Significance
Severe heat stress causes massive protein loss by aggregation ultimately causing cell death. Cellular survival relies on protein disaggregation mediated by the Hsp70-ClpB (Hsp100) bichaperone system in most bacteria. Pseudomonas aeruginosa additionally codes for two stand-alone ClpG disaggregases, which had been acquired by horizontal gene transfer by the species and most abundant clone C strains, respectively. These ClpG disaggregases largely contribute to the resolution of protein aggregates to confer superior heat tolerance partially replacing the DnaK-ClpB system.
Keywords: protein disaggregation, AAA+ protein, heat tolerance, mobile genetic element, Hsp100
Abstract
AAA+ disaggregases solubilize aggregated proteins and confer heat tolerance to cells. Their disaggregation activities crucially depend on partner proteins, which target the AAA+ disaggregases to protein aggregates while concurrently stimulating their ATPase activities. Here, we report on two potent ClpG disaggregase homologs acquired through horizontal gene transfer by the species Pseudomonas aeruginosa and subsequently abundant P. aeruginosa clone C. ClpG exhibits high, stand-alone disaggregation potential without involving any partner cooperation. Specific molecular features, including high basal ATPase activity, a unique aggregate binding domain, and almost exclusive expression in stationary phase distinguish ClpG from other AAA+ disaggregases. Consequently, ClpG largely contributes to heat tolerance of P. aeruginosa primarily in stationary phase and boosts heat resistance 100-fold when expressed in Escherichia coli. This qualifies ClpG as a potential persistence and virulence factor in P. aeruginosa.
Maintenance of protein homeostasis is essential for all living organisms from bacteria to humans. Crucial components of the protein quality control system are diverse and include degradation and refolding factors that are preventing the accumulation of misfolded proteins (1, 2). Severe stress conditions can overwhelm these protective activities, leading to massive aggregation of cellular proteins. The loss of proteins by aggregation impedes multiple cellular processes and can ultimately lead to cell death. Additionally, protein aggregates might cause toxicity by depleting cellular factors, including molecular chaperones. Cellular survival during aggregation causing adverse growth conditions therefore relies on the solubilization of aggregated proteins (3). This specific protein quality control activity is particularly important for bacteria, fungi, and plants as they are routinely exposed to heat (4, 5).
The major front line against severe heat stress is formed by ring-forming, hexameric AAA+ disaggregases that exert an ATP-fueled translocation activity enabling them to extract single polypeptides from the aggregate by threading them through their central pore. Tandem AAA+ domains, mediating ATP binding and hydrolysis, provide the energy for the disaggregation process. Notably, all characterized AAA+ disaggregases do not function on their own but strictly require cooperation with partner proteins. These partners (also called adaptors) target AAA+ disaggregases to the aggregate surface and concurrently stimulate AAA+ ATPase and threading activities. Variable N-terminal (N) and inserted middle (M) domains of AAA+ disaggregases mediate specific binding partners and are therefore essential for protein disaggregation.
The central disaggregation machinery of most bacteria, fungi, and plants is formed by an aggregate-binding Hsp70 protein (DnaK in bacteria) that acts as a partner of central homologous disaggregases: ClpB (bacteria), Hsp104 (fungi), or Hsp101 (plants). This bichaperone system is most important for conferring heat tolerance (4–8). Some Gram-positive bacteria (e.g., Bacillus subtilis) lack a ClpB homolog and protein disaggregation is taken over by the AAA+ chaperone ClpC (9, 10). ClpC activity essentially depends on cooperation with adaptor proteins, which function mechanistically similar to Hsp70 by recruiting substrates and stimulating ClpC ATPase activity (11, 12). The role of Clp disaggregases extends beyond heat tolerance though, being involved in e.g., virulence, persistence, oxidative stress, and antibiotic resistance (6, 7, 13–15). Although the molecular basis of virulence phenotypes mainly remains to be unraveled, ClpB has been shown to be required for type VI secretion system functionality in Francisella tularensis (16).
Pseudomonas aeruginosa is a highly successful opportunistic pathogen, which causes a broad range of infections (17). A worldwide dominating group of P. aeruginosa strains is the clone C cluster, with members prevalent in the environment and in a broad spectrum of acute and chronic diseases (18–22). Although extensive inter- and intraclonal genome variability has been observed, the genetic determinants underlying the successful spread of P. aeruginosa clone C strains are poorly understood.
The P. aeruginosa core genome and a transmissible locus for protein quality control 1 (TLPQC-1) present in clone C strains code for two horizontally transferred AAA+ disaggregases, termed ClpG/ClpGGI (this work and ref. 23). TLPQC-1 and ClpG homologs have previously been shown to confer heat resistance in various environmental and clinically relevant bacteria (23, 24). Here we show that ClpG/ClpGGI work independently from partner proteins in contrast to previously characterized AAA+ disaggregases. ClpG/ClpGGI exhibit high disaggregation activity in vitro, confer superior heat tolerance to P. aeruginosa clone C strains, and upon expression in Escherichia coli boost protein disaggregation and survival during severe heat stress independent of aggregate-binding Hsp70. Our work leads to the hypothesis that horizontal transfer of ClpG accompanied the development of the species P. aeruginosa to increase resistance toward adverse stress conditions, and acquisition of ClpGGI contributed to the worldwide transmission of the clone C population in patients and the aquatic habitat.
Results
P. aeruginosa ClpG, a AAA+ Protein, Was Acquired by Horizontal Gene Transfer.
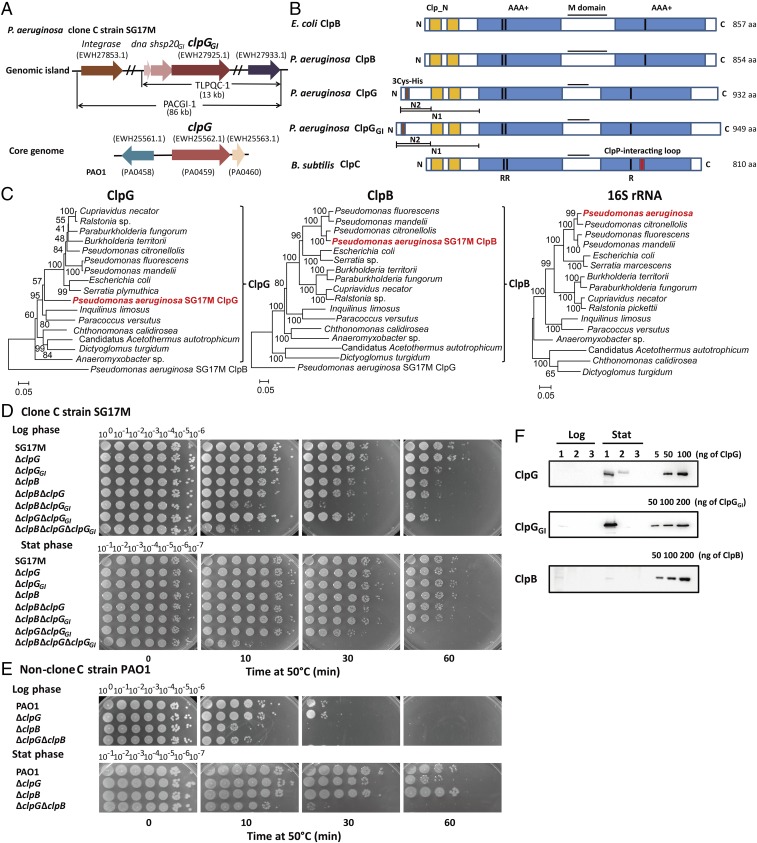
P. aeruginosa clone C strains harbor the so far uncharacterized AAA+ proteins ClpG and ClpGGI, which are phylogenetically distant from other functionally characterized AAA+ chaperones of Gram-negative and -positive bacteria (SI Appendix, Fig. S1). ClpG is encoded on the core genome of P. aeruginosa, while ClpGGI is encoded by TLPQC-1, a part of the Pseudomonas aeruginosa clone C genomic island 1 (PACGI-1) (Fig. 1A) (25).
Fig. 1.
