Significance
RNA molecules fold into functional structures as they are being synthesized by transcription. Here, we developed a single-molecule fluorescence resonance energy transfer (FRET) assay that permits real-time monitoring of the cotranscriptional folding of RNA molecules. This single-molecule FRET assay makes it possible to carry out controlled studies of the two main physical mechanisms that regulate cotranscriptional RNA folding: transcriptional pausing and transcriptional speed.
Keywords: thiamine pyrophosphate riboswitch, single-molecule FRET, cotranscriptional folding
Abstract
Because RNAs fold as they are being synthesized, their transcription rate can affect their folding. Here, we report the results of single-molecule fluorescence studies that characterize the ligand-dependent cotranscriptional folding of the Escherichia coli thiM riboswitch that regulates translation. We found that the riboswitch aptamer folds into the “off” conformation independent of its ligand, but switches to the “on” conformation during transcriptional pausing near the translational start codon. Ligand binding maintains the riboswitch in the off conformation during transcriptional pauses. We expect our assay will permit the controlled study of the two main physical mechanisms that regulate cotranscriptional folding: transcriptional pausing and transcriptional speed.
A key principle of biology is that structure determines function. Functional RNA working as regulators, ribozymes, and scaffolds in large molecular complexes should fold properly to function properly. Several factors, such as salts, metabolites, and temperature, can influence RNA folding by changing its thermodynamic energy landscape (1–3). The functional roles RNAs play, however, may be transient (4). This means RNAs may not have enough time to find the global minimum of the folding energy landscape before they function. In vivo, RNAs fold sequentially from the 5′-end to the 3′-end as they are synthesized. Their folding is also influenced by transcriptional pauses, which are ubiquitous (5). It is therefore possible that the functional structure of RNAs may be influenced more strongly by transcription kinetics than by the global energy landscape of RNA folding (6–9).
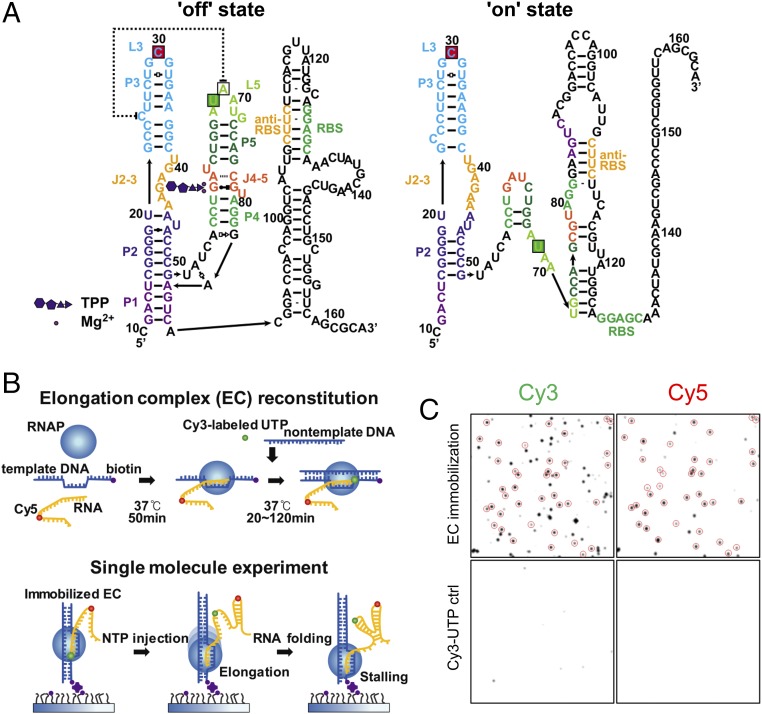
Thiamine production in bacteria, fungi, and plants is controlled by an RNA structure called the thiamine pyrophosphate (TPP) riboswitch (10–13). The TPP riboswitch exists in the untranslated regions of messenger RNAs, where it regulates their splicing, transcription, or translation by coupling TPP binding to a structural transition of the riboswitch (11). As an example, the translation regulation model of the Escherichia coli thiM riboswitch is shown in Fig. 1A (12, 14). In the TPP-bound form, TPP is encapsulated in the aptamer region, which is composed of the P1–P5 helices. In this state, translation is repressed (the “off” state) because the ribosome binding site (RBS) is inaccessible to ribosomes. In the TPP-unbound form, the disrupted P1 helix base-pairs with an anti-RBS sequence, and translation begins (the “on” state).
Fig. 1.
