Significance
The cyclin D1 proto-oncoprotein is a crucial regulator of cell cycle progression, and excessive cyclin D1 expression and/or activity is a hallmark of many human cancers. The SCFFBXO31 ubiquitin ligase complex is responsible for targeting cyclin D1 for ubiquitination and degradation, thereby regulating its turnover after DNA damage. In this work, we report the structural, biophysical, and functional characterization of the interaction between cyclin D1 and Skp1–FBXO31. We find that the F-box protein FBXO31 targets cyclin D1 for phosphorylation-independent ubiquitination by recognizing and binding to its extreme C terminus. This knowledge enhances our understanding of the molecular mechanism of FBXO31-mediated cyclin D1 proteolysis following genotoxic stress and may facilitate efforts to identify a novel molecular target for anticancer drug discovery.
Keywords: Skp1–FBXO31–cyclin D1 structure, ubiquitin system, cell cycle
Abstract
Ubiquitin-dependent proteolysis of cyclin D1 is associated with normal and tumor cell proliferation and survival. The SCFFBXO31 (Skp1–Cul1–Rbx1–FBXO31) ubiquitin ligase complex mediates genotoxic stress-induced cyclin D1 degradation. Previous studies have suggested that cyclin D1 levels are maintained at steady state by phosphorylation-dependent nuclear export and subsequent proteolysis in the cytoplasm. Here we present the crystal structures of the Skp1–FBXO31 complex alone and bound to a phosphorylated cyclin D1 C-terminal peptide. FBXO31 possesses a unique substrate-binding domain consisting of two β-barrel motifs, whereas cyclin D1 binds to FBXO31 by tucking its free C-terminal carboxylate tail into an open cavity of the C-terminal FBXO31 β-barrel. Biophysical and functional studies demonstrate that SCFFBXO31 is capable of recruiting and ubiquitinating cyclin D1 in a phosphorylation-independent manner. Our findings provide a conceptual framework for understanding the substrate specificity of the F-box protein FBXO31 and the mechanism of FBXO31-regulated cyclin D1 protein turnover.
The eukaryotic ubiquitin–proteasome system has long been recognized as an integral part of cellular protein turnover and homeostasis, and, in many cases, plays a pivotal role in the regulation of normal and cancer-related cellular processes (1). In particular, ubiquitin-mediated proteolysis provides a mechanism of temporal enforcement and coordination of cell cycle transitions by working in concert with cyclin-dependent kinases (Cdks) to regulate the levels of various cyclin proteins and Cdk inhibitors (2). D-type cyclins encoded by three closely related genes (cyclins D1, D2, and D3) are major downstream targets of extracellular signaling pathways that transduce mitogenic signals to the core oscillatory cell-cycle engine (3). Through their association with and activation of Cdk4 and Cdk6, as well as sequestration of the Cdk inhibitors p21 and p27, D-type cyclins serve as key activators of G1 phase reentry and progression in response to mitogenic stimuli (4). Cyclin D1, the most intensively investigated D-type cyclin, is frequently deregulated in cancer, and its overexpression directly contributes to genomic instability and tumorigenesis (5). Altered ubiquitination and stability of cyclin D1 are largely responsible for its elevated levels in tumor cells and were found to be a biomarker of cancer phenotype and disease progression (6). Thus, a deeper understanding of regulatory mechanisms of ubiquitin-dependent cell cycle control may potentially assist in development of novel anticancer therapies.
Ubiquitin–protein ligases (i.e., E3s) recognize and interact with target proteins, thereby controlling substrate specificity for ubiquitination reactions. The multimeric SCF (Skp1–Cul1–F-box protein) complexes are the key ubiquitin–protein ligases that have been linked to cell cycle regulation (7). While interacting with the adaptor protein Skp1 through the N-terminal F-box motif, the F-box protein components of the SCF complexes recognize specific substrates through their variable C-terminal protein–protein interaction domains, including WD40 repeats (FBXW subfamily), leucine-rich repeats (LRRs; FBXL subfamily), and other different or unknown domains (FBXO subfamily) (8). Four F-box proteins, Skp2 (9), FBXW8 (10), FBXO4 (11), and FBXO31 (12), are involved in regulation of cyclin D1 ubiquitination and subsequent degradation. A recent genetic study of these F-Box proteins has led to the view that other unidentified ubiquitin ligases also appear to control cyclin D1 accumulation (13). Although Skp2 and FBXW8 possess the respective LRR and WD40 repeat domains, FBXO4 and FBXO31 belong to the third class of F-box proteins, whose C-terminal domains exhibit no sequence homology to WD40 or LRR repeats (14). The crystal structure of the Skp1–FBXO4 complex reveals that FBXO4 adopts an unexpected small GTPase-like fold (15, 16). Whereas Skp2, FBXO4, and FBXW8 are involved in the normal cell cycle-dependent oscillation of cyclin D1, FBXO31 is a DNA damage-induced checkpoint protein that promotes cyclin D1 degradation and subsequent G1 cell-cycle arrest in response to genotoxic stress (12). FBXO31 was first identified as a candidate tumor suppressor gene in several cancers, and its expression is down-regulated in breast cancer cell lines and primary breast tumors as well as in hepatocellular carcinoma (17, 18). In addition to cyclin D1, five other FBXO31 substrates have been identified (19–23). Despite its clear roles in different cellular processes, the mechanism whereby FBXO31 renders substrate specificity to the SCF complex remains largely unknown.