The Hsp100 protein ClpG substantially contributes to heat tolerance of P. aeruginosa. (A) Genomic context of ClpG and ClpGGI located on the core genome and on the TLPQC-1 part of PACGI-1 in P. aeruginosa SG17M (23). Locus: dna (EWH27923.1), shsp20GI (EWH27924.1). (B) Domain organization of E. coli ClpB, B. subtilis ClpC, and P. aeruginosa ClpG/ClpGGI proteins. The Hsp100 proteins are composed of two ATPase domains (AAA-1 and AAA-2), an N-terminal domain (Clp_N), and an inserted middle (M) domain. Clp_N2 of ClpG/ClpGGI includes a potential Zn2+-binding motif composed of three cysteines and a histidine residue. (C) ClpG of P. aeruginosa has a unique position within the phylogenetic tree of ClpG homologs deviating from the phylogenetic position of ClpB, which is in congruence with the 16S RNA phylogenetic tree. (D and E) Heat tolerance of clone C P. aeruginosa SG17M (D) or nonclone C P. aeruginosa PAO1 (E) and indicated mutant cells in logarithmic (log) and stationary (stat) growth phase. Cells were grown at 20 °C and subjected to lethal heat shock at 50 °C for 10, 30, and 60 min. Cellular viabilities were determined by spotting serial dilutions (100–10−6) on LB plates. (F) Western blot analysis of ClpG, ClpGGI, and ClpB protein levels in logarithmic and stationary growth phase. 1: SG17M wild type; 2: ΔclpG, ΔclpGGI, or ΔclpB deletion mutant; and 3: ΔclpB ΔclpG ΔclpGGI triple deletion mutant. Purified proteins serve as standards for protein amount.
ClpG and ClpGGI exhibit 63% sequence identity and 76% sequence homology and share the same domain organization (Fig. 1B and SI Appendix, Fig. S2). They harbor two AAA+ domains, an N1 N-terminal domain, an M domain inserted into the first AAA+ domain, and a C-terminal extension (Fig. 1B and SI Appendix, Fig. S2). The N1 N-terminal domain can be further divided into two subdomains. The N2 N-terminal subdomain is unique to ClpG/ClpGGI and includes a putative zinc binding motif containing three conserved cysteines and a single histidine residue. The N-terminal subdomain N shows sequence and structural homology to N domains of other Hsp100 protein family members (ClpA, ClpB, and ClpC). The ClpG/ClpGGI M domains are predicted to form a coiled-coil structure, similar to ClpB and ClpC M domains (26, 27). ClpG/ClpGGI lack the conserved tripeptide sequence L/I/V-G-F/L in the second AAA+ domain, which mediates interaction with the peptidase ClpP in ClpC (Fig. 1B and SI Appendix, Fig. S2) (28) and are therefore expected to work independently from ClpP, similar to ClpB, the major disaggregase in E. coli (29). A ClpB homolog is also encoded on the P. aeruginosa core genome with 45/62% and 47/64% sequence identity/similarity to ClpGGI and ClpG, respectively.
The phylogenetic clustering of ClpG and ClpGGI is different from the major bacterial disaggregase ClpB and, in contrast to ClpB, does not reflect the species relationship represented by 16S rRNA alignment (Fig. 1C and SI Appendix, Fig. S3A). This suggests that P. aeruginosa acquired clpG and clpGGI by horizontal gene transfer. Accordingly, close homologs of ClpG present in individual Pseudomonas species strains are located at a different position on the genome and such is ClpG of E. coli strain KTE154 located on a genomic island (SI Appendix, Fig. S3B), indicating frequent acquisition of ClpG homologs by horizontal gene transfer in proteobacteria.
The phylogenetic distribution of ClpG and ClpGGI is restricted, compared with widespread ClpB, as these proteins are only found in selected Gram-negative bacteria (Fig. 1C and SI Appendix, Fig. S3A). Furthermore, ClpG is specific to the species P. aeruginosa within the Pseudomonas genus, while ClpGGI seems to be a clone rather than a species-specific protein present in isolates of unrelated genera of Gram-negative bacteria, suggesting its recent acquisition. Summing up, ClpG/ClpGGI proteins form a subgroup of bacterial AAA+ chaperones that have been acquired by a set of Gram-negative bacteria via horizontal gene transfer.
ClpG Confers Superior Heat Tolerance to P. aeruginosa Strains.
To elucidate physiological functions of ClpG and ClpGGI, we constructed single and double mutants in the P. aeruginosa SG17M clone C strain. As the most closely related AAA+ chaperones ClpB, ClpC, and the biochemically uncharacterized ClpG homolog ClpK contribute to survival during nonlethal heat stress (4, 9, 24, 30), we tested the mutants for alteration in heat tolerance. For comparison, we additionally generated a P. aeruginosa ΔclpB mutant. As the dna-shsp20GI-clpGGI operon of TLPQC-1 of the aquatic isolate SG17M is largely expressed during stationary phase (25), we compared survival of cells in logarithmic and stationary growth phases upon lethal heat shock from 20 °C to 50 °C (Fig. 1 D and E).
Both ClpGGI and ClpB contribute to heat tolerance in logarithmic growth, while enhanced heat sensitivity is observed in the ΔclpB ΔclpGGI double deletion strain. Subsequent deletion of clpG further increased heat sensitivity in the triple mutant. Low levels of ClpGGI and ClpB production can be detected in the logarithmic growth phase, consistent with the moderate approximately 10-fold decrease in heat tolerance phenotype of the single mutants (Fig. 1F). In contrast, in stationary phase, only ΔclpG ΔclpGGI cells showed pronounced 100-fold reduced survival 60 min after heat shock, indicating a dominant role of ClpG/ClpGGI proteins (Fig. 1D). Additional deletion of clpB in ΔclpG ΔclpGGI cells, however, further increased heat sensitivity, suggesting that ClpB can partially compensate for ClpG function. Western blot analysis indicated substantial production of ClpGGI during stationary phase and >10-fold lower expression of ClpG, but barely detected ClpB production (Fig. 1F). These findings are in agreement with the pronounced and redundant roles of ClpGGI and ClpG in stationary phase heat tolerance. Together these findings indicate distinct, but overlapping contributions of ClpG/ClpGGI and ClpB proteins to bacterial heat tolerance in logarithmic and stationary growth phase. Inducing the expression of either ClpG or ClpGGI in single copy from the araC promoter at the chromosomal Tn7 site was sufficient to restore heat shock tolerance to wild-type levels in the ΔclpB ΔclpG ΔclpGGI triple deletion mutant (SI Appendix, Fig. S4); thus, the function of ClpB and ClpG chaperones in thermotolerance is interchangeable.
Since various bacterial AAA+ chaperones cooperate with the peptidase ClpP in protein quality control, we tested whether ClpG/ClpGGI function in heat tolerance depends on ClpP (SI Appendix, Fig. S5). P. aeruginosa SG17M ΔclpP cells were by far not as heat sensitive as ΔclpG ΔclpGGI mutant cells. Additionally, single copy Tn7-based genomic expression of either clpG or clpGGI was sufficient to restore heat tolerance in the ΔclpG ΔclpGGI ΔclpP triple deletion mutant to ΔclpP cell levels (SI Appendix, Fig. S5). This demonstrates that ClpG/ClpGGI function independently from ClpP in thermotolerance, consistent with the absence of the ClpP interaction motif L/I/V-G-F/L in ClpG/ClpGGI chaperones (Fig. 1B and SI Appendix, Fig. S2).
To generalize the roles of ClpG and ClpB in heat tolerance, we constructed respective ΔclpB and ΔclpG mutants in the well-investigated nonclone C P. aeruginosa reference strain PAO1, harboring ClpG, but not ClpGGI. Equally as in SG17M, the contribution of ClpB to heat tolerance was dominant during logarithmic growth, while clpG did not contribute (Fig. 1E). Notably, heat shock survival of the SG17M strain 30 min after heat shock is more than 103-fold higher compared with PAO1 (Fig. 1 D and E). As the survival rate of the SG17M ΔclpGGI mutant is still superior compared with wild-type PAO1, components in addition to ClpGGI must contribute to survival of clone C SG17M wild-type cells.
Analysis of heat tolerance of strain PAO1 during stationary phase revealed a major contribution of ClpG as deletion of clpG, but not clpB, enhanced heat sensitivity (Fig. 1E). Simultaneous deletion of clpB and clpG substantially increased heat sensitivity, again revealing overlapping and independent contributions of ClpB and ClpG chaperones to heat tolerance in P. aeruginosa cells.
ΔclpG/ΔclpGGI Mutant Cells Exhibit Increased Protein Aggregation and Defects in Protein Disaggregation.
Thermotolerance in bacteria and fungi is linked to protein disaggregation processes (31, 32) and any defect in this process will therefore cause increased protein aggregation at elevated temperatures. To test whether reduced heat tolerance can be correlated to increased protein aggregation, we first compared the amount and pattern of aggregated proteins in P. aeruginosa SG17M wild-type and AAA+ chaperone mutant cells after growth at the elevated temperature of 42 °C for 24 h (Fig. 2A and SI Appendix, Fig. S6A). We observed slightly increased protein aggregation in ΔclpB and ΔclpGGI cells; however, a significant increase in protein aggregation required simultaneous deletion of clpG/clpGGI copies and clpB correlating with the high heat sensitivity of the triple mutant (Fig. 1D). Isolated protein aggregates included a vast variety of cellular proteins. A 15-kDa protein particularly elevated in the aggregate fraction of the ΔclpB ΔclpG ΔclpGGI triple mutant was identified by mass spectrometry as the small heat shock protein IbpA (SI Appendix, Table S1). sHsps coaggregate with misfolded proteins thereby representing sensitive aggregation reporters to underline the aggregation phenotype. Similarly, we found the bacterial Hsp90 chaperones HtpG to be enriched in the aggregate fraction of triple mutants (Fig. 2A and SI Appendix, Table S1). These findings indicate that ClpB and ClpG/ClpGGI have overlapping functions in preventing and/or reversing protein aggregation. Notably, we also observed single protein species that already strongly aggregated in ΔclpB (LasI) or ΔclpGGI (NuoCD) single deletion mutants (Fig. 2A and SI Appendix, Table S1). This suggests that DnaK-ClpB and ClpGGI exhibit, to a certain degree, substrate-specific activities.
