Significance
In mitophagy, damaged mitochondria are targeted for disposal by the autophagy machinery. PARKIN promotes signaling of mitochondrial damage to the autophagy machinery for engagement, and PARKIN mutations cause Parkinson’s disease, possibly because damaged mitochondria accumulate in neurons. Because regulation of PARKIN abundance and the impact on signaling are poorly understood, we performed a genetic screen to identify PARKIN abundance regulators. Both positive and negative regulators were identified and will help us to further understand mitophagy and Parkinson’s disease. We show that some of the identified genes negatively regulate PARKIN gene expression, which impacts signaling of mitochondrial damage in mitophagy. This link between transcriptional repression and mitophagy is also apparent in neurons in culture, bearing implications for disease.
Keywords: PARKIN, mitophagy, phosphoubiquitin, THAP11, genome-wide screen
Abstract
PARKIN, an E3 ligase mutated in familial Parkinson’s disease, promotes mitophagy by ubiquitinating mitochondrial proteins for efficient engagement of the autophagy machinery. Specifically, PARKIN-synthesized ubiquitin chains represent targets for the PINK1 kinase generating phosphoS65-ubiquitin (pUb), which constitutes the mitophagy signal. Physiological regulation of PARKIN abundance, however, and the impact on pUb accumulation are poorly understood. Using cells designed to discover physiological regulators of PARKIN abundance, we performed a pooled genome-wide CRISPR/Cas9 knockout screen. Testing identified genes individually resulted in a list of 53 positive and negative regulators. A transcriptional repressor network including THAP11 was identified and negatively regulates endogenous PARKIN abundance. RNAseq analysis revealed the PARKIN-encoding locus as a prime THAP11 target, and THAP11 CRISPR knockout in multiple cell types enhanced pUb accumulation. Thus, our work demonstrates the critical role of PARKIN abundance, identifies regulating genes, and reveals a link between transcriptional repression and mitophagy, which is also apparent in human induced pluripotent stem cell-derived neurons, a disease-relevant cell type.
Dysfunction of mitochondria is implicated in aging and human disease, including Parkinson’s disease. Mitochondrial functionality is assured by a hierarchical system of interdependent cellular quality-control mechanisms acting at molecular or organellar levels to allow rapid adaptation to mitochondrial stress and damage (1). The E3 ligase PARKIN (encoded by the PARK2 gene) takes on the role of a major sentinel that integrates multiple quality-control mechanisms ranging from facilitating proteasomal degradation of mitochondrial proteins to suppressing mitochondrial antigen presentation (2, 3).
PARKIN’s best-characterized role in mitochondrial quality control is in PINK1/PARKIN-mediated mitophagy where PARKIN acts in concert with the PINK1 kinase in the signaling of mitochondrial damage to the autophagy machinery. Herein, PINK1 accumulates on the surface of dysfunctional mitochondria and recruits and activates cytosolic PARKIN. Preformed ubiquitin chains on multiple mitochondrial surface proteins are extended by activated PARKIN and S65-phosphorylated by PINK1 (4–7). The accumulation of S65-phosphorylated ubiquitin (pUb) on mitochondria constitutes the signaling mechanism to engage the autophagy machinery for selective clearance of dysfunctional mitochondria (8). The efficiency of mitophagy appears to depend on the pUb signal: while PINK1 generated pUb from preformed chains in the absence of PARKIN can induce some mitophagy, the presence of PARKIN amplifies the accumulation of pUb via a feed-forward mechanism and enhances mitophagy (8, 9). Mutations in both PINK1 and PARKIN are a cause of familial Parkinson’s disease, suggesting that compromised mitophagy is an underlying feature (10, 11). Thus, the thorough understanding of how these proteins regulate mitophagy, and how these proteins are themselves regulated, is important for the further understanding of mitophagy and the pathogenesis of Parkinson’s disease.
Previous studies identified regulators of PINK1/PARKIN-mediated mitophagy employing RNAi screens with damage-induced mitochondrial translocation of overexpressed GFP-PARKIN as a mitophagy proxy (12–15). These efforts profoundly advanced the understanding of mitophagy regulation; however, little is known about how cells set the threshold for mitophagy to proceed. In this regard, cellular regulation of PARKIN abundance is of particular interest as it may represent a mechanism to tune the progression of mitophagy by impacting pUb accumulation to adapt to physiological state changes.
The CRISPR/Cas9 gene-editing technology as a screening tool appears to be superior to RNAi in most cases of lethality screens (16–18) and in phenotypic screens (19). Using cells expressing a PARKIN reporter protein from the endogenous PARK2 promoter and in which steady state PARKIN levels dictate the kinetics of pUb accumulation, a phenotypic genome-wide CRISPR/Cas9 pooled screen was performed and resulted in a list of 53 positive and negative regulators. We show that transcriptional repression negatively regulates endogenous PARKIN steady-state level. In particular, THAP11 depletion affects both PARKIN protein levels and pUb accumulation in multiple cell types. Finally, human induced pluripotent stem cell (iPSC)-derived inducible Neurogenin 2 (iNGN2) neurons in which THAP11 was targeted by CRISPR/Cas9 display de-repression of PARK2 transcription and enhanced pUb accumulation, demonstrating the impact of PARKIN-level regulation in a relevant cell type.
Results
PARKIN Levels Dictate Kinetics of pUb Accumulation.
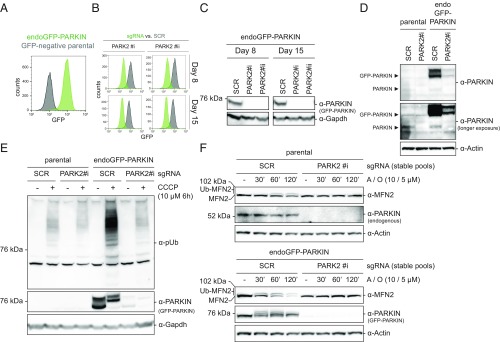
To assess the effects of cellular PARKIN abundance on downstream processes, we generated cellular models with different levels of PARKIN expression. HEK293-based JumpIN TI 293 cells expressing endogenous PARKIN were infected with lentiviruses for stable integration of Cas9 to enable gene editing and PARKIN depletion (parental cells) (Fig. S1A). Next, we aimed at generating cells with mildly elevated PARKIN expression additionally amenable to functional screening for physiological regulators of PARKIN. Thus, parental cells were transfected with a transgene encoding GFP-PARKIN under the control of a large PARK2 promoter fragment, which contains enhancer and repressor sites for physiological control of expression (20) (Fig. S1B). We selected a clone showing the lowest discrete GFP-signal in flow cytometry (endoGFP-PARKIN cells) (Fig. 1A). The fluorescence signal is below the detection threshold of conventional fluorescence microscopy, but is GFP-PARKIN–dependent because PARKIN-directed single guide RNAs (sgRNAs) resulted in the concomitant loss of GFP-PARKIN protein and GFP-fluorescence (Fig. 1 B and C). While endogenous PARKIN protein levels in the parental cells were near the detection threshold, GFP-PARKIN was readily detectable, likely reflecting multiple transgene insertions (Fig. 1D).