Ubiquitination of cyclin D1 is believed to be triggered by phosphorylation of a single threonine residue (Thr286) near the C terminus of the protein (24) by glycogen synthase kinase 3β (GSK3β) (25) and through a MAPK pathway (10, 12). Mutation of Thr286 to alanine (i.e., T286A) was shown to reduce cyclin D1 polyubiquitination and stabilize the protein in proliferating and quiescent fibroblasts (24). GSK3β-mediated phosphorylation of cyclin D1 Thr286 was also found to promote cyclin D1 nuclear export by facilitating its interaction with the nuclear exportin CRM1 (26). The highly stable cyclin D1 T286A mutant remains in the nucleus throughout the cell cycle (25). As cyclin D1 accumulates in the nucleus during G1 phase and exits into the cytoplasm as cells proceed into S phase (27), it is possible that the phosphorylated cyclin D1 at Thr286 is targeted for nuclear export for subsequent degradation in the cytoplasm. In this paper, we report crystal structures of the human Skp1–FBXO31core complex and this complex bound to a Thr286-phosphorylated C-terminal peptide of cyclin D1, revealing a zinc ion-binding, β-barrel–containing substrate-recognition domain in FBXO31 that binds to the extreme C terminus of cyclin D1 independent of phosphorylation at Thr286. Our in vitro biochemical studies show that the SCFFBXO31 ubiquitin ligase can ubiquitinate both unphosphorylated forms of cyclin D1 and its T286A mutant. Taken together, these studies provide fundamental structural insights into the FBXO31–cyclin D1 interaction and delineate a mechanism for SCFFBXO31-mediated ubiquitination of cyclin D1 in a phosphorylation-independent manner.
Results and Discussion
Crystallization and Structure Determination.
We produced the binary Skp1–FBXO31core complex by coexpressing a truncated human Skp1 protein (28) and a core fragment of human FBXO31 (residues 66–539) that lacks the N-terminal 65 residues preceding the F-box domain. The Skp1–FBXO31core construct was crystallized alone and bound to a 17-residue phosphorylated peptide corresponding to the extreme C-terminal region of cyclin D1 (residues 279–295; phosphorylated at Thr286). The structure of the Skp1–FBXO31core complex was determined at 2.7-Å resolution by the single-wavelength anomalous dispersion (SAD) method by using an Hg derivative (Table S1). The resultant electronic map allowed unambiguous tracing of the two polypeptide chains and the positioning of most side chains, except for a 59-residue disordered region in FBXO31 (residues 384–442). The final model of the binary structure contains two Skp1–FBXO31core molecules in the asymmetric unit. The crystal structure of the ternary Skp1–FBXO31core–cyclin D1 complex was solved at 2.7-Å resolution by molecular replacement by using the binary structure as a search model (Table S1). It also contains two Skp1–FBXO31core monomers, each bound to the phosphorylated cyclin D1 peptide.
The Skp1–FBXO31core Complex.
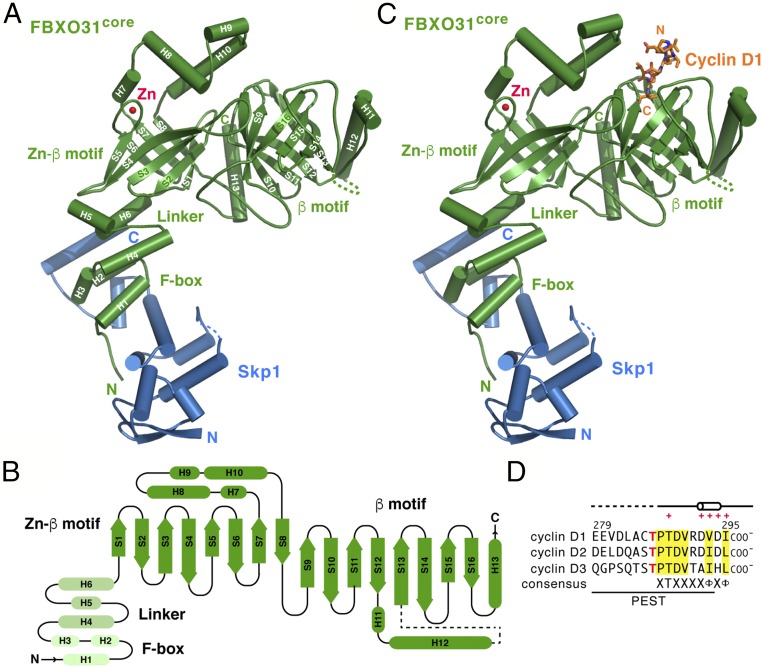
The Skp1–FBXO31core complex has an elongated structure that is highly analogous to the overall spatial arrangement of other Skp1–F-box protein complexes (29) (Fig. 1A). FBXO31core consists of an N-terminal F-box domain (residues 66–101; helices H1–H3), an α-helical linker domain (residues 102–146; helices H4–H6), and a C-terminal domain (residues 147–539; helices H7–H12 and strands S1−S13; Fig. 1 A and B). The F-box domain folds into the same three-helix cluster conformation as in other reported F-box protein structures (29). The molecular interaction between the F-box and Skp1 is also very similar to other Skp1–F-box protein complexes. The linker domain of FBXO31 forms a three-helix platform to position the C-terminal domain well away from Skp1 and the F-box domain. Our results reinforce the view that F-box proteins are key components responsible for positioning and orienting the substrate optimally for ubiquitination by the SCF-bound E2 (29).