Fig. 2.
Enhanced protein aggregation in P. aeruginosa SG17M cells lacking ClpB and ClpG/ClpGGI. (A) P. aeruginosa SG17M and mutants were cultured at 42 °C for 24 h and protein aggregates were isolated. Proteins that specifically aggregate in ΔclpGGI (NuoCD) and ΔclpB (LasI) cells are indicated. Aggregation of the HtpG and IbpA chaperones is most pronounced in the ΔclpB ΔclpG ΔclpGGI triple mutant. (B) P. aeruginosa SG17M and mutants were grown at 30 °C overnight (1) and incubated at 42 °C for 60 min (2). Next, cells were shifted to 30 °C for 60 min (3) and 120 min (4) for recovery. Protein aggregates were isolated at indicated steps and analyzed by SDS-PAGE. For semiquantitative comparison, the density of the entire lane was estimated by ImageJ with SG17M (A) or lane 1 of SG17M (B) arbitrarily set as 1. These experiments were repeated twice with similar results and one representative shown.
In an alternative approach, we shifted P. aeruginosa SG17M wild-type, ΔclpG ΔclpGGI, and ΔclpB ΔclpG ΔclpGGI mutant cells from 30 °C to 42 °C for 60 min and allowed for recovery upon subsequent incubation at 30 °C (Fig. 2B and SI Appendix, Fig. S6B). Protein aggregates were enhanced in ΔclpG ΔclpGGI and ΔclpB ΔclpG ΔclpGGI cells and subsequent heat shock increased the amount of aggregated proteins in all cells. Upon return to 30 °C, protein aggregates were partially removed and this process occurred with similar relative speed, although ΔclpB ΔclpG ΔclpGGI cells still contained a high aggregate load after a 120-min incubation. These findings support complementary activities of ClpB and ClpG/ClpGGI chaperones in P. aeruginosa protein quality control and point to a function of ClpG/ClpGGI in handling of protein aggregates.
ClpG Is a Potent Disaggregase Without Any Accessory Factor.
As ClpB and ClpG proteins seem to exert overlapping activities in P. aeruginosa cells, we wanted to compare their biochemical properties. As ClpGGI is more widespread than ClpG, and ΔclpGGI cells are more heat sensitive compared with ΔclpG (Fig. 1D), we focused our analysis largely on ClpGGI and subsequently confirmed key features for the ClpG counterpart.
We purified ClpGGI and first tested for nucleotide-driven hexamer formation as a common feature of AAA+ chaperones. ClpGGI oligomerization was analyzed by size exclusion chromatography (SEC) (SI Appendix, Fig. S7) and transmission electron microscopy (TEM) (SI Appendix, Fig. S8). In the absence of ATP, ClpGGI is mainly present as a monomer (SI Appendix, Fig. S7). ClpGGI oligomers formed upon addition of ATP (2 mM), although oligomerization was not complete under the given experimental conditions (SI Appendix, Fig. S7). Partial oligomer formation of ClpGGI was also observed in TEM (+ATPγS), although less efficient compared with E. coli or P. aeruginosa ClpB (SI Appendix, Fig. S8A). The electron density map of ClpGGI oligomeric particles revealed the formation of a hexameric ring structure composed of two layers, likely formed by AAA-1 and AAA-2 domains (SI Appendix, Figs. S8 B and C and S9). While ClpGGI has an extended N-terminal domain, the electron density map of ClpGGI is flattened in the N-terminal region and the map is likely restricted by a high mobility of the N-terminal extension. Fitting of the ClpGGI map with a ClpB structural model (Protein Data Bank ID code 4D2Q) (33) revealed missing density for ClpB M domains, which can be explained by the reduced size of the ClpGGI M domain, but also by the limited resolution of the map. Together these findings show that ClpGGI forms hexamers in an ATP-dependent manner as reported before for other AAA+ chaperones.
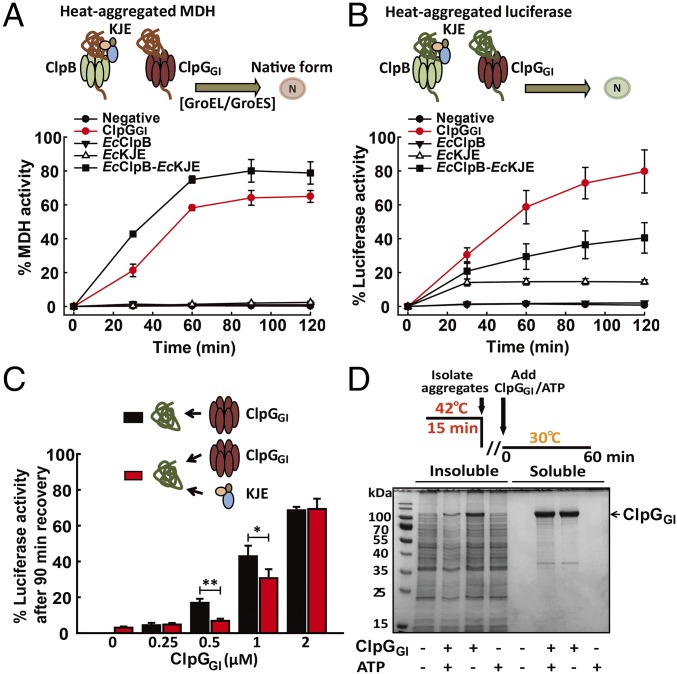
Next, we tested whether the complementary functions of ClpB and ClpGGI observed in vivo can be documented as complementary chaperone activities in vitro and therefore tested for disaggregation activity of ClpGGI. We monitored the disaggregation of heat-aggregated malate dehydrogenase (MDH), which can be efficiently rescued by the DnaK-ClpB bichaperone system. To couple the fast solubilization of MDH aggregates with fast MDH refolding, all assays were performed in the additional presence of the E. coli GroEL/GroES chaperone system. The GroEL/GroES system on its own does not exhibit any disaggregation activity and only accelerates refolding of soluble, unfolded MDH after its removal from protein aggregates (34). E. coli ClpB (EcClpB) did not exhibit disaggregation activity on its own and essentially required cooperation with the E. coli DnaK chaperone system (DnaK, DnaJ, GrpE: EcKJE). The EcClpB-EcKJE bichaperone system reactivated 80% of aggregated MDH in 60 min (Fig. 3A). A comparable disaggregation activity was determined for the P. aeruginosa bichaperone system (PaClpB-PaDnaK-EcJE) (SI Appendix, Fig. S10A). Also, PaDnaK and PaClpB were interchangeable with their E. coli counterparts (SI Appendix, Fig. S10A), demonstrating full disaggregation potential of P. aeruginosa ClpB and DnaK. Strikingly and in contrast to EcClpB and PaClpB, P. aeruginosa ClpGGI exhibited high disaggregation potential on its own and rescued heat-aggregated MDH with an activity comparable to the EcClpB-EcKJE bichaperone system (Fig. 3A).
Fig. 3.
ClpGGI shows potent disaggregating activity in vitro and ex vivo. (A and B) Refolding of heat-aggregated malate dehydrogenase (MDH) and luciferase by the E. coli DnaK-ClpB bichaperone system (EcClpB and EcKJE) or ClpGGI. The activities of native enzymes were set as 100%. (C) Refolding of aggregated luciferase was monitored in the absence or presence of EcKJE and increasing concentrations of ClpGGI (*P < 0.05; **P < 0.01). (D) Resolubilization of heat-aggregated P. aeruginosa proteins by ClpGGI ex vivo. Soluble P. aeruginosa SG17M crude extract from cells grown at 30 °C for 24 h was heat treated at 42 °C for 15 min. Insoluble protein aggregates were isolated and subsequently subjected to resolubilization by ClpGGI/ATP (2 mM) for 60 min. Insoluble and soluble protein fractions were separated and analyzed by SDS-PAGE.
