Significance
Highly reduced genomes from bacteria that are long-term beneficial endosymbionts of insects often show remarkable structural stability. Endosymbionts in insects diverged by tens or hundreds of millions of years often have genomes almost completely conserved in gene order and content. Here, we show that an endosymbiont in some cicadas has repeatedly and independently fractured into complexes of distinct genomic and cellular lineages present in the same host. Individual endosymbiont lineages, having lost many of the essential ancestral genes, rely on each other for basic function and together seem to provide the same nutritional benefits as the ancestral single symbiont. These cicada endosymbionts show genomic parallels to mitochondria and provide another example of how normally stable genomes can lose structural stability.
Keywords: nutritional endosymbiont, genome evolution, organelle, mitochondria, cicadas
Abstract
Bacterial endosymbionts that provide nutrients to hosts often have genomes that are extremely stable in structure and gene content. In contrast, the genome of the endosymbiont Hodgkinia cicadicola has fractured into multiple distinct lineages in some species of the cicada genus Tettigades. To better understand the frequency, timing, and outcomes of Hodgkinia lineage splitting throughout this cicada genus, we sampled cicadas over three field seasons in Chile and performed genomics and microscopy on representative samples. We found that a single ancestral Hodgkinia lineage has split at least six independent times in Tettigades over the last 4 million years, resulting in complexes of between two and six distinct Hodgkinia lineages per host. Individual genomes in these symbiotic complexes differ dramatically in relative abundance, genome size, organization, and gene content. Each Hodgkinia lineage retains a small set of core genes involved in genetic information processing, but the high level of gene loss experienced by all genomes suggests that extensive sharing of gene products among symbiont cells must occur. In total, Hodgkinia complexes that consist of multiple lineages encode nearly complete sets of genes present on the ancestral single lineage and presumably perform the same functions as symbionts that have not undergone splitting. However, differences in the timing of the splits, along with dissimilar gene loss patterns on the resulting genomes, have led to very different outcomes of lineage splitting in extant cicadas.
Genome reduction is a stereotypical response in endosymbiotic bacteria. At the outset of an intracellular association with a eukaryotic host, bacterial genomes go through a turbulent phase of gene inactivation, genome rearrangement, and genome reduction (1–4). In cases where the relationship persists because the bacterial infection is beneficial for the host, the genomic dynamism in the endosymbiont often transitions to a long period of genomic stasis (5–7). Many genomes from bacterial endosymbionts that stably associate with hosts, especially those of sap-feeding insects, have undergone few or no genome rearrangements for tens or hundreds of millions of years (8, 9). The genes on these tiny genomes converge to a stable set encoding functions to which further loss would presumably have strong negative consequences for the symbiosis (10, 11). The retained gene sets typically include genes involved in genetic information processing (genome replication, transcription, and translation) and some aspects of basic central metabolism, as well as biosynthesis of essential amino acids and vitamins that the host requires (12).
However, genome stability is not inevitable in endosymbionts that provide a benefit to the host. Mitochondrial genomes stably map as colinear 15- to 18-kb circular genomes in most animals (13, 14) but exist as multichromosome fragmented genomes in many eukaryotes (15), including lice (16), plants (17, 18), and excavate protists (19). Similarly, we have previously shown that, in some cicadas, one of their ancient bacterial endosymbionts, Candidatus Hodgkinia cicadicola (hereafter Hodgkinia), has fragmented into two (20) or numerous (21) genomic and cellular lineages that are present in the same host. Hodgkinia’s coresident bacterial endosymbiont, Candidatus Sulcia muelleri (hereafter Sulcia), remains as a single genomic lineage in all cicadas studied to date (20, 21). These two bacteria share many characteristics common in long-term endosymbionts and organelles, including strict maternal transmission and extreme genome reduction. One of the most striking differences between Sulcia and Hodgkinia is in their rates of evolution. The genome of Sulcia shows a very low substitution rate across a wide diversity of Auchenorrhynchan insects (a group that contains cicadas, spittlebugs, treehoppers, and planthoppers) (22, 23) while substitution rates in Hodgkinia genomes have been estimated to be 50 to 100 times higher than those in Sulcia (21). We hypothesized that this difference in substitution rate reflects a difference in mutation rate and that the high mutation rate in Hodgkinia combined with long and variable life cycles of cicadas allows complementary inactivating mutations to rise to high frequencies in some cicadas, which eventually splits the lineage (20, 21).
Detailed mechanistic inferences on Hodgkinia lineage splitting are hard to make from existing data. Here, we sought to measure the amount of lineage splitting in the cicada genus Tettigades, where Hodgkinia was first reported to fragment into a symbiotic complex (20), and asked a series of questions related to the timing and outcome of the process. Is it clock-like, or does it happen in bursts? Is it rare or common? Does splitting lead to predictably subfunctionalized gene sets on new lineages, or does chance seem to play a large role? We sampled Tettigades species over the course of three field seasons and used amplicon sequencing, comparative metagenomics, and microscopy to address these questions.
Results
Sampling the Tettigades Diversity in Chile.
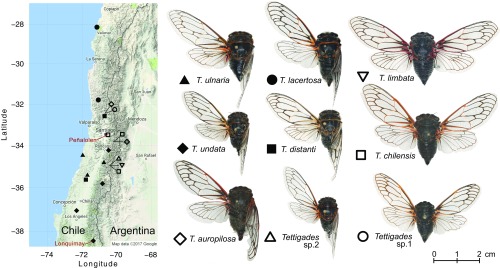
Apart from our 2014 study (20), the genus Tettigades has received little attention since its last taxonomic revision 60 years ago (24). Consequently, the availability of cicada material suitable for DNA-based work was limited. We spent three field seasons collecting cicadas across Chile, which, along with Argentina, is the native range of Tettigades (24). From nearly 1,000 Tettigades specimens, we identified 19 populations that were geographically, genetically, or morphologically distinct from each other (Fig. 1 and Dataset S1). Based on cicada morphology, our collections represented nine species, including two that have not been previously described (Fig. 1). Phylogenetic analysis of partial mitochondrial cytochrome oxidase I (COI) gene sequences indicated that our collection represents seven Tettigades clades (Fig. 2A). Six of these clades correspond to morphologically identified species while the seventh consists of specimens that were classified as three species based on morphology (the chilensis-sp. 1-auropilosa clade in Fig. 2A). Phylogenetic analysis of partial sequences of three protein-coding genes of the Sulcia symbiont of these cicadas reconstructed the same seven clades (SI Appendix, Fig. S1). However, we note that deep relationships among Tettigades species are not well-supported in any dataset, making our estimate of seven clades somewhat arbitrary. Genomic data for hosts and symbionts also fail to fully clarify the details of these relationships (SI Appendix, Fig. S2 A and B).
Fig. 1.
The collection sites of the experimental cicada populations and the morphology of the sampled Tettigades species. Symbols provided next to species names are shown on the map to indicate sites that each species was collected from. Two sites from where we sampled >15 individuals per population are indicated with red font. Representative specimens for each species were mounted and individually imaged using the Macropod system (Macroscopic Solutions LLC).