Fig. 1.
Crystal structures of Skp1–FBXO31core and its complex with the cyclin D1 peptide. (A) Ribbon diagram of the Skp1–FBXO31core complex, with the secondary structure elements of FBXO31 labeled. Skp1 and FBXO31 are shown in blue and green, respectively. The bound Zn2+ ion is depicted as a red sphere. Dashed lines indicate disordered regions. (B) Topology diagram of FBXO31core. Cylinders and arrows represent α-helices and β-strands, respectively. (C) Ribbon diagram of the Skp1–FBXO31core complex bound to the cyclin D1 peptide (orange “licorice sticks”). (D) Sequence alignment of the cyclin D1 peptide (residues 279–295) used in crystallization and its corresponding cyclin D2 (residues 273–289) and cyclin D3 (residues 276–292) peptides. Their C-terminal carboxylate groups are indicated. Cylinder indicates the 310-helix (residues 292–294), dashed lines disordered regions, and red crosses the residues of cyclin D1 that contact FBXO31. Residues conserved among D-type cyclins are highlighted in yellow. The phosphorylated Thr286 in cyclin D1 and its corresponding residue in cyclin D2 (Thr280) and cyclin D3 (Thr283) are highlighted in red. The predicted PEST-like sequence motif is underlined.
The C-terminal domain of FBXO31 is composed of two distinct α/β mixed motifs, each adopting a highly curved eight-stranded antiparallel β-sheet structure (Fig. 1 A and B). The two β-sheets are twisted with each strand in a right-handed helical manner to form a classic up-and-down continuous β-barrel by a succession of +1 connections, in which the first and last strands associate to close the barrel. The N-terminal β-barrel motif (residues 147−290; helices H7−H10 and strands S1−S8) sits on the triple-helix platform formed by the linker domain and covered by a four-helix lid at the opposite end. Interestingly, a zinc ion is found at the edge of the N-terminal β-barrel connecting to the four-helix lid and binds to two cysteine and two histidine residues (Fig. 1A). Hereafter we refer to the N-terminal β-barrel motif as the Zn-β motif. Unlike the covered Zn-β motif, the C-terminal β-barrel motif (residues 291−539; helices H11−H13 and strands S9−S16) forms a largely open-end barrel surrounded by three helices (hereafter called the β-motif), in which one end hubs the substrate-binding cavity. As far as we know, the overall folding topology of the C-terminal domain of FBXO31 has not been seen before.
The Skp1–FBXO31core–Cyclin D1 Complex.
The structure of the Skp1–FBXO31core–cyclin D1 complex reveals that the extreme C-terminal nine-residue region (residues 287−295) of cyclin D1 adopts a helical conformation and binds to the β-motif of FBXO31, in which the C-terminal carboxylate group of the peptide inserts into the open cavity in the β-barrel (Fig. 1 C and D). (No interpretable electron density was observed for the N-terminal eight residues including the phosphorylation site.) The association between cyclin D1 and FBXO31 buries a total of ∼570 Å2 of solvent-accessible surface on the two proteins, or ∼49% of total surface area of the cyclin D1 peptide. Cyclin D1 peptide binding does not induce significant conformational changes in FBXO31, with FBXO31 in the binary and ternary complex structures superimposing with a Cα rmsd of 0.36 Å.
The C-Terminal Domain of FBXO31.
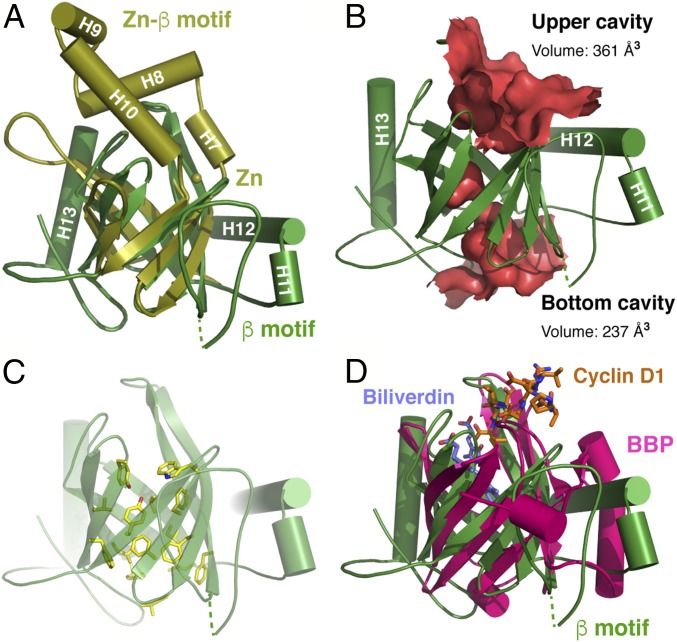
Two β-barrels in the C-terminal domain of FBXO31 are arranged ∼65° apart and can be superimposed onto each other with a 2.1-Å rmsd over 73 Cα atoms (Fig. 2A). The major differences in the two FBXO31 β-motifs are present within more mobile extended structures and loop regions. In the Zn-β motif, the H7, H8, H9, and H10 helices connecting the S7 and S8 strands, along with the extended loop between the S3 and S4 strands, form a lid that folds back to cap the open cavity at the end of the barrel, whereas the other end of the barrel is closed off by the H4, H5, and H6 helices from the linker domain. By contrast, the β-barrel of the β-motif has a well-defined cavity at each end, and the H11 and H12 helices wrap around the upper side of the barrel (Fig. 2B). The C-terminal α-helix H13 situated between the Zn-β motif and the β-motif serves as a molecular strut to support the backsides of the two barrels. Both β-barrels in these motifs are largely amphipathic, with polar and charged residues on the outside and nonpolar side chains buried on the inside. A cluster of aromatic side chains (Phe305, Tyr309, Phe321, Phe467, Phe488, Phe496, Phe498, and Trp500) form a twisted ladder in the central core of the β-barrel in the β-motif (Fig. 2C).