To substantiate this stand-alone disaggregation activity of ClpGGI, we used heat-aggregated luciferase as alternative substrate. ClpGGI was again highly active without assistance by additional factors and even showed a twofold increased yield of disaggregation/refolding of aggregated luciferase compared with EcClpB-EcKJE (Fig. 3B). A robust, stand-alone luciferase reactivation activity was also determined for P. aeruginosa ClpG, confirming the potent and independent disaggregation activity of this AAA+ chaperone (SI Appendix, Fig. S10B).
EcKJE initiates protein disaggregation by the bichaperone system by recruiting and activating EcClpB at the aggregate surface. As ClpG can act on protein aggregates on its own, we tested whether DnaK and ClpG can cooperate at the aggregate surface or whether they compete for binding to the aggregated substrate. To this end, we compared the luciferase disaggregation activity of ClpGGI at different concentrations (0.25–2 μM) either alone or in the presence of EcKJE (1 μM DnaK, 0.2 μM DnaJ, 0.1 μM GrpE), which exhibits only low level disaggregation activity in the absence of EcClpB (Fig. 3C). EcKJE reduced reactivation of heat-aggregated luciferase by ClpGGI if ClpGGI was present at lower concentration (0.5 μM) and a comparable inhibitory effect was noticed in the presence of PaKEcJE (Fig. 3C and SI Appendix, Fig. S10C). Increasing the ClpGGI concentration to 2 μM restored efficient luciferase reactivation in the presence of EcKJE and proceeded with the same disaggregation rate as determined in the absence of EcKJE (Fig. 3C). These findings indicate that the DnaK chaperone system and ClpGGI do not cooperate in protein disaggregation but instead compete for binding to the aggregate surface. The DnaK-ClpB bichaperone system and ClpG/ClpGGI therefore form independent disaggregase systems operating on the same substrate.
Next, we tested whether the uncovered disaggregation activity of ClpGGI is promiscuous and active toward aggregated proteins from P. aeruginosa SG17M. For this purpose, a lysate of soluble proteins was derived from P. aeruginosa stationary phase cells, subjected to heat denaturation and protein aggregates were isolated (Fig. 3D). Aggregates were incubated with and without ClpGGI in the absence and presence of ATP. We observed a spectrum of solubilized protein species for ClpGGI + ATP, although some minor bands were also observed without ATP, likely representing ClpGGI degradation products present in the ClpGGI sample. The ATP-dependent disaggregation activity of ClpGGI was most evident from the significant reduction in protein aggregate levels compared with control reactions (Fig. 3D and SI Appendix, Fig. S10D). We infer that ClpGGI resolublizes aggregates of heat-denatured proteins with a broad specificity.
ClpGGI Can Replace DnaK-ClpB and Hsp104 Disaggregase Functionality.
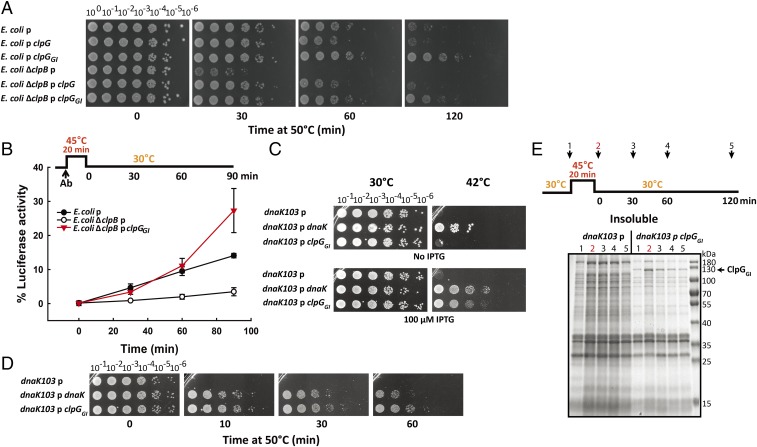
To demonstrate that the identified ClpG/ClpGGI disaggregation activity is operative in diverse genetic backgrounds (35), we tested whether ClpGGI can replace ClpB and Hsp104 activities in E. coli and Saccharomyces cerevisiae cells. We first expressed ClpG and ClpGGI in E. coli wild-type and ΔclpB mutant cells and tested for heat tolerance upon temperature upshift from 30 °C to 50 °C (Fig. 4A). E. coli ΔclpB cells were less heat tolerant compared with wild-type cells and viability was 104-fold reduced 60 min after heat shock, in agreement with literature (4). Expressing either ClpG or ClpGGI in ΔclpB cells fully restored thermotolerance, demonstrating that ClpG/ClpGGI can entirely take over ClpB disaggregase function in vivo. Notably, expressing ClpGGI in E. coli wild-type cells conferred superior thermotolerance, increasing the fraction of viable cells 100-fold after heat shock (120 min) compared with wild-type cells harboring an empty control plasmid (Fig. 4A).
Fig. 4.
ClpGGI restores disaggregase function in the absence of E. coli ClpB and DnaK. (A) E. coli wild-type and ΔclpB cells expressing clpG or clpGGI were grown at 30 °C and shifted to 50 °C. Cellular viabilities were determined by spotting serial dilutions (100–10−6) of cells on LB plates. (B) E. coli wild-type and ΔclpB cells (with and without clpGGI) expressing YFP-luciferase were grown to midlogarithmic growth phase at 30 °C. Protein synthesis was stopped by addition of erythromycin and cells were shifted to 45 °C for 20 min followed by a recovery period at 30 °C. Luciferase activity was determined prior to (set to 100%) and post heat shock and at the indicated time points during cellular recovery. p = pJN105; p clpG, p clpGGI = respective genes cloned in pJN105 (A and B). (C) Serial dilutions of E. coli dnaK103 mutant cells expressing dnaK or clpGGI were spotted on LB plates with and without 100 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubated at 30 °C and 42 °C. (D) E. coli dnaK103 mutant cells expressing dnaK or clpGGI were grown at 30 °C in the presence of 250 μM IPTG and shifted to 50 °C. Cellular viabilities were determined by spotting serial dilutions (10−1–10−6) of cells on LB plates. (E) E. coli dnaK103 mutant cells harboring an empty vector (p) or expressing clpGGI (p clpGGI) were grown at 30 °C in the presence of 250 μM IPTG and shifted to 45 °C for 20 min followed by a recovery period at 30 °C. Soluble and insoluble protein fractions were isolated at the indicated time points and analyzed by SDS-PAGE (SI Appendix, Fig. S11). p = pUHE21; p dnaK, p clpGGI = respective genes cloned in pUHE21 (C–E).
In a complementary approach we assessed the ability of ClpGGI to reactivate aggregated proteins in E. coli ΔclpB cells using thermolabile YFP-luciferase as disaggregation reporter (Fig. 4B). YFP-luciferase produced at 30 °C in E. coli wild-type and ΔclpB cells additionally expressed ClpGGI. After a heat shock to 45 °C in the presence of the protein synthesis inhibitor erythromycin and subsequent incubation at 30 °C, E. coli wild-type cells recovered 15% of luciferase activity within 90 min, whereas ΔclpB cells only allowed for 3% luciferase refolding. This ClpB-specific resolubilization defect of ΔclpB cells could be rescued by ClpGGI expression (Fig. 4B). Here, 30% of YFP-luciferase was refolded within 90 min, exceeding the reactivation potential of E. coli wild-type cells. This confirms that the presence of the alternative ClpGGI disaggregation system increases the ability of E. coli cells to rescue aggregated proteins.
As ClpB cooperates with the accessory chaperone DnaK, we tested the effect of clpGGI expression in E. coli dnak103 mutant cells, which synthesize a truncated nonfunctional DnaK protein (36). clpGGI expression restored the temperature-sensitive growth phenotype of dnaK103 mutant cells at 42 °C (Fig. 4C). Furthermore, clpGGI expression reestablished heat tolerance upon 50 °C heat shock to dnaK103 mutant cells, which otherwise lost viability within 10 min (Fig. 4D), again with an efficiency similar to plasmid-encoded DnaK wild type. The presence of ClpGGI in dnaK103 cells reduced the amount of protein aggregates isolated at 30 °C and allowed for aggregate removal during a subsequent recovery period after heat shock to 45 °C, in contrast to dnaK103 control cells harboring an empty vector (Fig. 4E and SI Appendix, Fig. S11). Together these findings substantiate a DnaK-independent disaggregation activity of ClpGGI, which can compensate for the broad protein folding defects associated with loss of functionality of the DnaK (Hsp70) partner chaperone.