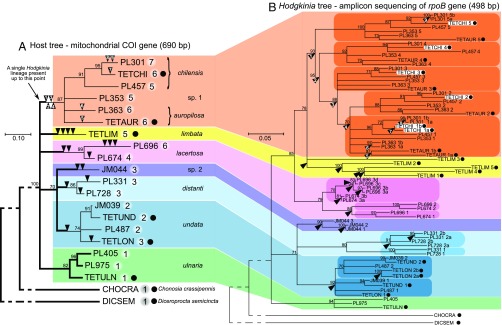
Fig. 2.
Maximum likelihood phylogenies of cicadas from the genus Tettigades (A) and of their Hodgkinia symbionts (B), based on partial sequences of mitochondrial cytochrome oxidase I (COI) gene and RNA polymerase subunit B (rpoB) gene, respectively. The phylogenies have been constrained based on multigene phylogenies for samples with genomes sequenced, which are indicated by black dots following leaf labels (SI Appendix, Fig. S2). Bootstrap values of 70% or more are shown as percentages above the nodes, and nodes with lower support have been collapsed. Colors represent species or species groups in which the ancestral state was a single lineage of Hodgkinia. In the host tree, thick branch lines indicate hosts, or the times in their evolutionary history when they hosted only a single Hodgkinia genotype, and numbers after leaf labels represent the number of distinct Hodgkinia rpoB genotypes in that sample, based on genomic data (when available) or rpoB amplicon data. Black arrowheads indicate Hodgkinia splitting events, as opposed to codiversification with hosts. The splits have been individually numbered in the chilensis-sp.1-auropilosa clade to more clearly show the inferred order of events. In the Hodgkinia tree, colored ovals identify clades that include strains from all sampled hosts in a species group, indicating that the first Hodgkinia split happened in the common ancestor of all sampled hosts from that clade. Note that Hodgkinia strains from a single cicada specimen occur in multiple places in the symbiont tree, as illustrated by TETCHI (shown inside white rectangles).
Hodgkinia Complexes Result from Multiple Independent Splits of a Single Ancestral Symbiont Lineage.
The genomic diversity of Hodgkinia in the 19 representative specimens of Tettigades spp. was estimated using amplicon sequencing of the Hodgkinia RNA polymerase subunit B (rpoB) gene. Preliminary low-pass metagenomic sequencing indicated that rpoB was present on most (likely all) Hodgkinia genomes from Tettigades spp., and therefore the number of rpoB sequence types could be used as a conservative proxy for the number of genomic lineages. We found that the 19 representative cicadas hosted between one and seven distinct Hodgkinia rpoB genotypes (Fig. 2A). Phylogenetic analysis showed that these genotypes cluster into seven distinct and well-supported groups corresponding to the seven cicada clades (Fig. 2B and SI Appendix, Fig. S2D)—no symbiont clades are shared among these cicada clades. This pattern indicates that the last common ancestor of Tettigades spp. hosted a single Hodgkinia genotype and that the symbiont splitting occurred only after the diversification of the cicada genus into clades. These results also show that symbiont splitting happened independently in six out of seven of these clades.
We then sequenced and fully assembled 21 Hodgkinia genomes from five representative host specimens, along with six new cicada mitochondrial and Sulcia genomes (Dataset S2). Symbiont genomes from Tettigades ulnaria, Tettigades undata specimen TETUND, and Diceroprocta semicincta were reported previously (20, 25). We compared the endosymbiont metagenomes of eight cicada specimens in total: six from the genus Tettigades, as well as two outgroups (Datasets S1 and S2). As expected, the Sulcia genomes were all colinear, encoded nearly identical sets of genes, and displayed >99% median nucleotide identity across protein-coding genes (Dataset S3). In contrast, Hodgkinia from all other Tettigades specimens was represented by several distinct genomes (Fig. 3). These genomic data confirm that our amplicon assay accurately reflected Hodgkinia diversity, with one exception: in Tettigades undata specimen TETLON, rpoB genotypes that differed at a single nucleotide position out of 498 sequenced (99.8% identity) appeared to represent polymorphism within a single lineage rather than genomes of distinct lineages. Interestingly, rpoB genotypes that differed at two nucleotide positions (99.6% identity) in Tettigades chilensis specimen TETCHI corresponded to distinct, recently diverged genomes (TETCHI1a and TETCHI1b) (Fig. 2B and SI Appendix, Fig. S2D) with very different gene contents.
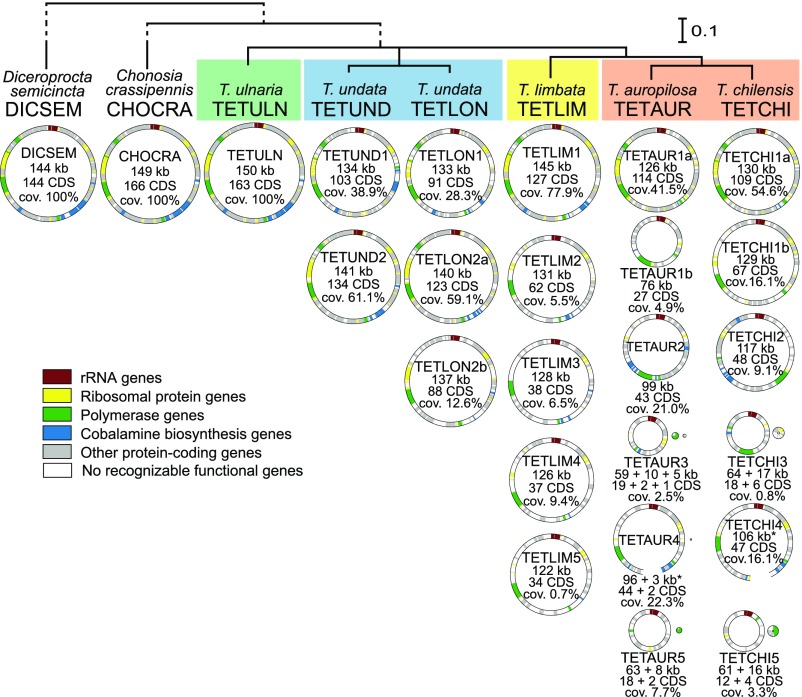
Fig. 3.
Circular diagrams of Hodgkinia genomes from eight cicada species. Host phylogeny is based on 15 mitochondrial genes (SI Appendix, Fig. S2), and host clades where Hodgkinia splits have happened independently are indicated with colored rectangles. All Hodgkinia genomes found in a specimen are represented by circles with diameter corresponding to size; protein-coding and rRNA genes classified as functional are indicated on each circle. In two cases, TETAUR4 and TETCHI4, circles are not closed because of unusual arrangements of chromosomes (SI Appendix, Fig. S11). Basic statistics are provided for each genome, including genome coverage as the proportion of total coverage of all genomes in a complex. See Dataset S5 for more detailed information.
The Timing of Hodgkinia Splits Varies by Millions of Years in Different Cicada Clades.