Fig. 2.
The C-terminal domain of FBXO31 features two β-barrel–containing motifs. (A) Superimposition of the Zn-β and β-motifs of FBXO31, colored in olive and bright green, respectively. (B) Molecular surface representation of the inner cavities of the β-barrel in the FBXO31 β-motif defined by HOLLOW (36). Volumes were calculated with CASTp (37). (C) Licorice-stick representation of side chains of the aromatic and hydrophobic residues lining the inner cavities of the β-barrel in the FBXO31 β-motif. (D) Superimposition of the FBXO31 β-motif (green) with the BBP protein (32) (magenta; Protein Data Bank ID code 1BPP). Cyclin D1 (orange) and the biliverdin IXγ chromophore (purple) are shown as licorice sticks.
Structural similarity search by Dali (30) for the Zn-β and β-motifs individually has identified small extracellular proteins in the lipocalin family that bind and transport largely small hydrophobic molecules (31). Despite the low degree of overall sequence conservation, lipocalin proteins share a conserved core fold characterized by a single eight-stranded up-and-down β-barrel. For example, the β-motif of FBOX31 can be superimposed onto the β-barrel structure of the bilin-binding protein (BBP) bound to the biliverdin IXγ chromophore with an rmsd of 3.5 Å for 72 Cα atoms, even though they share only ∼10% sequence identity (32) (Fig. 2D). Interestingly, the pigment molecule occupies a central cleft at the upper end of the BBP barrel, in a manner similar to the binding of the cyclin D1 peptide to the β-motif of FBXO31. Thus, FBXO31 possesses a previously uncharacterized type of substrate recognition domain in SCF E3s.
Molecular Interactions Between Cyclin D1 and FBXO31.
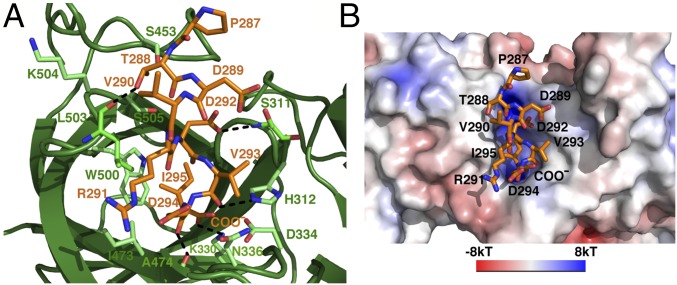
The cyclin D1 peptide binds to the upper cavity of the β-barrel in the β-motif of FBXO31 extending from strands 9 and 16 at one side of the surface to the center of the cavity (Fig. 3A and Fig. S1). The middle portion of the peptide (residues 289−292) adopts a turn-like conformation so that the peptide chain stays on top of the barrel, whereas a 310 helix formed by residues 292−294 is tucked into the cavity. Nearly all of the FBXO31 contacts are made by the last four C-terminal cyclin D1 residues (residues 292−295), and all eight strands of the C-terminal β-barrel of FBXO31 participate in interactions with cyclin D1. The side chain of Asp292 and the backbone carbonyl groups of Val293 and Asp294 bind the rim of the barrel with their contacts partially solvent-exposed. Ile295 and the C-terminal carboxylate group insert the furthest into the hub of the barrel, making intermolecular contacts in a mostly buried environment. The cyclin D1-binding cavity in FBXO31 is lined by hydrophobic residues on one side and polar residues on the other side (Fig. 3B and Fig. S1). The C-terminal carboxylate group of cyclin D1 is well positioned to make bifurcated hydrogen bonds with side chains of Lys330 and Asp334 and also receives a water-mediated hydrogen bond with Thr343 and Leu472. Consequently, the side chain of cyclin D1 Ile295 fits into a hydrophobic pocket flanked by side chains of Tyr309, Ile473, Trp500, and Leu503. These packing interactions rationalize the preference of a hydrophobic C-terminal residue in the recognition sequence.
Fig. 3.
Molecular interactions between cyclin D1 and FBXO31. (A) Close-up view of the FBXO31–cyclin D1 interface shows interacting residues of FBXO31 (green) and cyclin D1 (orange). Hydrogen bonds are shown as black dashed lines. (B) Molecular surface representation of the FBXO31 region involved in cyclin D1 binding colored according to the local electrostatic potential. The cyclin D1 residues are labeled.