Fig. 1.
endoGFP-PARKIN cells display PARKIN level-dependent pUb accumulation. (A) Clonal JumpIN Cas9+ cells carrying a stable transgene driving GFP-PARKIN expression from 4.5 kb of the endogenous PARK2 promoter (endoGFP-PARKIN; green) or not (GFP-negative parental cells; gray) were assessed for GFP-dependent fluorescence by flow cytometry. (B and C) endoGFP-PARKIN cells display near-complete CRISPR/Cas9-mediated GFP-PARKIN depletion. Cells expressing either an SCR sgRNA or one of two different PARK2-specific sgRNAs (#i, #ii) were assessed for GFP-dependent fluorescence (B) and GFP-PARKIN protein levels (C) 8 or 15 d postinfection. (D) Comparison of GFP-PARKIN protein levels in endoGFP-PARKIN cells to endogenous PARKIN. Protein lysates from stable cell pools expressing either an SCR sgRNA or a PARK2-specific sgRNA (#i) were assessed by WB analysis using indicated antibodies. (E) PARKIN-dependent pUb accumulation in JumpIN cells treated with CCCP. endoGFP-PARKIN cells or parental JumpIN cells expressing PARK2-specific or SCR sgRNAs for 8 d were treated with 10 µM CCCP or vehicle for 6 h, and lysates probed with the antibodies indicated. (F) PARKIN-dependent MFN2 ubiquitination and degradation in parental cells (Upper) or endoGFP-PARKIN cells (Lower). WB analysis using indicated antibodies of stable cell pools expressing PARK2-specific or SCR sgRNAs treated with A/O for the time indicated. WB, Western blot.
We then examined the downstream effects of PARKIN-level alterations by assessing the accumulation of pUb upon mitochondrial damage in parental and endoGFP-PARKIN cells, additionally infected with either a PARK2-specific or a control [scrambled (SCR)] sgRNA. After treatment of parental cells transduced with control viruses with the mitochondria-damaging agent carbonyl cyanide m-chlorophenyl hydrazone (CCCP) for 6 h, α-pUb signals appeared that were absent in DMSO-treated samples (Fig. 1E), indicating the initiation of mitophagy. pUb accumulation in endoGFP-PARKIN cells was robustly induced to a level higher than in parental cells, and this accumulation was abrogated in cells additionally targeted for PARK2. GFP-PARKIN migrated slower on SDS/PAGE in mitochondrial damage conditions (Fig. 1E), reflecting PARKIN activation, where the GFP-tag serves as a pseudo-substrate (21). Time-course analyses revealed PARKIN-dependent pUb accumulation at early time points of CCCP or antimycin A/oligomycin treatment (A/O; both trigger PARKIN-dependent mitophagy) (Fig. S1 C and D). Varying levels of PARKIN affected pUb accumulation to a lesser extent after 24 h of treatment (Fig. S1 C and D). GFP-PARKIN levels declined during mitochondrial damage, likely reflecting proteasomal degradation (22) (Fig. S1 C and D). These data indicate that PARKIN levels are limiting for pUb accumulation during the early phase of mitochondrial damage in JumpIN cells. While we observed mitochondrial damage- and PARKIN-dependent decline of the common mitophagy marker TOM20 in endoGFP-PARKIN cells, degradation rates were at low levels (Fig. S1C). This is in agreement with other studies observing significantly lower rates for cells expressing endogenous PARKIN compared with those overexpressing PARKIN, with low rates likely reflecting the “endogenous” situation in human patients with Parkinson’s disease, in whom it takes decades to develop the disease (23). The established PARKIN-dependent ubiquitination and proteasomal degradation of MFN2 (23) showed similar dependency on PARKIN levels in mitochondrial damage conditions, thereby further highlighting the importance of cellular PARKIN level (Fig. 1F). Thus, the generated cell line expresses functional GFP-tagged PARKIN from its endogenous promoter that is limiting for pUb accumulation and is amenable to fluorescence-based genetic screening.
A Phenotypic Genome-Wide CRISPR Knockout Screen Identifies Regulators of GFP-PARKIN Steady-State Level.
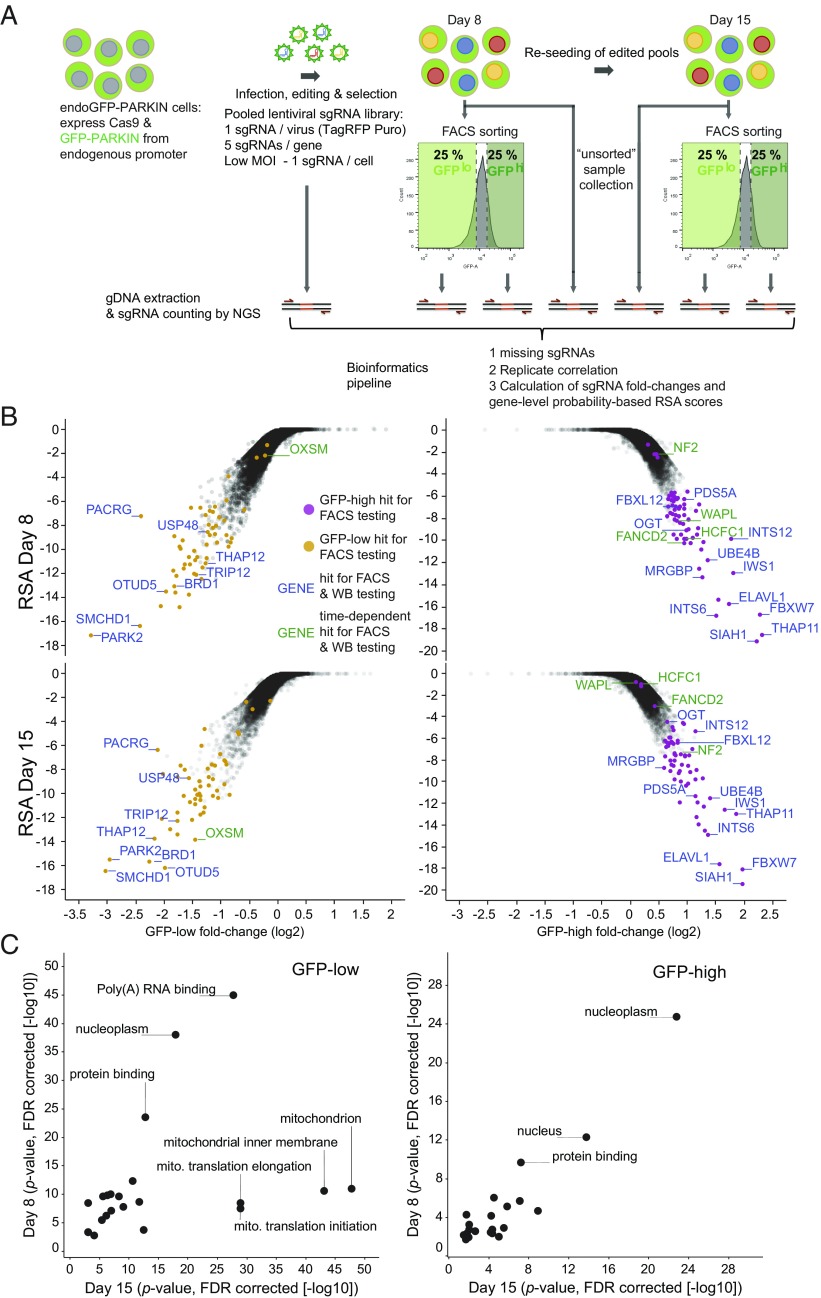
To identify transcriptional and posttranslational regulators of cellular PARKIN steady-state levels, we used the endoGFP-PARKIN cells in a genome-wide CRISPR/Cas9 knockout screen using GFP-fluorescence changes as a phenotypic read-out, similar to DeJesus et al. (19) (Fig. 2A). Cells were infected with a pooled lentiviral sgRNA library covering 18,360 genes (on average, five sgRNAs/gene) at a sufficiently low multiplicity of infection to bias for integration of a single lentiviral sgRNA cassette per cell. Following selection and purity assessment of infected pools using lentiviral puromycin resistance and TagRFP expression, respectively, pools were sorted for cells carrying the lowest 25% and the highest 25% of the GFP-fluorescence peak (“GFP-low” and “GFP-high” cell populations). The GFP-fluorescence profile of infected cells is similar to that of noninfected cells due to an overall low abundance of cells with phenotypes of interest; however, GFP-low and -high populations are enriched for cells expressing sgRNAs that negatively and positively regulate GFP-PARKIN, respectively (Fig. S2A). Cell sorting was performed at two time points, 8 and 15 d postinfection, to account for sgRNA-specific editing kinetics and time-dependent modulation of knockout-associated biological processes. Together with an “unsorted” sample to assess presort sgRNA library coverage, sorted GFP populations were subjected to genomic DNA extraction and subsequent sgRNA counting by next-generation sequencing (Fig. 2A).