Comparison of the cicada and Hodgkinia trees (Fig. 2 and SI Appendix, Fig. S2) allowed us to infer the order and timing of Hodgkinia splits. Each clade contains splits that happened in the common ancestors of all studied specimens from that clade and have codiversified with, and are shared among, all extant cicadas in that clade. All clades also contain splits that occurred after the ancestral lineage of each cicada clade diversified and are thus only present in one characterized individual from that clade. Using the chilensis-sp. 1-auropilosa clade as an example, splits labeled 1 to 4 in Fig. 2B occurred in the ancestral lineage of this group, resulting in five symbiont lineages in the last common ancestor of all extant cicadas in this clade. After diversification within the clade, additional splits occurred in the ancestors of specimens TETAUR (split 5), PL363 (split 6), TETCHI (split 7), and PL301 (splits 8 and 9), resulting in between five and seven rpoB genotypes per cicada specimen (Fig. 2B). Similar patterns were seen in all other Tettigades clades.
In the absence of fossil records or other reliable calibration points, we estimated the divergence times of Tettigades clades and Hodgkinia splits using previously published estimates of the rate of evolution of the COI gene in insects (26, 27) and Australasian cicadas (28, 29), corresponding to 0.011 to 0.012 substitutions per base per million years. The average uncorrected pairwise genetic distance between T. ulnaria and cicadas from all other Tettigades clades was 0.0953 (SD 0.0089), corresponding to a divergence time of 4.14 ± 0.39 Mya. This estimate coincides with the latest major period of accelerated uplift of the Central Andean Range (30), which makes sense, as Tettigades spp. are commonly found at higher elevations (24). To estimate the divergence times for subsequent splitting events in the genus Tettigades, we placed a conservative age range window of 5 to 3 My on the node between TETULN and TETCHI. Using alignments of all mitochondrial genes, we estimate that the seven major Tettigades clades diverged between 4 and 3 Mya (SI Appendix, Fig. S3A). Further, we estimate that T. chilensis and Tettigades auropilosa diverged ∼2 Mya and that T. undata specimens TETUND and TETLON diverged ∼1 Mya. Analysis of the concatenation of all universally retained Hodgkinia genes (SI Appendix, Fig. S3B) revealed large differences in the timing of symbiont splitting events. In chilensis-auropilosa and limbata clades, Hodgkinia splits started soon after host clades diverged, and subsequent splits were separated by hundreds of thousands of years. In contrast, the single shared split in the undata species clade took place ∼1 Mya, more than 2 My after the clade emerged. We estimate that the most recent Hodgkinia splits in T. chilensis specimen TETCHI and T. undata specimen TETLON occurred ∼0.3 Mya. We note that, due to a lack of Tettigades fossils or other independent calibration points, the confidence intervals for the absolute dates in above estimates are broad (SI Appendix, Fig. S3). However, we are more confident in our estimates of the relative timing of splitting events, and thus in concluding that these splitting events occurred at very different times in different Tettigades clades.
Hodgkinia Genomes Within a Complex Differ Dramatically in Size, Organization, Gene Content, and Relative Sequencing Coverage.
We observed major differences among Hodgkinia complexes from different hosts, but also between individual genomes that make up a single complex within a cicada (Fig. 3 and Datasets S4 and S5). D. semicincta, Chonosia crassipennis, and T. ulnaria hosted a single Hodgkinia lineage with a gene-dense genome of 144 to 150 kb and conserved genome organization, representing the ancestral state (Fig. 3 and SI Appendix, Fig. S4). All other Tettigades samples had multiple distinct Hodgkinia genomes, with considerably lower coding densities than the single-lineage Hodgkinia (Fig. 3).
These genomes often differed dramatically from each other. In single cicadas, the maximum observed difference in Hodgkinia genome size was 1.8-fold (in TETAUR), the maximum difference in numbers of protein-coding genes was 6.8-fold (in TETCHI), and, remarkably, the maximum difference in genome coverage—which we have shown correlates with cellular abundance (20, 21)—was 114-fold (in TETLIM). In Hodgkinia complexes that have undergone more splitting events (T. chilensis-auropilosa and Tettigades limbata), individual genomes differed among each other to a greater extent, not only in these three categories but also in genomic structure (SI Appendix, Figs. S5–S9). The five Hodgkinia genomes from TETLIM and three Hodgkinia genomes from T. undata specimen TETLON were all colinear but included several large deletions relative to TETULN (SI Appendix, Figs. S5 and S6). However, the genome of one of the lineages from T. undata specimen TETUND (TETUND1) experienced a large inversion that was absent in its sister lineage from specimen TETLON (TETLON1) (SI Appendix, Figs. S6 and S7). This shows that the inversion occurred after the ancestral Hodgkinia split in the undata clade and was not the cause of the split as we had previously speculated (20). Finally, all Hodgkinia genomes from T. chilensis and T. auropilosa have experienced rearrangements and major deletions relative to TETULN (SI Appendix, Figs. S8 and S9). This was particularly clear for the smaller and less abundant genomes (Fig. 3).
In some cases, we found that a Hodgkinia genome has fractured into two or three distinct genomic “minicircles” (for example, TETAUR3 and TETAUR5) (Fig. 3, SI Appendix, Fig. S9, and Dataset S4). We infer that these differences reflect subcellular genome fragmentation rather than lineage-splitting events because sequencing coverages (Dataset S4), organization relative to the ancestral genome (SI Appendix, Figs. S8 and S9), gene contents (Dataset S6), and phylogenetic analyses of individual genes (SI Appendix, Fig. S10) suggest that these minicircles exist in the same cell. One 3-kb, high-coverage minicircle encoding a single protein-coding gene could not be assigned to any lineage (TETAURX) (Dataset S4). Also, we saw some instances of genomes existing as stable alternative forms (SI Appendix, Fig. S11). These phenomena have been observed previously in other endosymbiont genome projects (31, 32) and are not further discussed here.
Hodgkinia Genomes Within a Complex Are Highly Complementary in Gene Content.
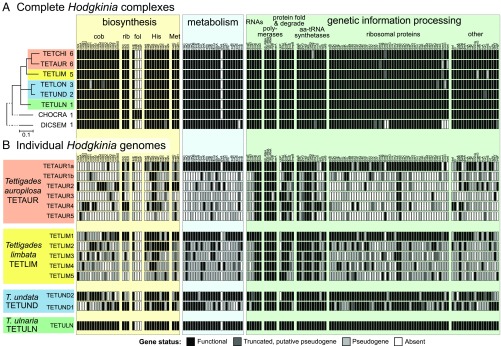
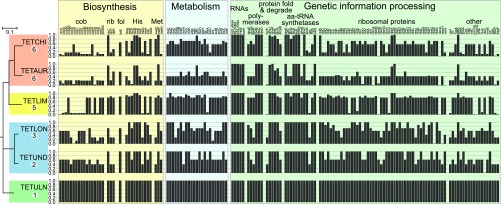
Next, we investigated the retention patterns of protein-coding genes in the Hodgkinia complexes from each cicada, as well as in genomes of individual lineages. We found that the total gene set encoded by each symbiotic complex is remarkably conserved (Fig. 4A and Dataset S6). As expected, the single Hodgkinia genomes from T. ulnaria, C. crassipennis, and, to a lesser extent, the divergent D. semicincta encoded very similar sets of nonhypothetical protein-coding genes. We also found the same patterns in Hodgkinia complexes that have resulted from splitting: The total collective gene set retained across all genomes was very similar to that of single Hodgkinia lineages (Fig. 4A). Despite this high level of total gene set conservation at the complex level, individual Hodgkinia genomes that comprise a complex within a single cicada differed dramatically in how many and which genes they encode (Fig. 4B and Dataset S6). In each cicada, one genome retained the majority of the ancestral gene set. Other genomes encoded many fewer genes, sometimes approaching only 10% of the ancestral set of 163 protein-coding genes. While a few translation-related genes were universally retained in all genomes—the ribosomal RNA operon (rrsA, rrlA, rrfA), tmRNA, three of the four RNA polymerase holoenzyme subunits (rpoB, rpoC, rpoD), a single ribosomal protein (rpsB), and five aminoacyl-tRNA synthetases (alaS, ileS, metG, pheT, valS)—most genes were present on only a subset of genomes (Fig. 4B). Each genome encoded at least two genes not present on any other genome, explaining why each individual lineage is retained.