The three residues preceding Ile295 contribute additional contacts to FBXO31. The γ-carboxylate group of Asp292 is hydrogen-bonded with the main chain NH of Ser311. The backbone carbonyl group of Val293 makes bidentate hydrogen bonds with side chains of His312 and Asn336, whereas the main chain carbonyl of Asp294 forms a hydrogen bond with the main chain NH of Ala474. In addition, the hydroxyl group of Thr288 is hydrogen-bonded with the main chain CO of Leu503. Alignment of 39 FBXO31 orthologs shows that surface residues involved in the interaction with cyclin D1 are conserved across species, whereas residues making direct contacts with cyclin D1 are invariant (Figs. S2 and S3).
Phosphorylation-Independent Cyclin D1 Binding and Ubiquitination.
The fact that the phosphorylated Thr286 does not participate in the cyclin D1–FBXO31 interaction in the ternary complex structure suggests that phosphorylation is not required for binding and ubiquitination of cyclin D1 by the SCFFBXO31 ubiquitin ligase. To test this hypothesis, we measured and compared the binding affinity of Skp1–FBXO31core for the Thr286-phosphorylated cyclin D1 peptide and the corresponding unphosphorylated peptide by using isothermal titration calorimetry (ITC; Fig. S4). As anticipated, the phosphorylated and unphosphorylated peptides bind to Skp1-FBXO31core with dissociation constants (Kd) of 3.5 ± 0.6 μM and 2.7 ± 0.4 μM, respectively. This result, in conjunction with the structural data, clearly implicates FBXO31 recognition of cyclin D1 in a phosphorylation-independent manner.
We next investigated the effects of the Thr286 phosphorylation on the ubiquitination of purified WT and mutant cyclin D1 proteins by using an in vitro assay reconstituted with purified human Uba1 E1, UbcH5a E2, Cul1–Rbx1, and Skp1–FBXO31. Because MAPK/ERK kinase plays an important role in phosphorylation of cyclin D1 after γ-irradiation (12), we therefore phosphorylated cyclin D1 by using the purified active recombinant ERK2 kinase. ERK2 phosphorylates cyclin D1 on Thr286 as well as on Thr156 and Ser219, two minor sites that also match the proline-directed phosphorylation site consensus. We confirmed phosphorylation levels and sites of cyclin D1 by tryptic digestion and LC-MS/MS (Fig. S5) and by immunoblotting with a cyclin D1 phospho-Thr286–specific antibody (Fig. 4A). As expected, addition of Skp1–FBXO31core to reaction mixtures containing the unphosphorylated cyclin D1 significantly stimulates formation of high molecular weight cyclin D1–ubiquitin conjugates (Fig. 4A, lane 5). The SCF ligase activity with FBXO31core is comparable with that of the control reaction in the presence of the full-length FBXO31 (named FBXO31FL; Fig. 4A, lane 4). Interestingly, the levels of cyclin D1 ubiquitination are largely similar between the Thr286-phosphorylated and the unphosphorylated proteins (lane 5 vs. lane 10). As GSK3β can also phosphorylate cyclin D1 on Thr286 (25), we used a constitutively active GSK3β mutant (i.e., S9A) to phosphorylate cyclin D1. The phosphorylated protein displays a similar ubiquitination level to that of the unphosphorylated cyclin D1 (Fig. S6).
Fig. 4.
In vitro ubiquitination of cyclin D1 by SCFFBXO31 does not require phosphorylation of cyclin D1 on Thr286. (A) In vitro ubiquitination of the untreated and ERK2-phosphorylated WT cyclin D1 proteins by Skp1–FBXO31FL and Skp1–FBXO31core. (Top) Cyclin D1 and cyclin D1–ubiquitin conjugates detected by Western blots with an anti-cyclin D1 antibody. (Bottom) Phosphorylation of Thr286 by Western blots with an anti-cyclin D1 phospho-Thr286 antibody. Asterisks indicate the likely high molecular weight cyclin D1 aggregates. (B) In vitro ubiquitination of the WT and the T286A mutant cyclin D1 by Skp1–FBXO31core. (C) In vitro ubiquitination of cyclin D1 by WT and mutant FBXO31core proteins. In B and C, cyclin D1 and cyclin D1–ubiquitin conjugates were detected by Western blots with an anti-cyclin D1 antibody.
We next sought to measure the effects of the T286A mutation in cyclin D1 ubiquitination by using our in vitro assay. We found that the T286A mutant is polyubiquitinated at a level similar to WT cyclin D1 (Fig. 4B, lane 4 vs. lane 8). Like the WT protein, the extent of ubiquitination of the T286A mutant is not affected by ERK2-mediated phosphorylation (Fig. S7A). Moreover, elimination of two minor phosphorylation sites (T156A/S219A) on cyclin D1 has no effect on its ubiquitination in the unphosphorylated and phosphorylated states (Fig. S7B).
As cyclin D1 binds and activates Cdk4 and Cdk6, its ability to complex with the kinase may impact its ubiquitination by SCFFBXO31. To address this question, we isolated specific binary complexes of the unphosphorylated and phosphorylated WT and T286A mutant cyclin D1 proteins with the purified Cdk6 by using gel-filtration chromatography and comparatively evaluated for their degree of ubiquitination by SCFFBXO31. The levels of the polyubiquitinated cyclin D1 bound to Cdk6 are similar to free cyclin D1 in the unphosphorylated and phosphorylated states (Fig. S8). Together, these data indicate that phosphorylation of Thr286 is not essential for the FBXO31-mediated ubiquitination of cyclin D1 alone and in complex with Cdk6.