Fig. 2.
A genome-wide CRISPR/Cas9 screen identifies genes affecting cellular GFP-PARKIN level. (A) Workflow of the screening process. endoGFP-PARKIN cells were transduced with a lentiviral sgRNA library targeting 18,360 genes with an average of five sgRNAs per gene in independent replicates (n = 2). Infection was performed at a low MOI to achieve one sgRNA/cell. After selection, the least (GFP-low) and most (GFP-high) GFP-fluorescent cells (25%) were FACS-sorted on day 8 and day 15 postinfection, and genomic DNA was recovered from the unsorted and sorted pools and subjected to NGS for sgRNA counting. sgRNA count data were processed to assess missing sgRNAs as a measure of library coverage as well as replicate correlation. Count ratios for individual sgRNAs from GFP-low and -high are expressed as fold-change values and plotted against gene-level probability-based RSA scores. (B) Analysis of GFP-low and -high samples reveals candidate hit genes. (Upper) Day 8 analysis. (Lower) Day 15 analysis. y axis: RSA scores; x-axis: sgRNA fold-changes in GFP-low vs. GFP-high (second strongest sgRNA). Candidate hit genes selected for individual confirmation are colored, and genes selected for WB analysis are labeled (Fig. 3B) (green labeling for time-point–specific genes). (C) Hypergeometric gene-set enrichment analysis was performed using the top 500-scoring genes of either the GFP-low or -high analysis. The P values denoting the significance of the enrichments from day 8 are plotted against day 15 (−log10). FDR, false discovery rate; gDNA, genomic DNA; MOI, multiplicity of infection; NGS, next-generation sequencing; WB, Western blot.
Our data showed high coverage of the sgRNA library with <0.25% missing sgRNAs across samples and a high correlation between replicates with r2 values >0.97 (Fig. S2B). Fold-change values were calculated from sgRNA counts in GFP-low and GFP-high populations as well as the “unsorted” population and the input library. Comparing fold-changes in the “unsorted” population, we observed that the essential gene set from KBM7 cells (24) was enriched among genes with strong negative fold-changes, suggesting that these genes are also essential in endoGFP-PARKIN cells and demonstrating efficient genome-wide editing in the pooled screening mode (Fig. S2C). Our data revealed additional candidate essential genes in these cells (Dataset S1).
For the gene-centric analysis of GFP-signals, we calculated redundant siRNA activity (RSA) scores as a probability-based measure of the performance of all sgRNAs of a gene with respect to the entire library (25). To account for effect size, RSA scores were plotted against fold-change values of the second strongest sgRNA (Fig. 2B and Dataset S2). As expected, PARK2 was recovered among the genes showing the strongest fold-change values and RSA scores in GFP-low cells at both time points, confirming efficient on-target editing (Figs. 1 B and C and 2B). Interestingly, sgRNAs targeting the PACRG gene—oriented head-to-head adjacent to PARK2 and sharing the same promoter region—were among the most strongly enriched in GFP-low cells, likely because the transgene promoter contains PACRG sgRNA targeting sequences (Fig. 2B). Thus, the phenotypic screen for regulators of GFP-PARKIN levels resulted in a high-quality dataset exemplified by the high coverage of the genome-wide sgRNA library and high correlation between replicates and of essential gene dropouts. Furthermore, the recovery of PARK2 among the strongest scoring genes suggests robust separation of signal from noise, a notion further supported by a comparable signal window in a GFP-based screen that used a similar setup (19).
A hypergeometric enrichment analysis of genes in GFP-low and -high cells revealed predominantly large gene sets in the 10 most significant sets on both time points, with the top-scoring sets comprising >1,000 genes (Fig. S2D). Enrichment of gene sets related to “nucleoplasm” and “nucleus” as well as other sets related to transcription and protein synthesis may point to GFP-PARKIN levels being particularly sensitive to modulation of biogenesis rather than turnover. “Fanconi anemia nuclear complex” represented one of the smallest enriched sets and has been linked to PARKIN and mitophagy (26). Half of the top 10 enriched processes in GFP-high cells showed similar P values for both time points; however, other processes were present only on day 8, indicating time-point–dependent scoring for some genes but not others (Fig. S2D). Indeed, in GFP-low cells, multiple gene sets related to mitochondrial function were enriched only on day 15, linking GFP-PARKIN level decline to mitochondrial dysfunction and suggesting detection of genes associated with different PARKIN-affecting processes at the two time points (Fig. 2C and Fig. S2D).
Individual sgRNA Testing of Screen Hits Results in a List of 53 Positive and Negative Regulators of GFP-PARKIN.
For downstream testing in lower throughput assays, gene scores were assigned (see Materials and Methods). Based on these scores, we selected genes for confirmation according to the following criteria: first, the top-scoring 25 genes from both GFP-low and -high cells from both time points were selected; second, genes in the top 100 common to both time points in the respective GFP population were selected; and finally, genes in the top 50 of one time point, but not in the top 1,000 of the other time points of the respective GFP population, were chosen as those genes display time-point dependency. In total, 114 genes were selected for confirmation and were considered as primary screen “hits” (Fig. 2B, marked genes).
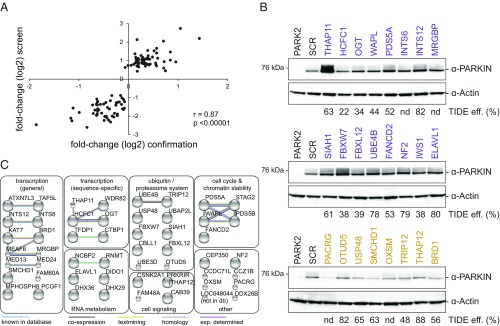
For individual confirmation of this list of genes, the strongest scoring sgRNA per gene was used to infect endoGFP-PARKIN cells individually, allowing for multiple integrations of lentiviral sgRNA expression cassettes. To avoid clonal effects, selected cell pools were analyzed for GFP-fluorescence by flow cytometry on days 8 and 15. sgRNAs selected from GFP-low cells exclusively resulted in GFP-signals lower than the SCR control (Fig. S3, gray highlighting). Likewise, sgRNA hits from GFP-high cells showed predominantly higher GFP-signals (Fig. S3). Comparing primary screen enrichment values for a given sgRNA with GFP-fluorescence changes in individual sgRNA experiments revealed a highly significant correlation, demonstrating the robustness of the screening approach (Fig. 3A).