Fig. 4.
The list of all nonhypothetical protein-coding and RNA (other than tRNA) genes identified in the genomes of Hodgkinia from DICSEM, CHOCRA, and Tettigades spp. The classification (apparently functional, pseudogenized, or absent) of each gene is based on the length of the ORF relative to that in the genome of TETULN Hodgkinia. (A) At the level of a Hodgkinia complex, the highest level functional state of a gene in any of the genomes of a complex is provided. Host phylogeny is based on 15 mitochondrial genes (SI Appendix, Fig. S2), the number of distinct Hodgkinia lineages in a given specimen is provided after each leaf label, and host clades where Hodgkinia splits have happened independently are indicated with colored rectangles. (B) At the level of individual Hodgkinia genomes from four complexes, representing three independent splitting events. cob, cobalamin; fol, folate; rib, riboflavin.
Some gene functional categories had distinctive retention patterns. Many of the genes encoding ribosomal proteins, or involved in general bacterial metabolism (having no direct relation to host nutrition), showed a tendency to be retained only on the most complete and abundant genome in a complex (Fig. 4B). Genes involved in biosynthesis of amino acids and vitamins, which are thought to be products that benefit the host (33), were also typically present on only one genome. However, these biosynthesis-related functional copies were typically scattered across genomes of various sizes and abundances. This was particularly clear for the 17 genes in the cobalamin biosynthesis pathway. In all Hodgkinia complexes (with the exception of TETLON, where cobH and cobN have experienced frameshifts and are likely pseudogenes), each cobalamin-related gene was retained only as a single functional copy. Copies of these 17 genes are scattered across nearly all genomes in a complex.
The similarity of gene sets among four pairs of sister genomes from T. chilensis and T. auropilosa, which descended from a lineage present in the ancestor of these two species, suggests that gene loss happened rapidly after the split but that these greatly reduced genomes have remained without further major changes for about 2 My (SI Appendix, Fig. S12). The patterns were less clear in T. undata, where the populations represented by the two specimens appear to have diverged relatively soon after the Hodgkinia split (SI Appendix, Figs. S3 and S12).
Gene Dosage Varies Widely Among Genes in Biosynthetic Pathways.
Maximal dosage of a gene, defined as the summed relative abundance of all apparently functional gene copies on all genomes in a Hodgkinia complex, occurs when all genomes from a population of cells in a cicada encode that gene (or when Hodgkinia exists as a single lineage, which by definition comprises 100% of the complex). Assuming no change to the size of the host tissue devoted to housing Hodgkinia and no change to Hodgkinia cell size, every split followed by gene nonfunctionalization in one of the lineages results in a lowering of the overall dosage of that gene. For example, the dosage of a gene retained on TETUND1 but lost on TETUND2 will be 39% because TETUND1 makes up 39% of the total Hodgkinia complex (Fig. 3). The overall effect of splitting will therefore be a reduction in the average dosage of genes that are not retained by all new lineages. These changes in dosage have the potential to decouple gene abundances for genes sharing a biochemical pathway but present on different lineages. We note that these differences can be dramatic: The most and the least abundant genomes in the TETLIM and TETCHI complexes differed in abundance by two orders of magnitude (Fig. 3 and Dataset S5).
We found that functional copies of biosynthesis-related genes tend to be scattered across genomes at different coverages, which results in highly variable dosage of genes involved in different steps of a biosynthetic process. For example, in all complexes, hisD and hisE were retained on most or all Hodgkinia genomes (dosage ≥92%), but the dosage of hisA varied among complexes from 41 to 100%, hisB varied from 16 to 100%, hisC varied from 5 to 87%, and so on (Fig. 5). We do not currently understand how drift and selection have influenced the likelihood of retention of these biosynthesis-related genes on a particular genome or the relative abundance of the genomes where particular genes have been retained. However, selection for even dosage may explain why, in Tettigades Hodgkinia complexes, the majority of genes involved in genetic information processing and metabolism are retained on the most abundant genome in each complex. It is plausible that the normal functioning of Hodgkinia lineages requires these genes to be relatively abundant at a complex level. However, a subset of these genes—and not the same ones in different hosts—were sometimes retained only on lower-abundance genomes (Figs. 4B and 5). We do not yet know if changes in gene expression can alleviate these massive gene dosage differences.
Fig. 5.
The summed dosage of nonhypothetical genes present in Hodgkinia complexes, calculated as the cumulative relative abundance of all genomes where the gene has been classified as functional. Maximal dosage is 100% (gene present and apparently functional in all genomes); minimal dosage is 0% (gene pseudogenized or deleted in all genomes).
Different Hodgkinia Cell Types Carry Distinct rRNA Copies.