In an additional approach, we analyzed whether ClpGGI can replace Hsp104 function in the propagation of amyloid-like protein aggregates, prions, in yeast cells. Prion fibril fragmentation, which leads to the production of smaller propagons that are transmitted to daughter cells, is executed by the Hsp104 disaggregase. To monitor propagation of the yeast prion states [PSI+] and [URE3], originating from amyloid-formation of the Sup35 and Ure2 proteins, we made use of the yeast reporter strains 779-6A and 1075. In these strains, the production of the Ade2 reporter protein required for adenine biosynthesis depends on the respective prion state. We expressed ClpGGI in the yeast reporter cells and concurrently inhibited the endogenous Hsp104 disaggregase by low concentrations of guanidine hydrochloride (GuHCl), a specific inhibitor of Hsp104 activity (37). We confirmed that ClpGGI disaggregation activity is only mildly affected by 5 mM GuHCl, in contrast to Hsp104, which is strongly inhibited (SI Appendix, Fig. S12A). On adenine-limiting medium, the yeast reporters form white colonies if the prion state is maintained but turn red upon prion loss because of the accumulation of a metabolite of the Ade2 substrate. Hsp104 inhibition led to formation of red reporter colonies, indicating prion loss, while the presence of ClpGGI changed colony color to pink (SI Appendix, Fig. S12B). This suggests a weak [PSI+] and [URE3] phenotype and thus partial ClpGGI activity in prion maintenance. On medium lacking adenine, loss of the prion state abolishes growth of the reporter strains. On such a medium, ClpGGI allowed for growth of the 1075, but not the 779-6A strain, again suggesting that ClpGGI can partially rescue Hsp104 functionality in yeast prion propagation. Notably, a comparable, partial activity in yeast prion propagation had been determined for E. coli ClpB, when coexpressed with cooperating E. coli DnaK in yeast prion reporter cells (38).
ClpG Overcomes Mechanistic Restrictions of ClpB, Rationalizing Its Factor-Independent Disaggregation Activity.
ClpG/ClpGGI show high stand-alone disaggregation activity, whereas ClpB strictly requires cooperation with its Hsp70 (DnaK) partner chaperone. ClpB has low basal ATPase activity and does not bind protein aggregates efficiently, while DnaK targets protein aggregates to ClpB to concurrently stimulate ClpB ATPase activity (39–42). We speculated that the partner-independent ClpG/ClpGGI and DnaK-dependent ClpB disaggregation activities must be reflected in different biochemical properties of the AAA+ chaperones. We therefore tested whether ClpG/ClpGGI and ClpB differ in (i) ATPase activities and (ii) their abilities to bind model protein aggregates.
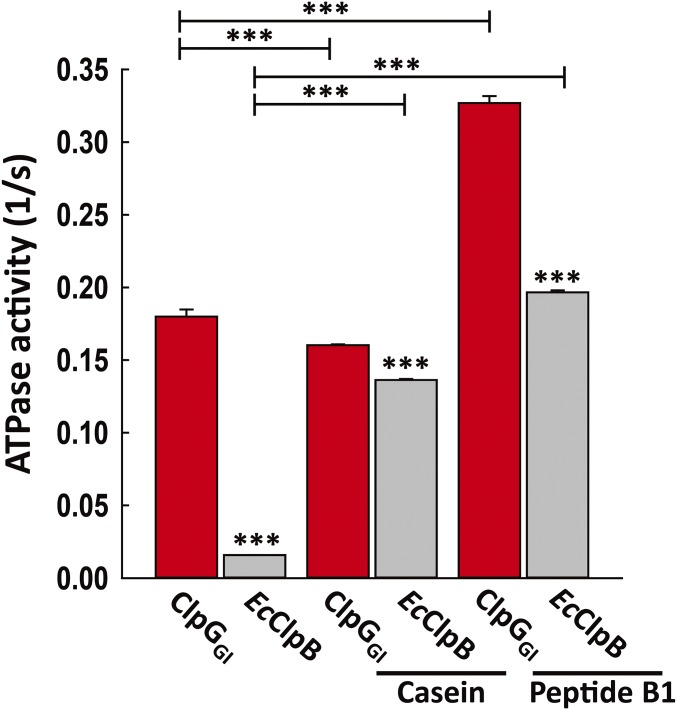
ClpGGI and ClpG show a 12-fold higher basal ATPase activity compared with EcClpB (Fig. 5 and SI Appendix, Fig. S13). ClpG/ClpGGI therefore do not require stimulation by a partner protein to reach increased ATPase activities. The ATPase activity is specific, as a double Walker B mutant in ClpGGI (ClpGGI E383A E723A) did not display ATPase activity (SI Appendix, Fig. S13). We next assessed substrate-stimulated ATPase activities by using casein and peptide B1 (AHAWQHQGKTLFISRKTYRIC) as these model substrates are directly recognized by ClpB without DnaK assistance. Casein binds to the N-terminal domain and the substrate processing pore site of ClpB, whereas peptide B1 exclusively interacts with the central ClpB pore site (43). Casein and peptide B1 stimulated ClpB ATPase activity by 8- and 12-fold, respectively (Fig. 5). ClpGGI ATPase activity was also increased by peptide B1 (1.8-fold) while casein showed no effect, indicating that ClpGGI and ClpB differ in substrate recognition in parts.
Fig. 5.
ClpGGI exhibits high basal ATPase activity and differs in substrate specificity. Basal and substrate-stimulated ATPase activities of EcClpB and ClpGGI were determined. The substrates, casein and peptide B1, enhanced the ATPase activity of ClpB, while ClpGGI activity was only further enhanced by peptide B1 (***P < 0.001; EcClpB compared with ClpGGI and ClpGGI/EcClpB ± substrate stimulation).
Difference in substrate binding can be likely attributed to differences between the N-terminal domains of ClpG/ClpGGI and ClpB. Its unique N1 domain might hinder ClpG/ClpGGI to interact with casein while enabling the binding to protein aggregates at the same time. To assess the role of the ClpG/ClpGGI N-terminal domains we either deleted the entire N-terminal region (ΔN1) or only N2, the very N-terminal, ClpG/ClpGGI-specific extension harboring a putative Zn2+-binding motif (ΔN2) (Fig. 6A). ClpG/ClpGGI wild-type and N-terminal deletion constructs were expressed at increased levels largely comparable for all constructs in P. aeruginosa ΔclpB ΔclpG ΔclpGGI mutant cells and analyzed for their ability to restore heat tolerance upon temperature upshift to 50 °C (Fig. 6B and SI Appendix, Fig. S14). All ClpG/ClpGGI N-terminal deletion variants failed to restore heat tolerance, indicating that the unique N-terminal extensions are essential for functionality (Fig. 6B and SI Appendix, Fig. S2). In contrast, deletion of the N-terminal domain of P. aeruginosa ClpB (ΔN-ClpB) still conferred heat tolerance, in agreement with former studies showing that the N-terminal domain of EcClpB is not essential for protein disaggregation (Fig. 6B) (44–47).
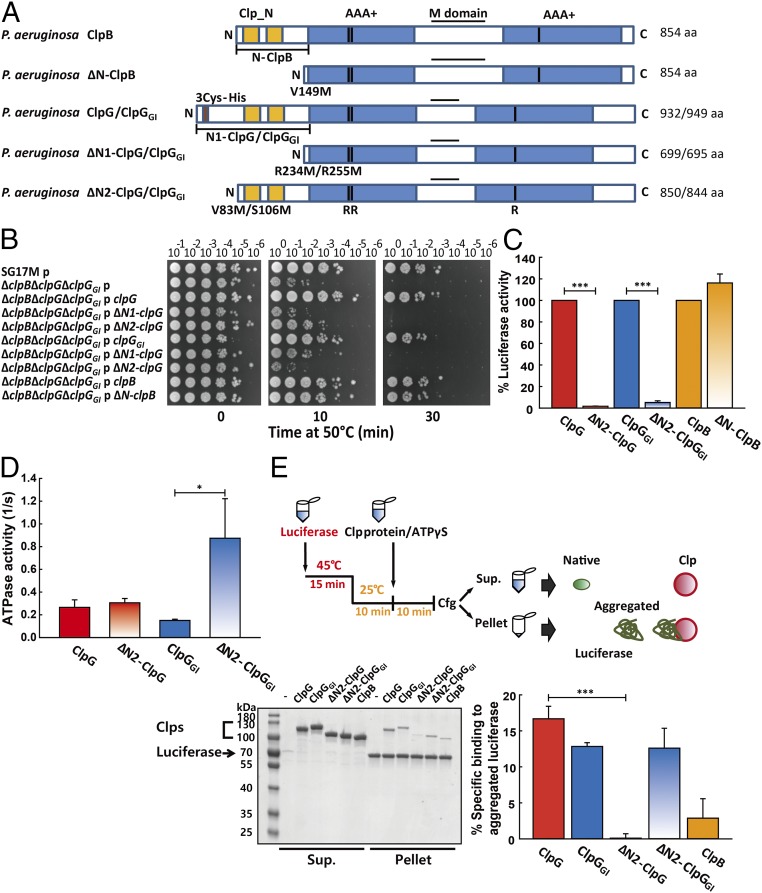
Fig. 6.