FBXO31 Determinants for Cyclin D1 Recognition.
Our structural and biochemical studies suggest that FBXO31 specifically recognizes and binds cyclin D1 in a manner independent of phosphorylation. To study the relation between the abilities of FBXO31 to interact with cyclin D1 and mediate its ubiquitination, we mutated three FBXO31 residues individually to alanine and tested the capacity of the resulting variants (S311A, H312A, and K330A) to promote cyclin D1 polyubiquitination in our in vitro ubiquitination assay (Fig. 4C). Alanine substitutions of His312 and Lys330, whose side chains are involved in hydrogen-bonding interactions with the respective Val293 and the terminal carboxylate group of the cyclin D1 peptide, result in significant loss of ubiquitination activity (Fig. 4C, lanes 6 and 7). In contrast, alanine replacement of Ser311, whose main chain amide hydrogen-bonds to cyclin D1 Asp292, has much less effect on the rate and extent of the FBXO31-mediated ubiquitination reaction (Fig. 4C, lane 5). This result supports our structure-based conclusion that the very C terminus of cyclin D1 in the absence of phosphorylation is essential for its recognition and ubiquitination by FBXO31.
Cyclin D1 Subcellular Localization and Stability.
To address whether the SCFFBXO31 complex mediates the phosphorylation-independent ubiquitination and degradation of cyclin D1 in mammalian cells, we transiently coexpressed fluorescent Venus-tagged cyclin D1 with or without mTurquoise-tagged FBXO31 in HeLa cells and measured the subcellular localization and degradation rates of WT cyclin D1 and the T286A mutant by using live-cell imaging techniques. In agreement with previous observations (25), WT Venus-cyclin D1 is localized in nucleus and cytoplasm, whereas the T286A mutant resides a slightly higher ratio in the nucleus (Fig. 5 A and B). As FBXO31 itself is distributed in nucleus and cytoplasm (Fig. S9), the coexpression of cyclin D1 with FBXO31 results in a higher nucleus/cytoplasm expression ratio of WT cyclin D1 but has no effect on the localization of the T286A mutant (Fig. 5A), presumably reflecting the greater activity of FBXO31 in the cytoplasm. To determine the effects of FBXO31 on the stability of the cyclin D1 protein, cells transfected with WT cyclin D1 and the T286A mutant or cotransfected with cyclin D1 and FBXO31 were treated with cycloheximide, and fluorescent signals in each individual cell were measured by time-lapse imaging (Fig. 5C). In the absence of FBXO31, WT cyclin D1 has an approximately twofold higher decay rate than the T286A mutant. Coexpression of FBXO31 with cyclin D1 causes a 10-fold increase in the degradation of WT and mutant cyclin D1 proteins, whereas the decay rate of WT cyclin D1 is approximately twofold of that of the T286A mutant. These results support the notion that FBOX31 promotes phosphorylation-independent degradation of cyclin D1.
Fig. 5.
Subcellular localization of cyclin D1 and FBXO31-mediated cyclin D1 degradation in HeLa cells. (A–C) Venus-tagged WT and T286A mutant cyclin D1 were transiently coexpressed with mock or mTurquoise-tagged FBXO31 in HeLa cells. SiR-Hoechst was added to the culture 48 h after transfection. More than 800 cells were scored for each measurement, and error bars represent the SD (*P < 0.05 and **P < 0.01). (A) Subcellular localization of cyclin D1 was quantified by segmenting images with the Hoechst signal, and the nuclear/cytoplasmic fluorescence intensity ratios were determined from triplicate wells by using Matlab scripts. (B) Representative fluorescent images showing subcellular localization of the Venus-tagged WT cyclin D1 and the T286A mutant. (Scale bars, 200 μm.) (C) Degradation rate of the Venus-tagged WT cyclin D1 and the T286A mutant in the absence or presence of FBOX31. Cycloheximide (100 µg/mL) was added to each well 48 h after transfection, and cells were imaged every 15 min for at least 3 h. Single-cell degradation analysis was conducted by using custom Matlab scripts, and the average rate of decay for each experimental condition for defined cell populations was calculated by fitting a linear decay curve (Movies S1–S12).
The Zinc-Binding Structural Motif of FBXO31.