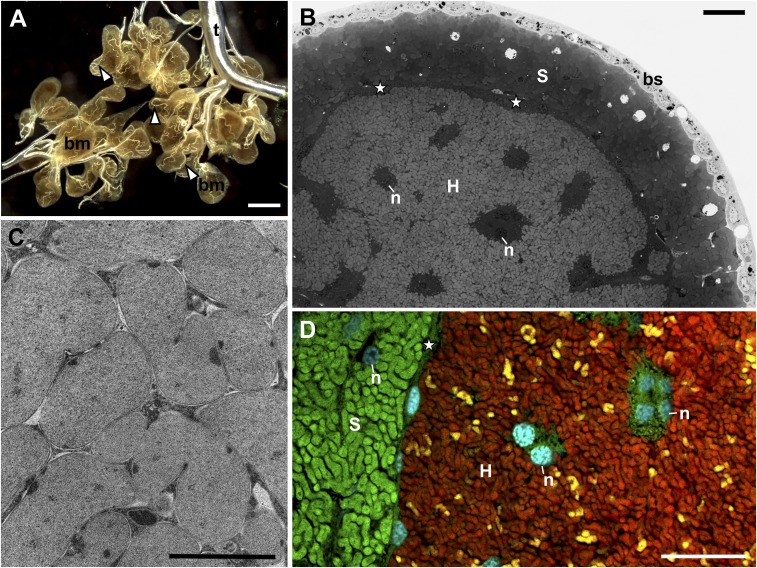
We used light, fluorescence, and transmission electron microscopy (Fig. 6) to look for systematic differences between cicadas hosting Hodgkinia of various complexities and to study the distribution of rRNA types in Hodgkinia cells. In all cicada species, Hodgkinia cells are irregularly shaped, delimited by triple membranes, and densely packed within the large syncytium that occupies the central part of each bacteriome lobe (34) (Fig. 6 A–C). We found no obvious ultrastructural differences among Hodgkinia cells from Tettigades species that differ in the level of complex fragmentation (SI Appendix, Fig. S13). We have previously demonstrated that genomes of different lineages are localized in distinct cell types in T. undata specimen TETUND, as well as in Magicicada tredecim (20, 21). Here, we extend this result in T. chilensis to rRNA molecules. Probes targeting a region of 16S rRNA conserved across Hodgkinia genomes stain all bacterial cells within the Hodgkinia-occupied syncytium, but probes specific to the rRNA variant encoded on one genome stain only a subset of cells (Fig. 6D). Furthermore, probes targeting rRNA variants encoded on distinct Hodgkinia genomes produce nonoverlapping signal (SI Appendix, Fig. S14). Together, these data indicate that (at least) genomes and rRNA molecules are not commonly exchanged among Hodgkinia cells within Tettigades individuals. Each cell type seems to possess a distinct genotype and ribotype.
Fig. 6.
The organization of a cicada bacteriome. (A) The tissue-level view of a dissected male bacteriome from Tettigades sp. 1, which is divided into lobes that are entwined with tracheoles (white arrowheads). bm, bacteriome; t, tracheal system; stereoscope microscope. (Scale bar: 500 μm.) (B) The internal organization of the bacteriome lobe of T. lacertosa, representative of the genus. The external layer consists of simple epithelium (bs, bacteriome sheath). The layer underneath consists of distinct bacteriocyte cells filled with Sulcia (S), and the central part of the bacteriome consists of a single large syncytium filled with cells of Hodgkinia (H). n, nucleus; star, external area of the syncytium that houses Hodgkinia. Light microscope, methylene blue staining. (Scale bar: 10 μm.) (C) Hodgkinia cells in the cytoplasm of the syncytium of Tettigades undata. Note close proximity among cells and their irregular shape. Transmission electron microscope. (Scale bar: 1 μm.) (D) Fluorescence microphotograph of the central part of the bacteriome lobe of T. chilensis specimen TETCHI. Cyan corresponds to the Hoechst, universal DNA stain. Green and red represent the signal of fluorescently labeled probes that target 16S rRNA of Sulcia and Hodgkinia, respectively. Yellow represents the signal of a probe specific to 16S rRNA of only one of Hodgkinia variants, TETCHI2. (Scale bar: 10 μm.) Annotations are the same as in B.
Differences in Hodgkinia Complexes Within Cicada Populations Suggest That Splits Are Ongoing.
Because different cicada populations that would normally be considered the same species in a morphological or COI-based sense can differ in Hodgkinia complex structure (e.g., T. chilensis represented by PL301, TETCHI, and PL457 in Fig. 2), we used amplicon sequencing to see if differences in rpoB genotypes could be found among individuals within a single cicada population. We first assayed two individuals from 12 of the 19 populations used for rpoB amplicon sequencing (Fig. 2A and Dataset S1). In 10 populations, both individuals contained identical sets of rpoB genotypes. However, in one case, rpoB genotypes differed at up to two nucleotide positions, and in another population one of the replicate specimens hosted an additional Hodgkinia genotype that was completely absent in the other specimen. These results indicate that significant Hodgkinia diversity might exist within a single population of a cicada species.
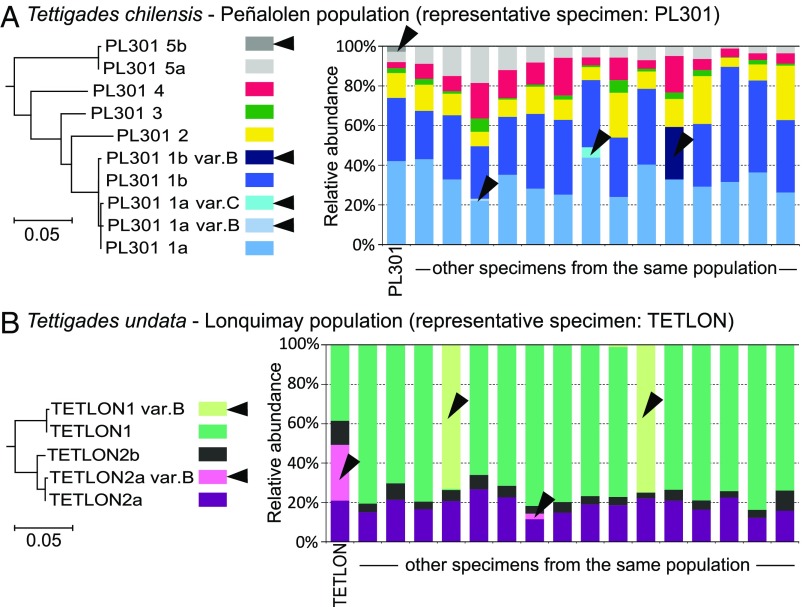
To further explore Hodgkinia lineage diversity within single cicada populations, we sequenced rpoB amplicons from additional specimens from two populations where we had at least 15 individuals (Fig. 7). We want to be clear that our interpretations of the following data, especially of the cases when only a small number of reads representing a given genotype are present in a specimen, are complicated by the possibility of amplicon sequencing artifacts. In both the T. chilensis population from Peñalolen and the T. undata population from Lonquimay (Fig. 1), most individual cicadas hosted a very similar set of Hodgkinia rpoB genotypes. Similar relative abundances of genomes across individuals suggest limited tolerance for variation. However, in both populations, we identified genotypes that varied by only one SNP and were present in not more than two host individuals and which either complemented or replaced a more widespread and very closely related genotype. In several cases, a genotype represented by many reads in some specimens was represented by only few reads in others, suggesting its presence at very low abundance. The genotype abundant in only some specimens may be due to single nucleotide variants of rpoB with no functional significance but could also indicate very recent or ongoing splits. In the case of the T. undata from Lonquimay (specimen TETLON), the rpoB polymorphism between the widespread genotype TETLON_2a and the rare genotype TETLON_2a_varB (Fig. 7B) seems only to reflect a point mutation with no further significance: Genome sequencing confirmed the polymorphism within the rpoB gene but revealed no evidence of substantial genomic variability. In contrast, the single polymorphic position between widespread and consistently abundant Hodgkinia genotypes PL301_1a and PL301_1b in T. chilensis from Peñalolen (Fig. 7A) appears to reflect genuine genome diversity that likely reflects a recent or ongoing split. Our attempts to assemble the Hodgkinia lineage showing this rpoB polymorphism from specimen PL301 resulted in a large number of short contigs, consistent with the idea that very similar but distinct symbiont variants were present in that specimen.