Fig. 3.
Confirmation of primary screen hit genes. (A) High correlation of individual sgRNA performance in the primary pooled screen and individual confirmation experiments. Day 8 fold-changes (log2) for the strongest scoring sgRNAs in the primary screen were plotted against fold-changes (log2) as determined in Fig. S3. Pearson’s correlation and corresponding significance values are shown. n = 108. (B) Lysates from endoGFP-PARKIN cells individually expressing the strongest scoring sgRNAs for 24 hits for 15 d were assessed for GFP-PARKIN levels and for CRISPR/Cas9 editing efficiencies by TIDE. (C) Fifty-three positive and negative regulators subjected to functional gene network analysis. Top and bottom 25 hits confirmed by flow cytometry on day 8 (Fig. S3), together with additional hits validated in B were used as input query for STRING database analysis of known and predicted protein-protein interactions (physical and functional). STRING gene annotations were used to group hits. The highest confidence cutoff was used (STRING V10.0). Large spheres, protein structure known; small spheres, no known protein structure. db, database; eff., efficiency; exp., experimentally; nd, not determined.
To confirm that the knockout of selected hits affects steady-state GFP-PARKIN abundance, we assessed protein levels for a subset of the hits. The best sgRNAs targeting 24 randomly selected hits, 16 from GFP-high and eight from GFP-low cells, were lentivirally delivered to endoGFP-PARKIN cells, and protein lysates were probed with PARKIN-specific antibodies 15 d postinfection (Fig. 3B). In most cases, CRISPR knockout of selected hits from GFP-high or GFP-low cells resulted in an increase or decrease, respectively, of GFP-PARKIN compared with the control, fully consistent with the data from FACS experiments (Fig. 3B). In some cases, the effects were mild, which can be partially explained by inefficient editing as assessed by the TIDE (Tracking of Indels by Decomposition) sequence trace composition assay (27) (Fig. 3B).
We defined a list of regulators by selecting the top-scoring 25 GFP-low and -high hits in the day 8 FACS analysis (those genes show a very high correlation with the primary screen, r = 0.94) and additionally included genes that were tested in Western blot analyses. This resulted in a list of 53 regulators, which we subjected to STRING functional gene network analysis to reveal their biological function and association with each other (Fig. 3C and Dataset S2) (28). STRING analysis allowed grouping of hits in several categories covering a range of biological processes and revealed genes that have not been confidently associated with each other in previous studies as well as others known to be in functional gene networks (Fig. 3C). The most-populated categories contained genes linked to general and sequence-specific transcription and suggests an important role in PARKIN steady-state–level control. Genes linked to the categories general transcription, RNA metabolism, as well as cell cycle and chromatin stability also scored in the gene-set enrichment analysis of the screen and suggest on a genome-wide scale that PARKIN levels are sensitive to changes in these processes (Fig. S2D).
THAP11 Is Part of a Functional Network Mediating Transcriptional Repression of PARK2.
A specific effect on PARKIN levels may be exerted by genes in the “ubiquitin/proteasome system” and the “sequence-specific transcription” category, which includes a functional network comprising THAP11, HCFC1, OGT, and WDR82 (Fig. 3C). HCFC1 and WDR82 were robustly identified in the primary screen on day 8, and both THAP11 as well as OGT scored significantly at both time points in the primary screen and in confirmation experiments (Figs. 2B and 3B and Fig. S3). While WDR82 was identified in the GFP-low cells, increased GFP-PARKIN levels upon targeting THAP11, HCFC1, or OGT suggests that these genes collaborate in the negative regulation of GFP-PARKIN levels by repressing transcription from the endogenous PARK2 promoter.
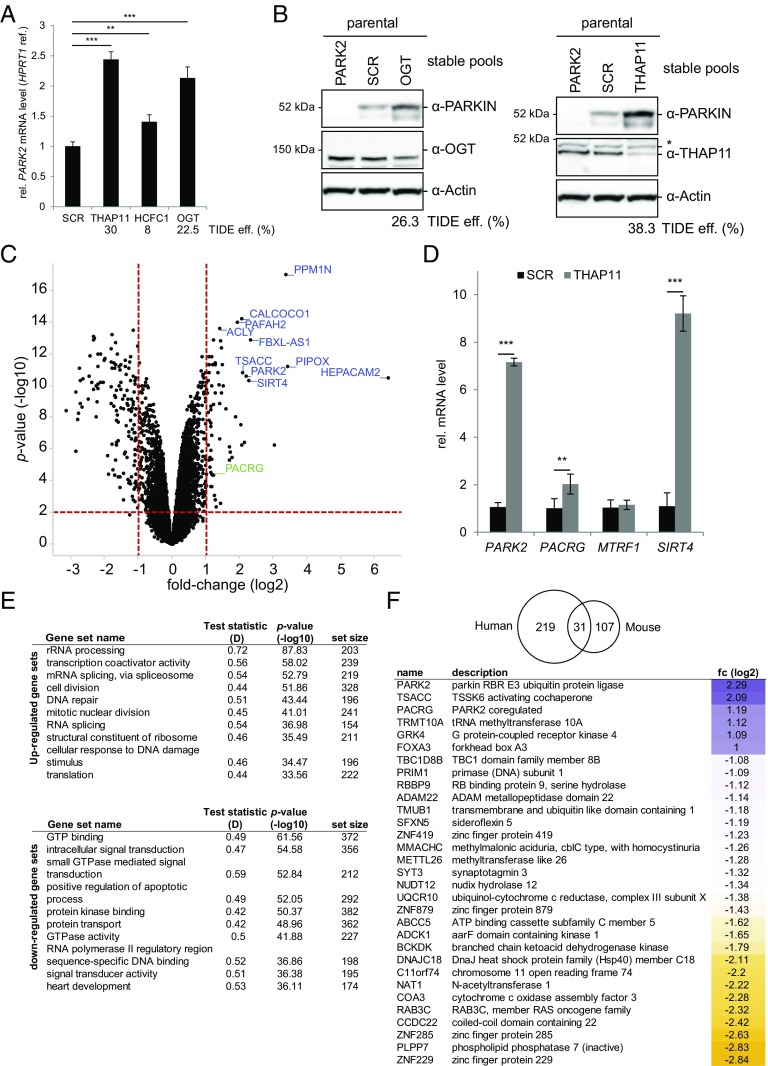
To test this hypothesis, parental cells expressing only endogenous PARKIN were targeted for THAP11, HCFC1, or OGT, selected and cultured beyond day 15 to generate stable cell pools, and subjected to qPCR to measure PARK2 mRNA levels. A significant increase in PARK2 mRNA levels was observable upon CRISPR knockout of any one of the genes targeted with THAP11 and OGT sgRNAs increasing PARK2 mRNA more than twofold (Fig. 4A). The small increase in HCFC1-targeted stable cells reflects a very low knockout percentage as assessed by TIDE, likely due to drift in the stable pool and consistent with the time-point–dependent scoring of HCFC1 in the primary screen (Fig. 4A). A more pronounced increase in PARK2 mRNA levels was observable on day 8 postinfection compared with stable cells (Fig. S4A). Regardless, the change in transcript levels was also reflected in protein levels, with endogenous PARKIN accumulating in stable cells lacking OGT or THAP11 (Fig. 4B). Thus, transcriptional de-repression leads to an increase in the steady state of endogenous PARKIN protein levels.