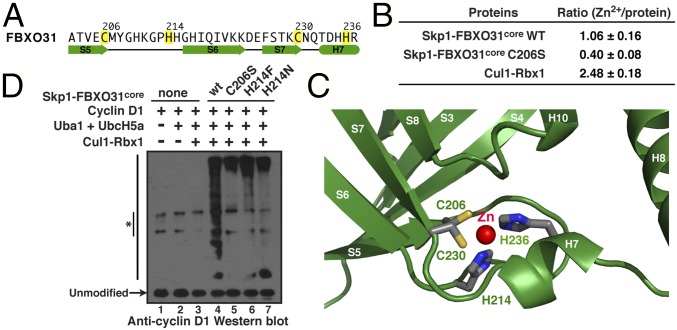
The FBXO31 structure reveals a previously unknown, unique CHCH-type zinc-binding site in its Zn-β motif (Fig. 6A and SI Materials and Methods). The identity of the metal ion was confirmed by inductively coupled plasma-optical emission spectrometry (ICP-OES) analysis of multielement contents of the WT and mutant FBXO31core proteins. WT Skp1–FBXO31core from four different protein preparations has a [Zn2+]/protein ratio of 1.06 ± 0.16, whereas the ratio decreases to 0.40 ± 0.12 by mutation of Cys206 to serine (i.e., C206S; Fig. 6B). As a control, the Cul1–Rbx1 component of the SCF E3 complex that contains a three zinc-containing RING finger has a [Zn2+]/protein ratio of 2.48 ± 0.18. Amino acid spacing between the ligands of the zinc ion (Cys-X7-His-X15-Cys-X5-His, where X represents a variable amino acid) is quite different from the consensus sequences of other zinc fingers (33). Cys206 and His214 at the end of the S5–S6 loop form a lasso-like structure, whereas Cys230 and His236 lie at the end of the S7 strand and the C-terminal turn of the short H7 helix, respectively (Fig. 6C). The side chains of Cys206 and His235 form a sandwich-like structure into which the imidazole ring of His214 fits, where the carbonyl oxygen of Cys206 forms a hydrogen bond with the amine nitrogen of His214. The four zinc ligands are invariant in all FBXO31 orthologs (Fig. S2).
Fig. 6.
The zinc-binding structural motif of FBXO31. (A) The sequence of the zinc-binding motif in FBXO31 with the four zinc ligands highlighted in yellow. Cylinders and arrows indicate α-helices and β-strands. (B) ICP-OES analysis shows the ratio of Zn2+ to the WT and mutant FBXO31core proteins. The ratios were calculated as the average and SD from four independent measurements. (C) Close-up view of the zinc-binding site of FBXO31 shows the coordinating residues as licorice sticks. (D) In vitro ubiquitination of cyclin D1 by the WT and mutant FBXO31core proteins. Cyclin D1 and cyclin D1–ubiquitin conjugates were detected by Western blots with an anti-cyclin D1 antibody. Asterisks indicate the likely high molecular weight cyclin D1 aggregates.
The role of zinc binding in tethering the β-barrel of the Zn-β motif of FBXO31 suggests that Zn2+ helps rigidify the antiparallel β-sheet surrounded by connecting loops (Fig. 6C). If so, one would anticipate that the folding and activity of FBXO31 would be compromised when its Zn2+ binding is abolished. To address this issue, we generated seven single-point mutants of FBXO31 (C206A, H214A, C230A, H236A, C206S, H214F, and H214N), four of which fail to express in Escherichia coli or form insoluble aggregates. C206S, H214F, and H214N mutants were produced, albeit at a much lower level than the WT protein. Although they tend to aggregate as judged by gel-filtration chromatography, the purified mutant proteins are fully active in vitro with regard to their ubiquitination activity toward cyclin D1 (Fig. 6D, lane 5–7 vs. lane 4). This result implicates zinc binding in facilitating the proper folding of FBXO31.
FBXO31 Binds both Cyclin D2 and Cyclin D3.
Various combinations of D-type cyclins were shown to be expressed in different cell types (3). Cyclin D1, cyclin D2, and cyclin D3 have high degrees of sequence conservation (52–64% identity) and exhibit extensive homology in the extreme C-terminal region, including the signature threonine residue embedded in a PEST-like (proline-, glutamic acid-, serine-, and threonine-rich) sequence motif (5). The PEST motif is considered a characteristic property of short-lived proteins degraded via the ubiquitin–proteasome pathway (34). Three of the five cyclin D1 residues that directly interact with FBXO31 are variably conserved hydrophobic amino acids in cyclin D2 and cyclin D3 (Fig. 1D). Furthermore, the main chain atoms of the cyclin D1 peptide make significant contributions to its interactions with FBXO31 as described here earlier. FBXO31 is therefore likely able to recognize and bind to cyclin D2 and cyclin D3 in a manner analogous to cyclin D1. To this end, we measured binding affinities of Skp1–FBXO31core to the corresponding unphosphorylated C-terminal cyclin D2 (residues 273–289) and cyclin D3 (residues 276–292) peptides by using ITC. Indeed, we found that the cyclin D2 and cyclin D3 peptides bind Skp1–FBXO31core with Kd values of 3.0 ± 0.5 μM and 1.9 ± 0.5 μM, respectively, compared with a Kd of 2.7 ± 0.4 μM for the cyclin D1 peptide (Fig. S10).
Implications for FBXO31-Substrate Recognition and Cyclin D1 Regulation by SCFFBXO31.
In many of the well-characterized F-box protein–substrate interactions, F-box proteins recognize the internal sequences of their targets for rapid degradation. In our crystal structure of the Skp1–FBXO31core–cyclin D1 complex, the free carboxylate tail of cyclin D1 inserts into the open cavity on the surface of the FBXO31 C-terminal β-barrel motif. By restricting the major interactions to the extreme C-terminal four residues, FBXO31 appears to recognize and bind to its substrates based on these sequence determinants. Nonpolar residues (such as Ala, Val, Ile, or Leu) are likely preferred at the C-terminal position such as in the −2 position (i.e., two residues from the C terminus; Fig. 1D). A similar peptide recognition mechanism has been discovered in the PDZ domains that recognize their target proteins by incorporating the extreme C-termini of the target into a β-sheet in PDZ through antiparallel β-augmentation (35). Nonetheless, it is possible that FBXO31 can recognize internal sequence motifs present in its other substrates (20, 21). Further studies are needed to fully elucidate the molecular basis for substrate selectivity and specificity by FBXO31.