Fig. 7.
The diversity and relative abundance of Hodgkinia lineages in replicate specimens from single populations of (A) Tettigades chilensis (represented by specimen PL301) and (B) T. undata (represented by specimen TETLON), based on amplicon sequencing of rpoB gene. Colors represent different unique rpoB genotypes. The genotypes that are found in only a small proportion of specimens are indicated with arrowheads. Maximum likelihood trees for all genotypes from a population reveal that the genotypes found in a small proportion of specimens differ from widespread genotypes at a single nucleotide position (99.8% identity). They may indicate either single-nucleotide polymorphisms of little functional significance, or ongoing Hodgkinia splits. Preliminary genome data suggest that, in at least some instances, genotypes that differ at a single position do represent recent splitting events.
Discussion
Six Independent Origins of Hodgkinia Complexes as a Natural Experiment.
Our amplicon (Fig. 2) and genomic data (Fig. 3) show that Hodgkinia lineage splitting happened independently in each of the undata, distanti, sp. 2, lacertosa, limbata, and auropilosa-sp. 1-chilensis clades of the genus Tettigades. Because none of these six groups share any splits, they can be considered independent natural experiments that help us understand the process and the outcomes of Hodgkinia lineage splitting.
Some patterns are conserved among independently evolved Hodgkinia complexes. All genomes in all complexes universally encode a few genes involved in transcription and translation (Fig. 4B). In every complex, the most abundant lineage retains the majority of genes involved in basic cellular processes, such as central metabolism and genome replication, transcription, and translation (Fig. 4B), although there is a general trend toward the dominant lineage simply encoding more genes than any other genome in the complex. These gene retention patterns suggest that there is very strong selection to retain some genes involved in core informational processes, perhaps due to dosage or protein targeting effects. These patterns, combined with our amplicon data (Figs. 3 and 7), also indicate that new split lineages tend to be “spun off” from the dominant lineage and that one of the resulting new lineages often rapidly degenerates into a genome that encodes many fewer genes, is smaller, and is much less abundant. Supporting this model, in the five Tettigades Hodgkinia complexes that were fully characterized, we found no cases where two subdominant lineages are sister to each other. However, among cicadas characterized using rpoB amplicon sequencing only, there was one case (Tettigades lacertosa specimen PL696) (Fig. 2) where, following the first Hodgkinia split, both resulting lineages underwent subsequent splits.
But, in many ways, the differences among cicada clades are more informative than the similarities. The first and most obvious difference is in the number of lineages that make up a complex (Fig. 3). This value positively correlates with the time since the first split in a clade, which is three times longer in the cases of limbata and chilensis-auropilosa clades (>3 My; 5 to 7 Hodgkinia lineages) than in the case of the undata clade (∼1 My; 2 to 3 lineages) (Figs. 2 and 3 and SI Appendix, Fig. S3). This suggests that the initiation of splitting begets more splitting and that symbiotic complexity cannot be ratcheted back once started. But our data also indicate that splitting is not necessarily clock-like once started. Splits in the limbata clade, as well as shared splits in the chilensis-auropilosa clade, happened in a relatively short succession (a few hundred thousand years apart) and, in both cases, resulted in five lineages that stably persisted for >2 My. Only some of the chilensis and auropilosa populations have experienced additional splits over the last 2 My (Fig. 2 and SI Appendix, Fig. S3).
The second major difference, related to the number of lineages that comprise a complex, is the variation in gene retention in genomes that differ in abundance, and consequently in dosage of individual genes (Fig. 5). The number of copies present in a complex can differ by as much as two orders of magnitude even for genes with similar functions (e.g., ribosomal proteins), or involved in different steps of a single biosynthetic process. Independently evolved complexes differ dramatically in which genes have low dosage due to splitting and gene inactivation on the abundant genome.
The third major difference is in the genomic structure of the split lineages. For example, in T. limbata clade, the five Hodgkinia genomes are all colinear with each other and with the ancestral single Hodgkinia from T. ulnaria (SI Appendix, Fig. S5). In contrast, despite a similar time since the first split, the six Hodgkinia genomes of T. chilensis have undergone several genomic rearrangements, losing much of the colinearity with the ancestral single Hodgkinia from T. ulnaria (SI Appendix, Fig. S8). These data suggest that the frequency of inactivating mutations that lead to splitting (for example, genome rearrangements vs. point mutations) may be different in different cicada lineages. However, it may just be that Hodgkinia is more prone to rearrangements in some cicada lineages than others.
Overall, these natural experiments point to a large role for random chance in the evolution of Hodgkinia complexity. While the starting point in different Tettigades clades was the same—a single Hodgkinia lineage very similar to that of the present-day T. ulnaria symbiont—differences in the timing of the first split event, in whether and when additional splits were triggered, as well as in underlying mutational mechanisms, have resulted in wildly different genomic outcomes in extant cicadas.
The Apparent Importance of Gene Product Transport for Symbiotic Complexes.
Similar to mitochondria and chloroplasts, endosymbiotic bacteria that have undergone high levels of genome reduction are thought to rely on host proteins to function (35–38). The smallest Hodgkinia genome from Tettigades spp., TETCHI5, encodes only 16 proteins and 5 RNAs, on par with a typical insect mitochondrial genome (13 protein-coding, 2 rRNA, and 22 tRNA genes) (13). This extreme level of reduction of individual genomes, combined with the conservation of the ancestral Hodgkinia gene set at the complex level and complementarity among genomes with regard to which genes they retain, shows that individual lineages must rely not only on the host, but also on each other, to function. One striking example is the production and assembly of Hodgkinia ribosomes. A ribosome requires both rRNA and ribosomal proteins for normal rRNA folding, assembly, and function (39). While each Hodgkinia genome encodes an rRNA operon, the ribosomal proteins tend to only be encoded on the most abundant genome in a complex (Fig. 4B). Because our fluorescence microscopy shows that rRNA is not commonly exchanged among cells (Fig. 6D and SI Appendix, Fig. S14), the ribosomal proteins must be shuttled between Hodgkinia cell types so that each cell type can build functional ribosomes. Alternatively, the ribosomes in cells with degenerate genomes may function poorly or not at all. Similarly, genes that encode enzymes that act on genomes—which we have shown are not shared among cell types (20, 21), such as dnaE (the DNA polymerase III alpha subunit), dnaQ (the DNA polymerase III epsilon subunit), and rpoA (the RNA polymerase alpha subunit)—are lost by various Hodgkinia lineages (Fig. 4B). However, these Hodgkinia cell types contain genomes that must be replicated and transcribed. Thus, it appears that the products of genes involved in essential bacterial cellular functions need to be transported among Hodgkinia cells in a complex (20). The mechanisms supporting this transport may overlap with those for transport of nucleus-encoded gene products to organelles and endosymbionts (36, 38, 40, 41), but, at this time, they are unknown.
Why Does Splitting Happen, and Why only in Some Tettigades Species?