Fig. 4.
THAP11 is part of a functional gene network mediating transcriptional repression of endogenous PARKIN in JumpIN cells. (A) THAP11, HCFC1, and OGT negatively regulate PARK2 mRNA level. PARK2 mRNA level relative to HPRT1 was determined by qPCR on total RNA extracted from stable cell pools of parental JumpIN cells expressing nontargeting (SCR), THAP11- (second strongest sgRNA from screen), HCFC1-, or OGT-specific sgRNAs (strongest sgRNA in screen); n = 3; error bars indicate SDs. **P < 0.01, ***P < 0.001. Average CRISPR/Cas9 editing efficiencies were assessed by TIDE. (B) Targeting of either OGT (Left) or THAP11 (Right) negatively affects endogenous PARKIN protein level. Stable pools of parental cells as in A were analyzed by WB using antibodies indicated. *Nonspecific cross-reaction. CRISPR/Cas9 editing efficiencies were assessed by TIDE. (C and D) Parental cells were infected with THAP11-specific (second strongest sgRNA in screen) or SCR sgRNAs in triplicates for 15 d. Cell extracts were subjected to RNAseq (C), qPCR (D), or WB to assess THAP11 depletion and PARKIN protein elevation (Fig. S4B). In C, transcript fold-changes of THAP11-targeted vs. SCR samples (x-axis, log2) were plotted against P values (y axis, −log10). For hit calling, thresholds were set to <−1 and >1 log2 fold-changes and P < 0.01. The 10 genes most strongly repressed are labeled in blue. PACRG is also repressed by THAP11 (green). (D) qPCR using Taqman probes indicated, scored relative to GAPDH or HPRT1 (PACRG) mRNA, normalized to SCR control. n = 3; error bars indicate SDs. **P < 0.01, ***P < 0.001. (E) Gene-set enrichment analysis suggests modulation of diverse biological processes upon THAP11 depletion. Enrichment analysis for RNAseq using a weighted one-sided Kolmogorov-Smirnov test; test statistic D and negative logarithm (base 10) of adjusted P values are reported. The 10 most enriched sets among up-regulated (Upper) or down-regulated (Lower) transcripts are shown. (F) Comparison of the RNAseq dataset from THAP11-depleted JumpIN cells with data from Poché et al. (29), showing numbers of transcripts regulated by THAP11 in either one or both experiments [cutoff <−1 and >1 fold-changes (log2); P < 0.01]; genes commonly regulated by THAP11 are listed. eff., efficiency; fc, fold-change; ref., reference transcript; rel., relative; WB, Western blot.
Genome-Wide Investigation of THAP11-Mediated Transcriptional Regulation Identifies Transcripts with Diverse Functions and the PARK2 Genomic Locus as a Prime Target.
Recently, Poché et al. (29) reported that THAP11 depletion results in elevated PARK2 mRNA levels (among other genes) in the developing mouse retina, which was hypothesized to be caused by down-regulated mitochondrial genes followed by mitochondrial dysfunction in these samples. Data from our screen, however, suggest that targeting mitochondrial genes results in decreased PARKIN levels and motivates further investigation of THAP11-dependent transcripts in JumpIN cells (Fig. 2C and Fig. S2D). We therefore analyzed the gene expression profile of THAP11-depleted JumpIN cells using RNAseq (Fig. S4B). On day 15 postinfection with THAP11-directed sgRNAs, the expression of 92 genes was enhanced and 158 genes repressed compared with the control sgRNA-infected cells (more than twofold change in absolute value, 1% false discovery rate; Fig. 4C and Dataset S3). PARK2 was found among the most strongly up-regulated genes.
Gene-set enrichment analysis revealed diverse biological processes associated with the modulated genes (Fig. 4E). More specifically, significantly enriched gene sets for up-regulated transcripts are mostly implicated in protein biosynthetic processes as well as in cell division and DNA repair. In contrast, down-regulated transcripts showed significant enrichment of diverse gene sets related to GTPase-mediated signal transduction as well as positive regulation of apoptosis and protein kinase binding. Mitochondria-related transcripts were not enriched upon THAP11 depletion. While we confirmed some of the mitochondria-related transcripts reported to be down-regulated in mouse samples, other transcripts were unaffected or even oppositely regulated in JumpIN cells, including MTRF1 and SIRT4, respectively, which were confirmed by qPCR (Fig. 4D and Fig. S4C) (29). These findings suggest that THAP11 depletion has different effects in the developing mouse retina and human JumpIN cells, which is supported by a limited overlap of these data sets (Fig. 4F).
While most of the identified transcripts seem to be specific to either one of the two data sets, a set of 31 transcripts was commonly affected in the two studies, with PARK2 being the most strongly up-regulated (Fig. 4F). This common set also includes PACRG, which is adjacent to PARK2 in the genome sharing a bidirectional promoter, and was confirmed by qPCR (Fig. 4D). Unbiased analysis of the promoter sequences of the 31 commonly regulated genes revealed the most significant motif located primarily between −200 and 0 nucleotides from predicted transcription start sites (Fig. S4D). This sequence motif is present in the PARK2-PACRG promoter (Fig. S4E) and almost identical to the THAP11 motif identified in mouse retinal cells (29), confirming that THAP11 is a key regulator of this set of genes in the two disparate cell types. Targeting the motif in the PARK2 promoter in endoGFP-PARKIN cells using specific sgRNAs resulted in a bimodal GFP-fluorescence profile, with most cells showing either elevated or decreased GFP-PARKIN compared with the control, indicating the importance of the motif in PARKIN expression (Fig. S4 E and F). The bimodal profile is consistent with the notion that, depending on the CRISPR, PARK2 repression is inhibited by specific motif mutations in cells with elevated GFP-PARKIN, while PARK2 expression is inhibited by extensive promoter editing in cells with decreased GFP-PARKIN levels. Taken together, RNAseq revealed diverse THAP11-dependent transcripts in JumpIN cells and indicates that the PARK2 promoter is a prime target for THAP11-mediated regulation.
THAP11-Mediated Transcriptional Repression of PARK2 Regulates pUb Accumulation in Neuronal Cells.