Single-molecule fluorescence cotranscriptional folding assay. (A) Model for TPP-induced structural transition of the E. coli ThiM riboswitch. The fluorophore-labeling positions for single-molecule studies are indicated by green and red boxes (green for Cy3 or Dy547, red for Dy647). (B) Experimental scheme. Dy647-labeled seed RNA was incubated with a template DNA strand and phage T7 RNAP in a tube for 50 min at 37 °C. To assemble a full EC, Cy3-labeled UTP and a nontemplate DNA strand were added to the tube and incubated for 20 min. ECs were immobilized on a polymer-passivated quartz surface using streptavidin–biotin interactions. Elongation was resumed by injecting NTP, while RNA folding was observed using a single-molecule FRET microscope. (C) Single-molecule fluorescence images of EC (Top) and control (ctrl) images of nonspecifically bound Cy3-UTP (Bottom). The colocalized Cy3 and Cy5 spots are enclosed by circles. The percentage of acceptor spots colocalized with donor spots was estimated as 57 ± 14% from 10 measurements.
How does TPP binding induce such an extensive structural transition of the riboswitch? An X-ray crystal structure of the aptamer region of the TPP riboswitch reveals how the riboswitch recognizes TPP with high ligand specificity (12, 15), but the mechanism coupling TPP binding to the structural transition remains unclear. Various techniques have been used to characterize both the ligand-free state of the TPP riboswitch aptamer and its structural transition upon ligand binding (14, 16–20). An optical trapping study reported that the aptamer forms a secondary structure before TPP binding and a tertiary interaction after TPP binding (17). A single-molecule fluorescence resonance energy transfer (FRET) study reported that TPP stabilizes the structure of the P1 stem (19). Despite the application of so many techniques, the mechanisms coupling TPP binding to the structural transitions of the TPP riboswitch are not yet clearly understood. To understand the regulation of the TPP riboswitch, it is necessary to visualize the cotranscriptional folding of the aptamer in the context of a full riboswitch (11, 21, 22).
Results
Single-Molecule FRET Assay for Cotranscriptional Folding.
To monitor cotranscriptional folding of the TPP riboswitch at the single-molecule level, we prepared an elongation complex (EC) loaded with doubly labeled RNA transcript as shown in Fig. 1B. First, we incubated Cy5-labeled seed RNA with a template DNA (Table S1) and phage T7 RNA polymerase (RNAP) in a tube for 50 min. Then, we assembled full ECs by incubating the ternary complexes with nontemplate DNA and Cy3-labeled UTP. We then immobilized these on a polymer-passivated quartz surface using streptavidin–biotin interactions. The comparison of fluorescence images with and without ECs shows a low level of nonspecific binding and a high efficiency of EC assembly (Fig. 1C). To resume elongation, we added NTP while single-molecule fluorescence images were being taken. In this experimental scheme, transcription stalls upon encountering streptavidin on the quartz slide, providing a means to control the length of the RNA transcript involved in folding by changing the length of the DNA templates. The Cy3 and Cy5 labeling positions used for the experiments are shown in Fig. 1A. The dissociation constant (Kd) value of the dye-labeled riboswitch was similar to the Kd value for an unlabeled TPP riboswitch based on isothermal calorimetry measurements (23, 24), indicating that dye labeling does not significantly hinder TPP binding. We also confirmed that high FRET occurs when TPP binds the riboswitch (24).
Cotranscriptional Folding of the TPP Riboswitch Aptamer.
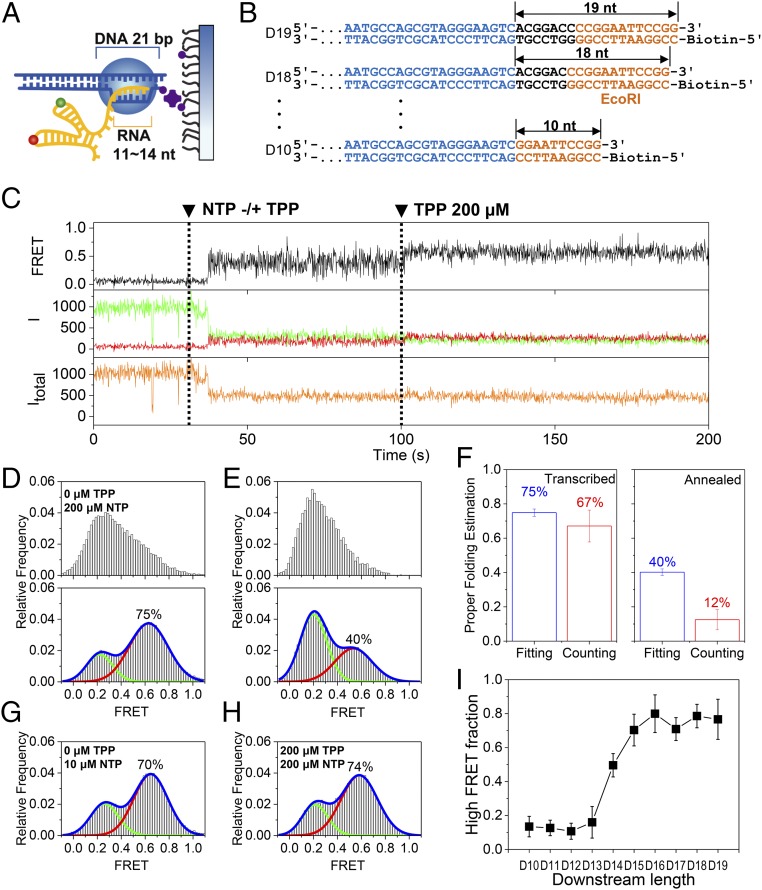
We first studied the cotranscriptional folding of the aptamer region. Previous structural and biochemical studies revealed that T7 RNAP encapsulates a 21-bp DNA duplex and an 11- to 14-nt RNA transcript (25) (Fig. 2A), but we could not predict how many nucleotides of the RNA transcript would be exposed outside RNAP when transcription stalls. To ensure that we have an RNA transcript whose aptamer region is completely exposed when transcription stalls, we prepared a series of DNA templates with varying lengths downstream from the aptamer coding region (D10–D19 in Fig. 2B).