We have previously speculated that the process of Hodgkinia lineage splitting is nonadaptive for the cicada host and perhaps deleterious in some cases (20, 21). As the Hodgkinia symbiotic complex becomes more fragmented, there is likely an increased cost to maintain and transmit a larger number of cell types, especially in very complicated cases, such as in Magicicada tredecim (21). Along with this increased complexity, the overall gene dosages can become quite uneven (42) (Fig. 5), which, together with the compartmentalization of biosynthetic pathways into distinct cells, may result in the Hodgkinia population becoming less efficient in making nutrients. Overall, it seems that each click of the ratchet-like process of splitting makes it increasingly difficult for the cicada to control its endosymbiont population. The question of whether splitting is driven mostly by neutral processes (i.e., increased mutational load due to long host lifecycles and/or host population bottlenecks) or adaptive forces (i.e., cheating by individual Hodgkinia lineages) at the levels of Hodgkinia genome copies and cells remains unsolved (20, 21) but is an interesting area for future research (43).
The amount of published data on the biology of the genus Tettigades is extremely limited (44). This is a general problem for cicadas: They have long life cycles compared with other insects, their geographic distributions can be patchy, and they spend most of their lives underground. As a consequence, much of what we know about Tettigades has come from our last three field seasons, as well as inspection of scattered records and collections. While certainly not conclusive at this point, we noted that several Tettigades species carrying complex Hodgkinia—T. chilensis, auropilosa, limbata, sp. 1, lacertosa—only emerged at certain sites in large numbers every few years. These emergence patterns roughly parallel those of the long-lived North American periodical cicada genus Magicicada (45), which harbor extremely fragmented Hodgkinia (21, 42), and contrasts with observations for cicada species with relatively simple symbioses (D. semicincta, T. ulnaria, T. undata) that appear to emerge every year because of their short overlapping generation times. While definitive studies will take decades, these observations are consistent with host life cycle length, or perhaps population dynamics, playing a role in the complexity of its Hodgkinia population (21).
Where Does Splitting End?
The extremely fragmented symbionts in the genus Magicicada (21, 42) make it clear that Chilean cicadas are far from the biological limit of how complicated Hodgkinia complexes can become. The Magicicada data, combined with our data showing that splitting seems to be ongoing in some cicada clades, suggest that Hodgkinia lineage splitting may continue, at least in some Tettigades species, into the future. If, as we suspect, the fragmentation process is deleterious, especially at its more advanced stages, these cicadas may be at an increasing disadvantage to ecologically similar organisms with more efficient symbioses. Unless the process is stabilized, the ultimate fate for the entire symbiosis may be extinction (21). That is, unless the degenerating symbiont is replaced by a new, more efficient microorganism that would allow the symbiosis to claw itself out of the “rabbit hole” it has dug itself into (1, 22).
Methods
Details of all methods are provided in SI Appendix.
Adult cicada specimens were captured in Chile between 2006 and 2016. The detailed list is provided in Dataset S1. In most cases, more than one specimen was available per population. Representative specimens were identified based on morphological characters (ref. 24, and references therein). The identity of all specimens used in the study was confirmed by partial sequencing of the mitochondrial cytochrome oxidase I (COI) gene.
DNA extracted from dissected bacteriome tissue, typically from replicate specimens per population, was used for amplification and sequencing of COI and of three protein-coding genes of Sulcia. The diversity and phylogenetic relationships of Hodgkinia in experimental cicadas were estimated using amplicon sequencing of a 498-bp region of RNA polymerase subunit beta (rpoB) gene, conserved in all Hodgkinia lineages from Tettigades spp. for which we had genomes sequenced. Amplicon libraries were sequenced in a multiplexed 2 × 300-bp Illumina Miseq lane, and the data were analyzed using mothur v. 1.37.4 (46), along with a custom analysis procedure that allowed us to reliably detect Hodgkinia genotypes present in each sample.
We sequenced the bacteriome metagenomes of six cicadas on various Illumina platforms (Dataset S2): specimens representing five divergent populations from the genus Tettigades, as well as a species from a closely related genus, Chonosia crassipennis. Quality-trimmed reads were used for assembly using SPAdes versions 3.1.1 and 3.7.0 (47). PCR was used to close gaps between scaffolds and to verify the sequences of all rRNA operons, as well as to verify alternative arrangements of some genomes. We assembled mitogenomes, Sulcia genomes from six sequenced cicadas, and genomes of all Hodgkinia lineages from five of them. The quality of the final genomes was verified by mapping reads and manual inspection of the read alignments. Annotation was conducted by recursive searches for a manually curated set of protein-coding, rRNA, and noncoding RNA (ncRNA) genes from all previously characterized Hodgkinia or Sulcia lineages using HMMER 3.1b2 (48). Based on their length relative to the references, significant hits were classified as functional (≥85%), putative pseudogenes (≥60%), or pseudogenes. The results were supplemented by rRNA searches using RNAmmer v. 1.2 (49) and tRNA searches with tRNAscan-SE v. 1.23 (50). The genomes were compared and illustrated using custom Python and Processing scripts.
Phylogenies of the cicadas, as well as their symbionts, were based on alignments of all genes other than tRNA that had been classified as functional in all genomes characterized. Sequences were aligned using mafft v. 7.221 (51), in protein space for protein-coding genes followed by reverse translation to nucleotides. Phylogenetic analyses of the partitioned datasets were conducted using RAxML v. 8.2.9 (52) assuming the GTR+GAMMA model and with 100 bootstrap replicates. The resulting multigene mitochondrial, Hodgkinia, and Sulcia trees were used to constrain phylogenies for a larger set of samples, which were based, respectively, on partial sequences of COI, rpoB, or on a concatenation of partial sequences of three Sulcia genes. In all cases, we used RAxML with the GTR+GAMMA model, partitions corresponding to three codon positions, and 1,000 bootstrap replicates.
The analyses of Tettigades divergence times were conducted using PhyloBayes v. 4.1c (53), based on alignments of all mitochondrial or shared Hodgkinia genes for all Tettigades clades with metagenomes assembled and Chonosia crassipennis as an outgroup. Tree topologies were based on RAxML reconstructions (SI Appendix, Fig. S2), except that we collapsed nodes between species groups. The runs were calibrated using the conserved estimate of the divergence time for the TETULN-TETCHI node (5 to 3 Mya); they were based on the average pairwise divergence of the full-length COI gene between TETULN and other Tettigades specimens, and on previously published estimates of COI evolution rate for insects and cicadas (26, 28, 29). Three independent PhyloBayes runs, with 15,000 generations and 5,000-generation burn-in, were run for each gene set.
For transmission electron microscopy (TEM), partially dissected cicada tissues were fixed in the field and stored in 2.5% glutaraldehyde solution and then fully dissected, postfixed using 1% osmium tetroxide, and embedded in epoxy resin. Semithin sections (1 µm thick) were stained with 1% methylene blue in 1% borax and analyzed and photographed under a Nikon Eclipse 80i light microscope. Ultrathin sections (90 nm thick) for TEM studies were contrasted with lead citrate and uranyl acetate and examined using a Jeol JEM 2100 (Jeol) electron microscope. For fluorescent in situ hybridization (FISH), ethanol-preserved bacteriome tissue was fixed in 4% formaldehyde solution, embedded in paraffin, and sectioned to the thickness of 5 to 10 µm. Hybridizations with fluorescently labeled probes targeting unique regions of rRNA, as well as unlabeled helper oligos, were conducted as described previously (20).