To address whether THAP11-mediated transcriptional repression of PARK2 impacts pUb accumulation, we initially examined THAP11-depleted endoGFP-PARKIN cells. As before, THAP11 depletion resulted in elevated levels of GFP-PARKIN compared with the SCR control (Fig. 5A). Upon treatment with A/O for 6 h, there was an increased accumulation of pUb in cells lacking THAP11 compared with control cells (Fig. 5A). In line with previous time-course experiments, treatment for 24 h did not reveal obvious differences in pUb accumulation between PARKIN- and THAP11-depleted nor control samples (Fig. 5A and Fig. S1 C and D). Upon treatment with A/O, PINK1 stabilization was observable in samples from all genotypes (Fig. 5A). However, we noted declining levels of OGT in mitophagy-triggering conditions, with kinetics that appeared to be altered upon THAP11 depletion, opening up the possibility of additional THAP11- and OGT-mediated PARK2 control during mitophagy. By using a different THAP11-directed sgRNA and a different α-pUb antibody we confirmed an enhanced accumulation of pUb upon treatment with A/O for 6 h in immunofluorescence analyses and observed pUb clustering in the perinuclear region where mitochondrial aggregates accumulate at early stages of mitophagy (30) (Fig. 5B).
Fig. 5.
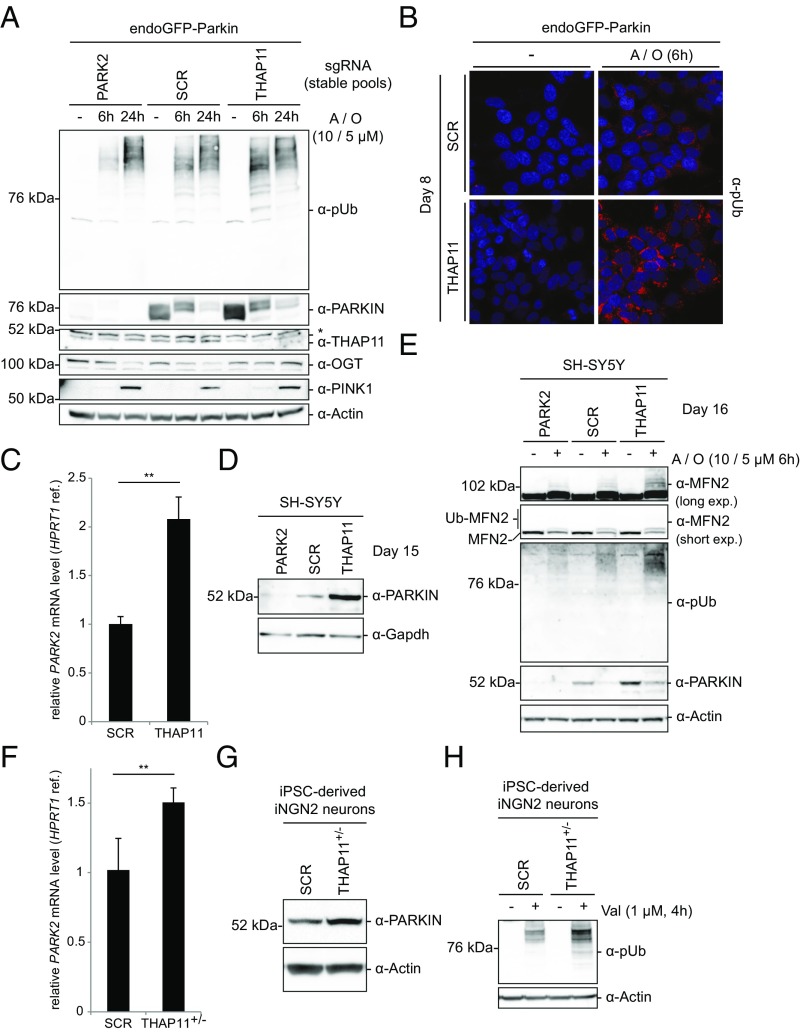
THAP11 modulates endogenous PARKIN abundance and damage-dependent mitophagy priming in endoGFP-PARKIN cells, neuroblastoma cells, and iPSC-derived iNGN2 neurons. (A) Stable pools of endoGFP-PARKIN cells expressing either THAP11-specific (second strongest sgRNA in screen), PARK2-specific, or nontargeting (SCR) sgRNAs were treated with A/O at concentrations shown or DMSO and analyzed at time points indicated by WB using indicated antibodies. *An unspecific cross-reaction of the THAP11 antibody. (B) THAP11 targeting positively affects damage-induced perinuclear pUb accumulation. endoGFP-PARKIN cells expressing THAP11-specific (strongest sgRNA in screen) or SCR sgRNAs for 8 d were treated with A/O or DMSO and analyzed by confocal immunofluorescence microscopy using pUb-specific antibodies (red). Nuclei were Hoechst stained (blue). (C) Extracts from SH-SY5Y Cas9+ cells expressing THAP11-specific (second strongest sgRNA in screen) or SCR sgRNAs for 15 d were analyzed by qPCR using probes indicated (C) or WB using antibodies indicated (D). For C, n = 3; error bars indicate SDs. (E) SH-SY5Y Cas9+ cells expressing sgRNAs as in C and D were treated with A/O or DMSO on day 16 postinfection and analyzed by WB using indicated antibodies. (F–H) iPSC iNGN2 THAP11+/− and iPSC iNGN2 SCR cells were differentiated for 10 d and extracts analyzed by qPCR using Taqman probes indicated (n = 4; error bars indicate SDs; **P < 0.01) (F), or by WB using antibodies indicated (G and H). In H, cells were treated with DMSO or valinomycin 4 h before harvesting. ref., reference transcript; WB, Western blot.
Given the relationship of PARK2 to Parkinson’s disease, where the primary cell types affected are neurons, we assessed the effects of THAP11 knockout on PARKIN levels and pUb accumulation in neuronal cell types. As in previous experiments, targeting THAP11 in SH-SY5Y neuroblastoma cells using the second-strongest sgRNA from the primary screen resulted in accumulation of both endogenous PARK2 mRNA and protein (Fig. 5 C and D). In addition, following mitochondrial damage, pUb accumulation was increased in cells where THAP11 was targeted, compared with cells expressing a control sgRNA (Fig. 5E). Notably, endogenous PARKIN protein levels declined upon mitochondrial damage, as observed in endoGFP-PARKIN cells (Fig. S1 C and D). While we only observed a mild effect on damage-induced MFN2 ubiquitination and degradation in PARK2-targeted samples, possibly due to other E3 ligases targeting MFN2 in mitophagy (31, 32), damage-induced ubiquitinated MFN2 was enhanced in neuroblastoma cells targeted for THAP11 compared with controls.
Finally, we addressed THAP11-mediated transcriptional repression of PARK2 and pUb accumulation in postmitotic human neuronal noncancer cells. THAP11 was targeted in human iPSCs carrying iNGN2 for rapid single-step induction of functional neurons (33) (iPSC iNGN2 cells) (Fig. S5 A and B). TIDE analysis of iPSC iNGN2 cells infected with viruses allowing expression of a THAP11-directed sgRNA revealed monoallelic editing in those cells (iPSC iNGN2 THAP11+/−) (Fig. S5C). To address the effect of THAP11 editing on PARKIN in postmitotic neurons, iPSC iNGN2 THAP11+/− cells or iPSC iNGN2 cells expressing a control sgRNA (iPSC iNGN2 SCR) were differentiated for 10 d. Transcriptional profiling of iNGN2 THAP11+/− and SCR neurons revealed highly similar expression levels of a panel of neuronal markers, indicating that monoallelic THAP11 CRISPR had no major effect on neuronal differentiation (Fig. S5D). However, both PARK2 mRNA and PARKIN protein levels were significantly increased in iNGN2 THAP11+/− neurons, demonstrating THAP11-mediated transcriptional repression in those cells (Fig. 5 F and G). Treating iNGN2 THAP11+/− neurons with the mitophagy trigger valinomycin for 4 h resulted in a pronounced increase in pUb accumulation compared with iNGN2 SCR neurons (Fig. 5H). Together, these data provide evidence that THAP11-mediated transcriptional repression occurs in differentiated human neuronal cells and demonstrate that PARKIN levels are limiting in an endogenous setting for early damage-induced pUb accumulation.