The unique N-domain extensions of ClpG/ClpGGI are essential for disaggregation activity. (A) Domain organization of ClpG/ClpGGI and ClpB. The generated N-terminal deletion variants of ClpG/ClpGGI (ΔN1 and ΔN2) and ClpB (ΔN) are indicated (B) P. aeruginosa SG17M ∆clpB ∆clpG ∆clpGGI expressing full-length, N-terminal truncated ClpG/ClpGGI or ClpB proteins from the vector pJN105 were subjected to heat shock at 50 °C for 10 min and 30 min. Cellular viabilities were determined by spotting serial dilutions (100–10−6) of cells on LB plates. p = pJN105. (C) Refolding of heat-aggregated luciferase was monitored in the presence of indicated chaperones. The activity of native luciferase in the presence of wild-type chaperones ClpG, ClpGGI, and ClpB after 90 min of refolding was set as 100%. (D) Basal ATPase activities of indicated ClpG/ClpGGI variants were determined. (E) Experimental scheme for monitoring binding of ClpG/ClpGGI and ClpB proteins to heat-aggregated luciferase. (Left) Heat-aggregated luciferase was incubated with the indicated chaperones in the presence of 2 mM ATPγS. Soluble (Sup.) and insoluble (pellet) fractions were analyzed by SDS-PAGE. (Right) Intensities of protein bands were quantified by ImageJ with the fraction (%) of chaperone present in the pellet fraction versus chaperone without luciferase aggregates calculated. Binding experiments were performed three times with a representative SDS-PAGE shown. Quantifications are based on three independent experiments (*P < 0.05; ***P < 0.001).
To further characterize the role of the unique N2 N-terminal extension, we purified and characterized ΔN2-ClpG/ΔN2-ClpGGI deletion mutants. The deletions did not affect global ClpG/ClpGGI protein structure as revealed by CD spectroscopy (SI Appendix, Fig. S15). Also ATP-dependent oligomerization of ΔN2-ClpGGI was similar to ClpGGI and ClpG (SI Appendix, Fig. S7). Consistent with the in vivo results, reactivation of aggregated luciferase by ClpG/ClpGGI in vitro was strongly reduced (ClpGGI) or entirely abrogated (ClpG) upon deletion of the N2 N-terminal extensions in contrast to ClpB (Fig. 6C and SI Appendix, Fig. S16).
The crucial role of the ClpG/ClpGGI N2 N-terminal extension for heat tolerance and protein disaggregation could be explained by the N-terminal domain to control ClpG/ClpGGI ATPase function or to target ClpG/ClpGGI to protein aggregates. Whereas basal ATPase activity of ΔN2-ClpG was comparable to ClpG, ΔN2-ClpGGI exhibited a 5.8-fold increased ATPase rate compared with ClpGGI, indicating that the N2 part of the N-terminal extension down-regulates ATPase activity (Fig. 6D). We next performed an in vitro binding assay with ClpG/ClpGGI wild-type and the N2 deletion variants using aggregated luciferase as substrate to test for N2 subdomain-mediated aggregate targeting. The binding of ClpG/ClpGGI to aggregated luciferase was monitored in the presence of nonhydrolyzable ATPγS, which stabilizes Hsp100-substrate interaction. As luciferase aggregates are insoluble, binding of ClpG/ClpGGI was monitored by separating soluble and insoluble protein fractions by centrifugation (Fig. 6E). We ensured that only very minor amounts of ClpG/ClpGGI and their N-terminal deletion variants are found in the pellet fraction in the absence of protein aggregates (SI Appendix, Fig. S17). We observed substantial binding of full-length ClpG/ClpGGI to both aggregated substrates and stronger binding compared with ClpB (Fig. 6E), confirming that ClpB requires DnaK as crucial accessory factor for aggregate binding (48). Binding to ClpG was entirely dependent on its unique N-terminal extension as ΔN2-ClpG no longer bound aggregated proteins (Fig. 6E). In contrast, ΔN2-ClpGGI binding to protein aggregates was not affected, unraveling an unexpected difference between ClpG and ClpGGI. Notably, ΔN2-ClpGGI still retained minor disaggregation activity in contrast to ΔN2-ClpG (Fig. 6C), consistent with its ability to still bind aggregated luciferase (Fig. 6E).
Together these finding provide a mechanistic rationale for why ClpG/ClpGGI act as potent, stand-alone disaggregases and unravel distinct functions of the unique N-terminal extensions in ATPase control in ClpGGI and aggregate targeting in ClpG rationalizing their crucial roles in protein disaggregation.
Discussion
In this work, we characterized the AAA+ chaperones ClpG/ClpGGI as stand-alone disaggregases, which confer superior heat tolerance to bacteria (Fig. 7). So far protein disaggregation and heat tolerance in bacteria have been predominantly linked to the AAA+ disaggregase ClpB, which forms a bichaperone system with the DnaK (Hsp70) partner chaperone. ClpB activity is crucial for thermotolerance in Gram-negative (4, 49, 50), but also Gram-positive bacteria (6, 51). The lack of ClpB in some Gram-positive bacteria is compensated by the presence of an alternative disaggregation system composed of ClpC and its adaptor proteins for aggregate targeting and ATPase activation (12, 30, 52, 53). While other bacterial AAA+ proteins (ClpE and ClpL) have also been implicated in protein disaggregation, a disaggregation activity has either not been documented or is very limited in vitro (51, 54–56). Here, we demonstrate that ClpG/ClpGGI overcome both mechanistic restrictions of ClpB and ClpC by exhibiting a high basal ATPase activity and directly binding to protein aggregates. These features allow for efficient ATP-dependent threading of aggregated proteins explaining why ClpG/ClpGGI function in protein disaggregation without assistance of accessory factors.
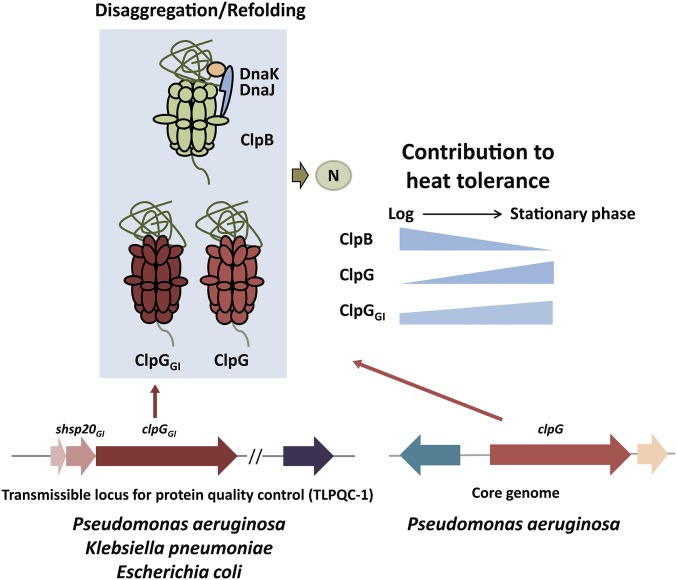
Fig. 7.
The ClpG/ClpGGI disaggregases increase disaggregation potential and heat resistance of P. aeruginosa SG17M. P. aeruginosa clone C strain harbors the classical disaggregating DnaK-ClpB bichaperone system and the ClpG and ClpGGI disaggregases (23). ClpG is located on the core genome, while ClpGGI is located on the TLPQC-1 locus as part of PACGI-1 genomic island. The contributions of the different disaggregating systems to heat tolerance differ at logarithmic and stationary growth. ClpB/ClpGGI are most important for heat resistance during logarithmic growth, while ClpG/ClpGGI dominate heat tolerance during stationary phase. Compared with ClpB, ClpG/ClpGGI have unique and extended N-terminal N1 domains to bind to substrates. TLPQC-1 encoding ClpGGI is also present in outbreak strains of K. pneumoniae and E. coli.
The unique N2 N-terminal extensions are characterized by distinct amino acid sequences, while ClpGGI harbors an additional insertion (SI Appendix, Fig. S2). This sequence diversity is reflected by functional diversity, as the N2 subdomain is essential for aggregate targeting of ClpG, while its counterpart regulates ClpGGI ATPase activity. Furthermore, the unique N2 N-terminal extensions likely interplay with the C-terminal “classical” Hsp100 N domains in ClpG/ClpGGI-mediated disaggregation. Hsp100 N domains can directly bind substrates (57) and coordination of both ClpG/ClpGGI N-terminal domains might be crucial for activity. Dissecting the interplay and specific roles of ClpG/ClpGGI N-terminal domains will be essential to understand the mechanistic basis of these stand-alone disaggregases. Such studies will also unravel whether substrate specificity of ClpG/ClpGGI is restricted to thermally aggregated proteins or extends to, e.g., irreversible oxidized proteins as it is the case for ClpB of Mycobacterium tuberculosis (14).