In contrast to the current model in which phosphorylation of cyclin D1 on Thr286 is required for its ubiquitin-mediated proteolysis by SCFFBXO31 (12), our structural and biochemical data clearly show that Thr286 phosphorylation is dispensable for cyclin D1 association with FBXO31 and ubiquitination by SCFFBXO31. Moreover, our cell-based experiments demonstrate that the cyclin D1-T286A mutant is localized more in the nucleus and FBOX31 can promote phosphorylation-independent degradation of WT cyclin D1 and the T286A mutant. These results reinforce the conclusion that phosphorylation of Thr286 directs nuclear export of cyclin D1 (25, 26) but is not associated with its subsequent ubiquitination and degradation by SCFFBXO31. FBXW8 (10) and FBXO4 (11) have been reported to recognize phosphorylated cyclin D1. It is also worthwhile to consider the possibility that phosphorylation of cyclin D1 on Thr286 acts to induce its ubiquitination by other ubiquitin ligases.
Materials and Methods
Proteins were expressed and purified by using standard methods. Crystallization was performed by using vapor diffusion in hanging drops. X-ray diffraction data were collected at the National Synchrontron Light Source (NSLS) beamline X29A and the NSLS-II beamline 17-ID-1. The binary structure was solved by SAD and the ternary structure by molecular replacement. ITC experiments were performed with a TA Nano ITC Low Volume calorimeter. In vitro cyclin D1 ubiquitination assays were performed by using purified proteins. Live-cell imaging analyses were done by using the ImageXpress MicroXL system. Details of all experimental procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank W. Shi at X29A beamline of NSLS and J. Jakoncic and A. Soares at 17-ID-1 beamline of NSLS-II for help with data collection; O. Vinogradova, X. Lin, and K. Nguyen for technical assistance with ITC experiments; and N. Ahn for her gift of the ERK2 expression plasmid. This work was supported by National Institutes of Health Grants GM099948 (to B.H.) and GM113141 (to X. Liu), and Connecticut Regenerative Medicine Research Fund Grant 15-RMA-UCHC-04 (to B.H.). The ImageXpress MicroXL was supported by National Center for Research Resources Grant S10 RR026680.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5VZT and 5VZU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708677115/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama KI, Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 3.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 4.Casimiro MC, Velasco-Velázquez M, Aguirre-Alvarado C, Pestell RG. Overview of cyclins D1 function in cancer and the CDK inhibitor landscape: Past and present. Expert Opin Investig Drugs. 2014;23:295–304. doi: 10.1517/13543784.2014.867017. [DOI] [PubMed] [Google Scholar]
- 5.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 6.Alao JP. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardozo T, Pagano M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 8.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1and cyclin D proteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okabe H, et al. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS One. 2006;1:e128. doi: 10.1371/journal.pone.0000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin DI, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanie T, et al. Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol Cell Biol. 2012;32:590–605. doi: 10.1128/MCB.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Hao B. Structural basis of dimerization-dependent ubiquitination by the SCFFbx4 ubiquitin ligase. J Biol Chem. 2010;285:13896–13906. doi: 10.1074/jbc.M110.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Z, et al. Structural basis of selective ubiquitination of TRF1 by SCFFbx4. Dev Cell. 2010;18:214–225. doi: 10.1016/j.devcel.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R, et al. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005;65:11304–11313. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- 18.Huang HL, Zheng WL, Zhao R, Zhang B, Ma WL. FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol Rep. 2010;24:715–720. doi: 10.3892/or_00000912. [DOI] [PubMed] [Google Scholar]
- 19.Vadhvani M, Schwedhelm-Domeyer N, Mukherjee C, Stegmüller J. The centrosomal E3 ubiquitin ligase FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS One. 2013;8:e57530. doi: 10.1371/journal.pone.0057530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson P, et al. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J Biol Chem. 2014;289:18514–18525. doi: 10.1074/jbc.M114.559930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, et al. F-box only protein 31 (FBXO31) negatively regulates p38 mitogen-activated protein kinase (MAPK) signaling by mediating lysine 48-linked ubiquitination and degradation of mitogen-activated protein kinase kinase 6 (MKK6) J Biol Chem. 2014;289:21508–21518. doi: 10.1074/jbc.M114.560342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malonia SK, Dutta P, Santra MK, Green MR. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci USA. 2015;112:8632–8637. doi: 10.1073/pnas.1510929112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeffery JM, et al. FBXO31 protects against genomic instability by capping FOXM1 levels at the G2/M transition. Oncogene. 2017;36:1012–1022. doi: 10.1038/onc.2016.268. [DOI] [PubMed] [Google Scholar]
- 24.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 25.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 28.Schulman BA, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flower DR, North AC, Sansom CE. The lipocalin protein family: Structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 32.Huber R, et al. Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution. J Mol Biol. 1987;198:499–513. doi: 10.1016/0022-2836(87)90296-8. [DOI] [PubMed] [Google Scholar]
- 33.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 35.Lee HJ, Zheng JJ. PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho BK, Gruswitz F. HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dundas J, et al. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.