholar]
- 22.Cao X, et al. A phosphorylation switch controls the spatiotemporal activation of Rho GTPases in directional cell migration. Nat Commun. 2015;6:7721. doi: 10.1038/ncomms8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehman HL, et al. Regulation of inflammatory breast cancer cell invasion through Akt1/PKBα phosphorylation of RhoC GTPase. Mol Cancer Res. 2012;10:1306–1318. doi: 10.1158/1541-7786.MCR-12-0173. [DOI] [PubMed] [Google Scholar]
- 24.Helms MC, et al. Mitotic-dependent phosphorylation of leukemia-associated RhoGEF (LARG) by Cdk1. Cell Signal. 2016;28:43–52. doi: 10.1016/j.cellsig.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arul N, Cho YY. A rising cancer prevention target of RSK2 in human skin cancer. Front Oncol. 2013;3:201. doi: 10.3389/fonc.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaiswal M, et al. Mechanistic insights into specificity, activity, and regulatory elements of the regulator of G-protein signaling (RGS)-containing Rho-specific guanine nucleotide exchange factors (GEFs) p115, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG) J Biol Chem. 2011;286:18202–18212. doi: 10.1074/jbc.M111.226431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aittaleb M, Gao G, Evelyn CR, Neubig RR, Tesmer JJ. A conserved hydrophobic surface of the LARG pleckstrin homology domain is critical for RhoA activation in cells. Cell Signal. 2009;21:1569–1578. doi: 10.1016/j.cellsig.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J Biol Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler AP, Ridley AJ. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp Cell Res. 2004;301:43–49. doi: 10.1016/j.yexcr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Bravo-Cordero JJ, et al. Spatial regulation of RhoC activity defines protrusion formation in migrating cells. J Cell Sci. 2013;126:3356–3369. doi: 10.1242/jcs.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirata E, et al. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J Cell Sci. 2012;125:858–868. doi: 10.1242/jcs.089995. [DOI] [PubMed] [Google Scholar]
- 32.Martin K, et al. Spatio-temporal co-ordination of RhoA, Rac1 and Cdc42 activation during prototypical edge protrusion and retraction dynamics. Sci Rep. 2016;6:21901. doi: 10.1038/srep21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abidi F, et al. Novel mutations in Rsk-2, the gene for Coffin-Lowry syndrome (CLS) Eur J Hum Genet. 1999;7:20–26. doi: 10.1038/sj.ejhg.5200231. [DOI] [PubMed] [Google Scholar]
- 34.Trivier E, et al. Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature. 1996;384:567–570. doi: 10.1038/384567a0. [DOI] [PubMed] [Google Scholar]
- 35.Lara R, Seckl MJ, Pardo OE. The p90 RSK family members: Common functions and isoform specificity. Cancer Res. 2013;73:5301–5308. doi: 10.1158/0008-5472.CAN-12-4448. [DOI] [PubMed] [Google Scholar]
- 36.Dunn GP, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brennan CW, et al. TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shang X, et al. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci USA. 2013;110:3155–3160. doi: 10.1073/pnas.1212324110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J, et al. Automated NMR fragment based screening identified a novel interface blocker to the LARG/RhoA complex. PLoS One. 2014;9:e88098. doi: 10.1371/journal.pone.0088098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark MJ, et al. U87MG decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lessey-Morillon EC, et al. The RhoA guanine nucleotide exchange factor, LARG, mediates ICAM-1-dependent mechanotransduction in endothelial cells to stimulate transendothelial migration. J Immunol. 2014;192:3390–3398. doi: 10.4049/jimmunol.1302525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi GX, Kaminski CN. Analysis of the Rit subfamily GTPase-mediated signaling and neuronal differentiation and survival. Methods Mol Biol. 2014;1120:217–240. doi: 10.1007/978-1-62703-791-4_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.