ClpG/ClpGGI are as potent as the DnaK-ClpB system in protein disaggregation, can complement the thermotolerance defect of E. coli ΔclpB and dnaK103 mutant cells, and together increase thermotolerance in P. aeruginosa SG17M clone C cells substantially. We even noticed higher disaggregation activity of the horizontally transferred ClpGGI toward aggregated luciferase in vitro and in vivo, potentially explaining why ClpGGI is promiscuous and functional in many bacteria, including E. coli, even compared with ClpG (Fig. 4) (23, 35).
As the well-established disaggregase ClpB, ClpG/ClpGGI-mediated protein disaggregation is linked to refolding of aggregated substrates, independent from the peptidase ClpP. This underlines the concept that thermotolerance largely relies on reactivation of essential proteins aggregated upon heat induction (3, 58).
What is the relationship between the coexisting disaggregation systems, DnaK-ClpB and ClpG/ClpGGI? We show here that both systems work independently from one another and exert largely overlapping and compensatory activities. Constitutive expression of either ClpB or ClpG/ClpGGI to higher levels largely restores heat tolerance of P. aeruginosa cells lacking the chromosomal copies of all disaggregases (Fig. 6B and SI Appendix, Fig. S4). The contribution of the three disaggregase systems to P. aeruginosa heat tolerance differ in logarithmic and stationary phase. Strong increase of ClpG/ClpGGI levels in P. aeruginosa clone C SG17M cells during stationary phase, likely enables ClpG/ClpGGI to outcompete DnaK, rationalizing their growth phase-specific contributions to heat tolerance (Fig. 7).
Despite superior biochemical performance, why is ClpG/ClpGGI not more widespread? No detrimental effect was observed upon overproduction of ClpG/ClpGGI, but expression of shsp20GI encoding the small heat shock protein in the dna-shsp20GI-clpGGI operon has been detrimental in E. coli. Furthermore, we can only speculate that, for example, the high intrinsic ATPase activity of ClpG/ClpGGI is hard to control. Alternatively, to covalently link the substrate binding domain to disaggregase functionality in the same protein may provide reduced regulatory flexibility. ClpG/ClpGGI might have developed in bacteria indwelling a demanding ecological niche in parallel to the Hsp70-ClpB system, or ClpG/ClpGGI is an evolutionary younger protein. Recent acquisition of ClpGGI homologs, often on plasmids and in more than one copy, by unconventional pathogens associated with industrial and clinical settings (see also below) suggests that certain man-made environments provide a strong selection pressure aiding predominantly the spread of the dna-shsp20GI-clpGGI core unit (59, 60).
Bioinformatic analysis indicates that both copies of the ClpG disaggregase, clpG and clpGGI, have been acquired by horizontal gene transfer. Growth benefits provided by mobile genetic elements have been so far largely linked to acquisition of antibiotic resistance, specific virulence factors, or the utilization of alternative carbon sources (e.g., degradation of aromatic compounds) (61, 62). Recently, a growth advantage provided by transmissible loci is becoming apparent: increased bacterial fitness during stress conditions (23, 25, 63). Exposure to elevated temperature represents one of the most adverse conditions for environmental bacteria, but is also relevant for pathogens in interaction with the host. Acquisition of the ClpG/ClpGGI disaggregases boosts protein quality control and increases viability of P. aeruginosa clone C during severe heat stress. The identification of NuoCD, subunits of the NAD quinolone oxidoreductase, as potential ClpGGI substrates (Fig. 2) suggests a role of ClpGGI in energy production through respiration. The ClpB disaggregase plays a role in virulence and host persistence of pathogenic and commensal bacteria (6, 7, 13–16). It remains to be tested whether a ΔclpG ΔclpGGI mutant exhibits phenotypes beyond heat sensitivity such as in microbial–host interactions. A potential function in virulence is emphasized as ClpG is uniquely encoded on the core genome of P. aeruginosa, the most important human nosocomial pathogen of the Pseudomonas genus. Remarkably, more clones of P. aeruginosa, such as clone J strains successful in acute and chronic infections and the aquatic habitat, additionally harbor ClpGGI on PACGI-1/TLPQC-1 (25), while the PA14 strain, which is lacking PACGI-1/TLPQC-1, is predominantly found in acute infections (22). The occurrence of ClpGGI extends beyond P. aeruginosa as ClpGGI is found in extremely heat-tolerant E. coli from food factories (2% of all strains), where ClpGGI and TLPQC-1 additively contribute to heat tolerance (35). Also 2/3 of Klebsiella pneumoniae strains from the clinical environment harbor ClpGGI (called ClpK in this species) suggesting that K. pneumoniae requires the extended heat tolerance phenotype to successfully colonize the clinical habitat (60). Infections caused by K. pneumoniae harboring clpGGI are, however, not characterized by increased virulence (60). This indicates heat tolerance to become an increasingly important virulence, persistence, or resistance factor, probably promoted by modern food production and medical sterilization procedures that involve heat treatment. Since ClpG/ClpGGI are AAA+ chaperones in P. aeruginosa clone C and other common clonal strains of different genera, they might represent a potential target to develop novel antimicrobial or antivirulence compounds and approaches.
Materials and Methods
Detailed information of all experimental procedures is provided in SI Appendix.
Strains, Plasmids, and Growth Condition.
The aquatic isolate P. aeruginosa SG17M was selected as the representative clone C strain. All strains, plasmids, and primers used in this study are listed in SI Appendix, Tables S2–S4. Growth conditions and construction of mutants and plasmids are described in SI Appendix.
Bioinformatic Analysis.
Protein sequences of ClpG (acc. no. EWH25562), ClpGGI (acc. no. EWH27925), and ClpB (acc. no. EWH24017) of P. aeruginosa SG17M (25) were used as queries to search for ClpG, ClpGGI, and ClpB homologs as described in SI Appendix.
Heat Shock Tolerance Assay.
P. aeruginosa cells incubated in LB broth with shaking at 20 °C were harvested from logarithmic (OD600 = 0.7) and stationary (OD600 = 2.5) phase. The cells were exposed to 50 °C for 10, 30, and 60 min and subsequently cell viability was determined by the spotting assay as described in SI Appendix.
In Vitro Disaggregating Activity Assay.
Disaggregating activity was determined with minor modification (64) as described in SI Appendix. MDH and luciferase (both from Roche) were used as model substrates.
In Vivo Chaperone Assay.
An E. coli MC4100 ΔclpB strain expressing YFP-luciferase and the respective chaperone was cultured in LB medium at 30 °C. To heat shock, cells were incubated at a nonlethal 45 °C for 20 min. Afterward, the cells were incubated at 30 °C for recovery and luciferase activity was measured at certain time points as described in SI Appendix.
ATPase Assay.
ATPase activity was determined by a coupled ADP monitoring/recycling reaction of pyruvate kinase (PK) and lactate dehydrogenase (LDH) as described in SI Appendix.
Prion Propagation Test.
Propagation of [PSI+] and [URE3] was monitored by their ability to promote expression of Ade2p using yeast strains, 779-6A (MATα, kar1-1, SUQ5, ade2-1, his3Δ202, leu2Δ1, trp1Δ63, and ura3-52) and 1075 (MATα, kar1-1, PDAL5::ADE2, his3Δ202, leu2Δ1, trp1Δ63, and ura3-52) (38) as described in SI Appendix.
Disaggregation of Proteins in Heat-Denatured Crude Extracts.
Disaggregation of proteins in heat-denatured crude extracts was performed with minor modifications (32) as described in SI Appendix.
Isolation of in Vivo Protein Aggregates.
In vivo protein aggregates were examined with minor modifications (65) as described in SI Appendix.
In Vitro Substrate Binding Assay.
The interaction between denatured luciferase and Clp protein variants was conducted as described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Daniel C. Masison (National Institute of Diabetes and Digestive and Kidney Diseases, NIH) for providing materials and valuable discussion for the prion propagation test, Johannes Gescher (Karlsruhe Institut für Technology) for providing the material for yeast cloning, Matthias P. Mayer [Center for Molecular Biology of the University of Heidelberg (ZMBH)] for help with CD measurements, and Regina Zahn (ZMBH) for technical assistance. Changhan Lee received a personal scholarship through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2014R1A6A3A03057742), and has been provided a travel grant from the Karolinska Institutet for a research visit to the University of Heidelberg. The project was funded by the Swedish Research Council for Medicine and Health (K2012-56X-22034-01-3), the Karolinska Institutet (U.R.), and by grants from the Deutsche Forschungsgemeinschaft (BB617/17-2 and MO 970/4-2) (to B.B. and A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712051115/-/DCSupplemental.