Discussion
Previous genetic screening efforts provided insights into the regulation of PINK1/PARKIN-mediated mitophagy (12–15). In these screens, high-level overexpression of GFP-PARKIN in cells naturally deficient of PARKIN, however, impeded the identification of regulators of PARKIN abundance. Here, we show that cellular PARKIN levels regulate pUb accumulation. In a screening campaign employing cells expressing GFP-PARKIN from the endogenous PARK2 promoter and CRISPR/Cas9 technology, we identified and confirmed hits resulting in a list of 53 positive and negative steady-state–level regulators. Our functional analysis of the transcriptional repressor THAP11 highlights the impact of endogenous PARKIN-level regulation on pUb accumulation in multiple cell types, including postmitotic iPSC-derived iNGN2 neurons.
It has been recently shown that PARKIN participates in a feed-forward pathway to amplify pUb accumulation during mitophagy (9). Our work reveals the critical role of cellular PARKIN levels in modulating the amplification of the pUb output: varying PARK2 copy number or endogenous PARKIN levels by means of THAP11 targeting revealed coupling of PARKIN levels and early pUb accumulation in a cell type-independent manner.
pUb is critical for the recruitment of autophagy receptors on ubiquitinated mitochondria (8). Concomitant with receptor recruitment, the TBK1 kinase is activated and TBK1-mediated receptor phosphorylation promotes receptor ubiquitin chain binding and further TBK1 activation, thereby revealing another feed-forward mechanism (8, 34, 35). The presence of two consecutive signal amplifications indicates the importance of rapid preparation of dysfunctional mitochondria for autophagosomal engulfment, and highlights the significance of PARKIN levels in the entire process, as efficient TBK1 activation is also dependent on PARKIN expression (8, 34, 35). Pathogenic mutations in TBK1 as well as PARK2 have been identified in amyotrophic lateral sclerosis-frontotemporal dementia and Parkinson’s disease, respectively, thereby encouraging speculation that low signal amplification upon mitochondrial damage is of pathophysiological importance (10, 36, 37). This speculation is further supported by a recent study reporting compromised pUb signaling in dopaminergic neurons containing PARK2 or PINK1 mutations (38). Future studies will reveal whether and how variants of hits identified in the current screen modulate disease risk—for example, TRIP12 (Figs. 2B and 3B and Fig. S3)—for which SNPs have been linked to Parkinson’s disease (39, 40).
Bioinformatic analysis of genes found in our screen identified gene sets related to transcription and protein biogenesis. This is consistent with the notion that PARKIN steady-state levels are particularly sensitive to the modulation of synthesis rather than turnover. Our analysis of THAP11 depletion phenotypes adds weight to this concept as released transcriptional repression is not compensated by protein turnover, resulting in PARKIN protein accumulation. Furthermore, our data suggest that removal of dysfunctional mitochondria by mitophagy is under substantial transcriptional control via PARKIN-level adjustment. Vice versa, PARKIN level appears to be linked to the functional state of mitochondria: PARKIN depletion was observed in acute mitochondrial damage conditions, and targeting of mitochondrial genes by CRISPR knockout likewise resulted in a decrease in GFP-PARKIN. Whether the latter is due to mitochondrial dysfunction-dependent triggering of mitophagy concomitant with PARKIN degradation remains speculative at this point (22). Thus, there appear to be two main processes accounting for cellular PARKIN levels: gene expression and mitochondrial dysfunction, respectively. It will be interesting to further investigate other hits which cannot be obviously attributed to these two processes, including proteins related to the ubiquitin/proteasome system, such as SIAH1 (Figs. 2B and 3 B and C and Fig. S3) present in Lewy bodies (41).
A previous chemogenomic screen identified epigenetic agents, drugs controlling cholesterol biosynthesis, and JNK inhibitors as chemical classes up-regulating PARK2 transcription (42). JNK family kinases and cholesterol biosynthesis genes did not score in the current screen, which may be explained by nonselectivity of compounds in that screen and different experimental setups. However, epigenetic regulators including the bromodomain proteins BRD1 and BRD8 were among confirmed hits supporting the notion that PARK2 transcription is regulated by this class of proteins (42). Concerning other confirmed hits, a subunit of the CK2 complex (CSNK2A1) and FBXW7 (Figs. 2B and 3 B and C and Fig. S3) were also identified in a previous genetic screen as positive regulators of mitophagy (13). In light of the present data, PARKIN depletion upon CK2 loss would be a logical explanation for the observed mitophagy defect.
The role of FBXW7 in mitophagy is unclear, and data presented here do little to clarify this situation, as FBXW7 appears to negatively regulate PARKIN, an observation at odds with its described role as a positive regulator of mitophagy. Clearly, further work is needed to understand the role of FBXW7 in mitophagy; however, FBXW7 and PARKIN appear to have roles beyond mitophagy, in cell cycle control. Both proteins interact to cooperate in cyclin E degradation and PARKIN regulates mitosis and genomic stability (43, 44). Interestingly, Lee et al. (44) showed that PARKIN levels change during the cell cycle, providing a possible explanation for the identification of cohesin-regulating genes in the screen performed here (Figs. 2B and 3 B and C and Fig. S3), which have been linked to cell cycle progression (45). Given the importance of PARKIN levels for pUb accumulation, it will be interesting to assess mitophagy at different stages of the cell cycle. Thus, the current data are of relevance not only for mitophagy research but also for research regarding cell cycle biology and cancer where PARK2 gene dosage reduction is a common event (43, 46).
THAP11 (also known as Ronin) contains a Thanatos-associated protein (THAP) domain, an atypical zinc finger motif with sequence-specific DNA binding activity, and was reported to be a transcriptional repressor (47). THAP11 requires HCFC1 (also known as HCF-1) to be functionally active, and HCFC1, in turn, is proteolytically matured by the O-GlcNAc transferase OGT (48, 49). Recruitment of an HCFC1-containing complex enables THAP11 to either up-regulate or repress target genes (49). Data presented here are in line with THAP11 acting in concert with HCFC1 downstream of OGT in the repression of PARK2 transcription. Furthermore, profiling of THAP11-depleted cells revealed both up- and down-regulated transcripts, consistent with a versatile role of THAP11 in transcriptional regulation (49). THAP11 was reported to be essential for mouse embryonic stem cell self-renewal and cell-growth regulation in human colon cancer cells (47, 49, 50). Data presented here indicate that THAP11 transcriptional programs are primarily cell- and/or species-specific. However, the identification of 31 THAP11-dependent genes in both mouse samples and human cells indicates some conservation of THAP11-mediated regulation and points to a THAP11 core transcriptional program that is not cell- and/or species-restricted. In this regard, THAP11-mediated regulation of PARK2, the gene most strongly repressed among the 31, is of particular interest as it represents a mechanism to regulate pUb accumulation in all cell types tested, and suggests that regulation of mitophagy is a core function of THAP11. Whether the spatially and developmentally restricted THAP11 expression contributes to time- and tissue-specific control of PARKIN, mitophagy, and concomitant processes remains an interesting question at this point (8, 51). Finally, both PACRG and PARK2 were reported to be up-regulated in response to oxidative stress and a ∼300-bp fragment of the common promoter has been determined to mediate the response (20). As this fragment contains the THAP11-binding site we are tempted speculate that THAP11-mediated repression of PARK2 (and PACRG) is released upon oxidative stress, a prominent susceptibility factor for the neurons affected in Parkinson’s disease.