Supplementary Material
Acknowledgments
We thank Filip Husník and Dan Vanderpool for valuable methodological advice; David Quammen for field work assistance; Ada Jankowska for technical assistance; and all members of the J.P.M. laboratory for helpful discussions. We thank Mario Enrique Elgueta Donoso (El Museo Nacional de Historia Natural), Ana M. Marino de Remes Lenicov (El Museo de La Plata), and Allen Sanborn (Barry University) for access to cicada collections; and Ben Price and Mick Web (National History Museum, London) and Jerome Sueur and Thiery Burgoin (Museum National d'Histoire Naturelle, Paris) for photographs of additional Tettigades types and nontypes. This work was supported by National Science Foundation Grants IOS-1256680 and IOS-1553529, National Aeronautics and Space Administration Astrobiology Institute Award NNA15BB04A, and National Geographic Society Grant 9760-15. C.S. and K.N. acknowledge funding from the Ernst Mayr Travel Grant, the Museum of Comparative Zoology (Harvard University), and the University of Connecticut.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Annotated genomes, next-generation sequencing reads, and Sanger sequences have been deposited in National Center for Biotechnology Information databases. The genomic data can be accessed through the Umbrella BioProject (accession no. PRJNA386376). Individual accession numbers are listed in Dataset S7.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712321115/-/DCSupplemental.
References
- 1.Husnik F, McCutcheon JP. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci USA. 2016;113:E5416–E5424. doi: 10.1073/pnas.1603910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manzano-Marín A, Latorre A. Settling down: The genome of Serratia symbiotica from the aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol. 2014;6:1683–1698. doi: 10.1093/gbe/evu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakeson KF, et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 2014;6:76–93. doi: 10.1093/gbe/evt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke GR, Moran NA. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latorre A, Manzano-Marín A. Dissecting genome reduction and trait loss in insect endosymbionts. Ann N Y Acad Sci. 2017;1389:52–75. doi: 10.1111/nyas.13222. [DOI] [PubMed] [Google Scholar]
- 6.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 7.Toft C, Andersson SGE. Evolutionary microbial genomics: Insights into bacterial host adaptation. Nat Rev Genet. 2010;11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 8.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 10.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 11.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 12.McCutcheon JP. The bacterial essence of tiny symbiont genomes. Curr Opin Microbiol. 2010;13:73–78. doi: 10.1016/j.mib.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cameron SL. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu Rev Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 14.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 2015;112:10177–10184. doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao R, Zhu X-Q, Barker SC, Herd K. Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol Evol. 2012;4:1088–1101. doi: 10.1093/gbe/evs088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z, Cuthbert JM, Taylor DR, Sloan DB. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc Natl Acad Sci USA. 2015;112:10185–10191. doi: 10.1073/pnas.1421397112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice DW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342:1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 19.Vlcek C, Marande W, Teijeiro S, Lukeš J, Burger G. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 2011;39:979–988. doi: 10.1093/nar/gkq883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Leuven JT, Meister RC, Simon C, McCutcheon JP. Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell. 2014;158:1270–1280. doi: 10.1016/j.cell.2014.07.047. [DOI] [PubMed] [Google Scholar]
- 21.Campbell MA, et al. Genome expansion via lineage splitting and genome reduction in the cicada endosymbiont Hodgkinia. Proc Natl Acad Sci USA. 2015;112:10192–10199. doi: 10.1073/pnas.1421386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett GM, Moran NA. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: An ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres BA. Revision del genero “Tettigades” Amy. et Serv. (Homoptera-Cicadidae) Revista del Museo de La Plata Nueva Serie. 1958;7:51–106. [Google Scholar]
- 25.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brower AV. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papadopoulou A, Anastasiou I, Vogler AP. Revisiting the insect mitochondrial molecular clock: The mid-Aegean trench calibration. Mol Biol Evol. 2010;27:1659–1672. doi: 10.1093/molbev/msq051. [DOI] [PubMed] [Google Scholar]
- 28.Marshall DC, et al. Inflation of molecular clock rates and dates: Molecular phylogenetics, biogeography, and diversification of a global cicada radiation from Australasia (Hemiptera: Cicadidae: Cicadettini) Syst Biol. 2016;65:16–34. doi: 10.1093/sysbio/syv069. [DOI] [PubMed] [Google Scholar]
- 29.Ellis EA, et al. Phylogeography of six codistributed New Zealand cicadas and their relationship to multiple biogeographical boundaries suggest a re‐evaluation of the Taupo Line. J Biogeogr. 2015;42:1761–1775. [Google Scholar]
- 30.Folguera A, et al. A review of Late Cretaceous to Quaternary palaeogeography of the southern Andes. Biol J Linn Soc Lond. 2011;103:250–268. [Google Scholar]
- 31.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sloan DB, Moran NA. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biol Evol. 2013;5:783–793. doi: 10.1093/gbe/evt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchner P. Studien an intracellularen symbionten v. die symbiontischen einrichtungen der zikaden. Z Morphol Oekol Tiere. 1925;4:88–245. [Google Scholar]
- 35.Morales J, et al. Development of a toolbox to dissect host-endosymbiont interactions and protein trafficking in the trypanosomatid Angomonas deanei. BMC Evol Biol. 2016;16:247. doi: 10.1186/s12862-016-0820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakabachi A, Ishida K, Hongoh Y, Ohkuma M, Miyagishima SY. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr Biol. 2014;24:R640–R641. doi: 10.1016/j.cub.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 37.McCutcheon JP. From microbiology to cell biology: When an intracellular bacterium becomes part of its host cell. Curr Opin Cell Biol. 2016;41:132–136. doi: 10.1016/j.ceb.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singer A, et al. Massive protein import into the early-evolutionary-stage photosynthetic organelle of the amoeba Paulinella chromatophora. Curr Biol. 2017;27:2763–2773.e5. doi: 10.1016/j.cub.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 40.Li HM, Chiu C-C. Protein transport into chloroplasts. Annu Rev Plant Biol. 2010;61:157–180. doi: 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- 41.Schleiff E, Becker T. Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- 42.Campbell MA, Łukasik P, Simon C, McCutcheon JP. Idiosyncratic genome degradation in a bacterial endosymbiont of periodical cicadas. Curr Biol. 2017;27:3568–3575.e3. doi: 10.1016/j.cub.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shitut S, Ahsendorf T, Pande S, Egbert M, Kost C. 2017. Metabolic coupling in bacteria. bioRxiv:10.1101/114462. [DOI] [PubMed]
- 44.Pirion A. Nota sobre la Tettigades chilensis. Rev Chil Hist Nat. 1929;33:308–311. [Google Scholar]
- 45.Williams KS, Simon C. The ecology, behavior, and evolution of periodical cicadas. Annu Rev Entomol. 1995;40:269–295. [Google Scholar]
- 46.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eddy SR. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagesen K, et al. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.