References
- 1.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Squires CL, Pedersen S, Ross BM, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 6.Chastanet A, Derre I, Nair S, Msadek T. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J Bacteriol. 2004;186:1165–1174. doi: 10.1128/JB.186.4.1165-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Oliveira NE, et al. clpB, a class III heat-shock gene regulated by CtsR, is involved in thermotolerance and virulence of Enterococcus faecalis. Microbiology. 2011;157:656–665. doi: 10.1099/mic.0.041897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krüger E, Völker U, Hecker M. Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance. J Bacteriol. 1994;176:3360–3367. doi: 10.1128/jb.176.11.3360-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krüger E, Witt E, Ohlmeier S, Hanschke R, Hecker M. The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol. 2000;182:3259–3265. doi: 10.1128/jb.182.11.3259-3265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirstein J, Dougan DA, Gerth U, Hecker M, Turgay K. The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J. 2007;26:2061–2070. doi: 10.1038/sj.emboj.7601655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlothauer T, Mogk A, Dougan DA, Bukau B, Turgay K. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc Natl Acad Sci USA. 2003;100:2306–2311. doi: 10.1073/pnas.0535717100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meibom KL, et al. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol Microbiol. 2008;67:1384–1401. doi: 10.1111/j.1365-2958.2008.06139.x. [DOI] [PubMed] [Google Scholar]
- 14.Vaubourgeix J, et al. Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe. 2015;17:178–190. doi: 10.1016/j.chom.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan L, Rodrigues PH, Bélanger M, Dunn W, Jr, Progulske-Fox A. The Porphyromonas gingivalis clpB gene is involved in cellular invasion in vitro and virulence in vivo. FEMS Immunol Med Microbiol. 2007;51:388–398. doi: 10.1111/j.1574-695X.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 16.Brodmann M, Dreier RF, Broz P, Basler M. Francisella requires dynamic type VI secretion system and ClpB to deliver effectors for phagosomal escape. Nat Commun. 2017;8:15853. doi: 10.1038/ncomms15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerr KG, Snelling AM. Pseudomonas aeruginosa: A formidable and ever-present adversary. J Hosp Infect. 2009;73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Rakhimova E, et al. Pseudomonas aeruginosa population biology in chronic obstructive pulmonary disease. J Infect Dis. 2009;200:1928–1935. doi: 10.1086/648404. [DOI] [PubMed] [Google Scholar]
- 19.Tielen P, et al. Genotypic and phenotypic characterization of Pseudomonas aeruginosa isolates from urinary tract infections. Int J Med Microbiol. 2011;301:282–292. doi: 10.1016/j.ijmm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Römling U, Wingender J, Müller H, Tümmler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer S, et al. Intraclonal genome diversity of the major Pseudomonas aeruginosa clones C and PA14. Environ Microbiol Rep. 2016;8:227–234. doi: 10.1111/1758-2229.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Soyza A, et al. EU FP7 funded COST Action BM1003 “Cell surface virulence determinants of cystic fibrosis pathogens” Developing an international Pseudomonas aeruginosa reference panel. MicrobiologyOpen. 2013;2:1010–1023. doi: 10.1002/mbo3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C, Wigren E, Lünsdorf H, Römling U. Protein homeostasis: More than resisting a hot bath. Curr Opin Microbiol. 2016;30:147–154. doi: 10.1016/j.mib.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Bojer MS, Struve C, Ingmer H, Hansen DS, Krogfelt KA. Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS One. 2010;5:e15467. doi: 10.1371/journal.pone.0015467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C, et al. A novel protein quality control mechanism contributes to heat shock resistance of worldwide-distributed Pseudomonas aeruginosa clone C strains. Environ Microbiol. 2015;17:4511–4526. doi: 10.1111/1462-2920.12915. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 27.Wang F, et al. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature. 2011;471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- 28.Kim YI, et al. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- 29.Mogk A, Kummer E, Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trentini DB, et al. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 2016;539:48–53. doi: 10.1038/nature20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 32.Mogk A, et al. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroni M, et al. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife. 2014;3:e02481. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlieker C, Tews I, Bukau B, Mogk A. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett. 2004;578:351–356. doi: 10.1016/j.febslet.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Gänzle M. Some like it hot: Heat resistance of Escherichia coli in food. Front Microbiol. 2016;7:1763. doi: 10.3389/fmicb.2016.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas JG, Baneyx F. Protein folding in the cytoplasm of Escherichia coli: Requirements for the DnaK-DnaJ-GrpE and GroEL-GroES molecular chaperone machines. Mol Microbiol. 1996;21:1185–1196. doi: 10.1046/j.1365-2958.1996.651436.x. [DOI] [PubMed] [Google Scholar]
- 37.Jung G, Jones G, Masison DC. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc Natl Acad Sci USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reidy M, Miot M, Masison DC. Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics. 2012;192:185–193. doi: 10.1534/genetics.112.142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oguchi Y, et al. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat Struct Mol Biol. 2012;19:1338–1346. doi: 10.1038/nsmb.2441. [DOI] [PubMed] [Google Scholar]
- 40.Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 42.Lee HW, et al. Heat shock protein 70 (HSP70) expression is associated with poor prognosis in intestinal type gastric cancer. Virchows Arch. 2013;463:489–495. doi: 10.1007/s00428-013-1461-x. [DOI] [PubMed] [Google Scholar]
- 43.Schlieker C, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- 44.Beinker P, Schlee S, Groemping Y, Seidel R, Reinstein J. The N terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J Biol Chem. 2002;277:47160–47166. doi: 10.1074/jbc.M207853200. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, Tek V, Akoev V, Zolkiewski M. Conserved amino acid residues within the amino-terminal domain of ClpB are essential for the chaperone activity. J Mol Biol. 2002;321:111–120. doi: 10.1016/s0022-2836(02)00591-0. [DOI] [PubMed] [Google Scholar]
- 46.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 47.Clarke AK, Eriksson MJ. The truncated form of the bacterial heat shock protein ClpB/HSP100 contributes to development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 2000;182:7092–7096. doi: 10.1128/jb.182.24.7092-7096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Acebrón SP, Martín I, del Castillo U, Moro F, Muga A. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 2009;583:2991–2996. doi: 10.1016/j.febslet.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 49.Eriksson MJ, Clarke AK. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:4839–4846. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simão RC, Susin MF, Alvarez-Martinez CE, Gomes SL. Cells lacking ClpB display a prolonged shutoff phase of the heat shock response in Caulobacter crescentus. Mol Microbiol. 2005;57:592–603. doi: 10.1111/j.1365-2958.2005.04713.x. [DOI] [PubMed] [Google Scholar]
- 51.Frees D, et al. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54:1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 52.Kummer E, et al. Bacterial and yeast AAA+ disaggregases ClpB and Hsp104 operate through conserved mechanism involving cooperation with Hsp70. J Mol Biol. 2016;428:4378–4391. doi: 10.1016/j.jmb.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Kirstein J, Molière N, Dougan DA, Turgay K. Adapting the machine: Adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 54.Suokko A, Poutanen M, Savijoki K, Kalkkinen N, Varmanen P. ClpL is essential for induction of thermotolerance and is potentially part of the HrcA regulon in Lactobacillus gasseri. Proteomics. 2008;8:1029–1041. doi: 10.1002/pmic.200700925. [DOI] [PubMed] [Google Scholar]
- 55.Miethke M, Hecker M, Gerth U. Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J Bacteriol. 2006;188:4610–4619. doi: 10.1128/JB.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park SS, et al. ClpL is a chaperone without auxiliary factors. FEBS J. 2015;282:1352–1367. doi: 10.1111/febs.13228. [DOI] [PubMed] [Google Scholar]
- 57.Rosenzweig R, et al. ClpB N-terminal domain plays a regulatory role in protein disaggregation. Proc Natl Acad Sci USA. 2015;112:E6872–E6881. doi: 10.1073/pnas.1512783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen SV, Harhay GP, Bono JL, Smith TP, Harhay DM. Genome sequence of the thermotolerant foodborne pathogen Salmonella enterica serovar Senftenberg ATCC 43845 and phylogenetic analysis of loci encoding increased protein quality control mechanisms. mSystems. 2017;2:e00190-16. doi: 10.1128/mSystems.00190-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jørgensen SB, et al. Heat-resistant, extended-spectrum β-lactamase-producing Klebsiella pneumoniae in endoscope-mediated outbreak. J Hosp Infect. 2016;93:57–62. doi: 10.1016/j.jhin.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 61.Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): What they do and how they work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 63.Krajewski SS, Joswig M, Nagel M, Narberhaus F. A tricistronic heat shock operon is important for stress tolerance of Pseudomonas putida and conserved in many environmental bacteria. Environ Microbiol. 2014;16:1835–1853. doi: 10.1111/1462-2920.12432. [DOI] [PubMed] [Google Scholar]
- 64.Mogk A, Deuerling E, Vorderwülbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 65.Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.