Materials and Methods
endoGFP-PARKIN cells were obtained by selecting a clone of JumpIN TI 293 cells stably expressing Cas9 (parental cells) additionally transfected with a pcDNA3.1-based plasmid expressing EGFP-PARKIN from 4.5 kb of the endogenous PARK2 promoter. For flow cytometry, endoGFP-PARKIN cells were harvested, resuspended in PBS supplemented with 2 mM EDTA, passed through a 40-µm filter, and analyzed on a BD FACSCanto II or a BD FACSAria Fusion. endoGFP-PARKIN cells transduced with lentiviruses were fixed using BD Cytofix (diluted 1:1 in PBS; BD Biosciences) according to the manufacturer’s instructions, resuspended in BD Stain buffer (BD Biosciences) supplemented with 2 mM EDTA, and passed through a 40-µm filter before analysis by flow cytometry. Detailed methodology for cell culture and cell line generation, pooled CRISPR screening, confirmation of individual sgRNAs, TIDE editing efficiency assays, qPCR, RNA sequencing, DNA motif enrichment analysis, cell reprogramming, iPSC maintenance and differentiation, generation of iNGN2 iPSCs, generation of iNGN2 SCR iPSCs and Thap11+/− iPSCs, differentiation of iNGN2 neurons, Western blotting, confocal immunofluorescence microscopy, and statistical analyses are described in SI Materials and Methods. No experiments requiring informed consent or institutional committee approval were performed during this study.
Supplementary Material
Acknowledgments
We thank Jean-Claude Martinou for advice and critical reading of the manuscript, Dominic Hoepfner for CRISPR screen support, Isabelle Fruh for iPSC characterization, Thierry Doll for cell reprogramming activities, Uwe Plikat for data infrastructure, and Tewis Bouwmeester as well as Beat Nyfeler for helpful discussions.
Footnotes
Conflict of interest statement: All authors are employees of, and may own shares of, Novartis Pharma AG.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711023115/-/DCSupplemental.
References
- 1.Rugarli EI, Langer T. Mitochondrial quality control: A matter of life and death for neurons. EMBO J. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matheoud D, et al. Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell. 2016;166:314–327. doi: 10.1016/j.cell.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 3.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazlauskaite A, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 7.Rose CM, et al. Highly multiplexed quantitative mass spectrometry analysis of ubiquitylomes. Cell Syst. 2016;3:395–403.e4. doi: 10.1016/j.cels.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ordureau A, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitada T, et al. Mutations in the Parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 11.Valente EM, et al. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am J Hum Genet. 2001;68:895–900. doi: 10.1086/319522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasson SA, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivatt RM, et al. Genome-wide RNAi screen identifies the Parkinson disease GWAS risk locus SREBF1 as a regulator of mitophagy. Proc Natl Acad Sci USA. 2014;111:8494–8499. doi: 10.1073/pnas.1321207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre V, et al. Genome-wide RNAi screen identifies ATPase inhibitory factor 1 (ATPIF1) as essential for PARK2 recruitment and mitophagy. Autophagy. 2013;9:1770–1779. doi: 10.4161/auto.25413. [DOI] [PubMed] [Google Scholar]
- 15.McCoy MK, Kaganovich A, Rudenko IN, Ding J, Cookson MR. Hexokinase activity is required for recruitment of Parkin to depolarized mitochondria. Hum Mol Genet. 2014;23:145–156. doi: 10.1093/hmg/ddt407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evers B, et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 17.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeJesus R, et al. Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. Elife. 2016;5:e17290. doi: 10.7554/eLife.17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YX, Muqit MM, Latchman DS. Induction of Parkin expression in the presence of oxidative stress. Eur J Neurosci. 2006;24:1366–1372. doi: 10.1111/j.1460-9568.2006.04998.x. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakovic A, et al. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: Study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J Biol Chem. 2013;288:2223–2237. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.König R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 26.Sumpter R, Jr, et al. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165:867–881. doi: 10.1016/j.cell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poché RA, et al. RONIN is an essential transcriptional regulator of genes required for mitochondrial function in the developing retina. Cell Rep. 2016;14:1684–1697. doi: 10.1016/j.celrep.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu M, et al. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell. 2013;24:1153–1162. doi: 10.1091/mbc.E12-08-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yun J, et al. MUL1 acts in parallel to the PINK1/Parkin pathway in regulating mitofusin and compensates for loss of PINK1/Parkin. Elife. 2014;3:e01958. doi: 10.7554/eLife.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter B, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirulli ET, et al. FALS Sequencing Consortium Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freischmidt A, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 38.Shiba-Fukushima K, et al. Evidence that phosphorylated ubiquitin signaling is involved in the etiology of Parkinson’s disease. Hum Mol Genet. 2017;26:3172–3185. doi: 10.1093/hmg/ddx201. [DOI] [PubMed] [Google Scholar]
- 39.Fung HC, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: First stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 40.Simón-Sánchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liani E, et al. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson’s disease. Proc Natl Acad Sci USA. 2004;101:5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasson SA, et al. Chemogenomic profiling of endogenous PARK2 expression using a genome-edited coincidence reporter. ACS Chem Biol. 2015;10:1188–1197. doi: 10.1021/cb5010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong Y, et al. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat Genet. 2014;46:588–594. doi: 10.1038/ng.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SB, et al. Parkin regulates mitosis and genomic stability through Cdc20/Cdh1. Mol Cell. 2015;60:21–34. doi: 10.1016/j.molcel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tedeschi A, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–568. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta A, et al. PARK2 depletion connects energy and oxidative stress to PI3K/Akt activation via PTEN S-nitrosylation. Mol Cell. 2017;65:999–1013, e7. doi: 10.1016/j.molcel.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dejosez M, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capotosti F, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 49.Dejosez M, et al. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010;24:1479–1484. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker JB, Palchaudhuri S, Yin H, Wei J, Chakravarti D. A transcriptional regulatory role of the THAP11-HCF-1 complex in colon cancer cell function. Mol Cell Biol. 2012;32:1654–1670. doi: 10.1128/MCB.06033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong G, et al. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111:3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuierer S, Roma G. The exon quantification pipeline (EQP): A comprehensive approach to the quantification of gene, exon and junction expression from RNA-seq data. Nucleic Acids Res. 2016;44:e132. doi: 10.1093/nar/gkw538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie ME, et al. limma Powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medina-Rivera A, et al. RSAT 2015: Regulatory sequence analysis tools. Nucleic Acids Res. 2015;43:W50–W56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey TL, Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128. doi: 10.1093/nar/gks433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.