Fig. 2.
Cotranscriptional folding of the aptamer. (A) Model EC stalled on a surface. An RNA transcript of 11–14 nt and a DNA template of 21 bp are protected by the EC. (B) Series of DNA templates with varying downstream lengths (black) after the region of the aptamer (blue). DNA length was altered by progressive deletion of the downstream part, while maintaining the orange region. DNA templates are named D# according to their downstream length (#). (C) Representative time traces of the single-molecule cotranscriptional experiment with D17. Fluorescence intensities of a donor and an acceptor are colored in green and red, respectively, while the corresponding FRET appears in black. The total intensity (Itotal) is shown in orange. NTP (200 μM) and TPP (200 μM) were injected at 30 s and 100 s, respectively (dashed lines). (D) FRET histograms before (Top) and after (Bottom) 200 μM TPP injection for D17. (E) FRET histograms before (Top) and after (Bottom) 200 μM TPP injection for the thermally annealed synthetic aptamer. (F) Estimation of properly folded RNA for cotranscriptionally folded D17 (Left) and thermally annealed aptamer (Right). Two different methods were used: fitting the FRET histograms in D, Bottom and E, Bottom to two Gaussian functions (blue bars) and counting TPP-responsive molecules (red bars). Error bars represent SD from three experiments, each with more than 50 molecules. (G) FRET histogram for D17 at 10 μM NTP concentrations during transcription. (H) FRET histogram for D17 in the presence of 200 μM TPP during transcription. In D, G, and H, the TPP and NTP concentrations indicate the values used during the elongation phase, but the histograms were made after the injection of 200 μM TPP. (I) Fraction of the high FRET population of D10–D19. Error bars represent SD from three experiments, each with more than 50 molecules.
Fig. 2C shows representative time traces obtained with D17, which is long enough to fully expose the aptamer region of the riboswitch at stalling, as is shown in Fig. 2I. After NTP injection, the total intensity dropped simultaneously with the FRET jump (Fig. 2C). Cy3 intensity is known to increase when a protein is positioned nearby (26), so we interpreted the Cy3 intensity drop upon NTP injection as the escape of the Cy3-labeled base from RNAP. After waiting 70 s for the RNA to elongate to the stalling site, we added 200 μM TPP to observe the effects of TPP binding. We observed an increase in FRET efficiency upon TPP injection, indicating that TPP successfully bound to the riboswitch (Fig. 2C). A comparison of FRET histograms before and after TPP injection reveals a large increase in the high FRET population in the presence of TPP (Fig. 2D). On the other hand, when we performed the same experiment with a thermally annealed synthetic aptamer, the TPP-induced increase in the high FRET population was much smaller (Fig. 2E). We then used two different approaches to estimate the relative population of properly folded (i.e., TPP-responsive) molecules. We fit the FRET histograms in the presence of TPP to two Gaussian functions (histograms in Fig. 2 D, Bottom and E, Bottom), and we counted the number of molecules with FRET jumps upon TPP injection (Fig. 2C). As Fig. 2F shows, the two approaches gave similar results for the cotranscriptionally folded aptamer (75% vs. 67%), but produced a significant difference for the thermally annealed RNA (40% vs. 12%). In the case of the thermally annealed RNA, a significant proportion of molecules did not respond to TPP, but contributed to the high FRET population (Fig. S1). These molecules contributed to the properly folded RNA estimate in the fitting approach, but not in the counting approach. In the case of cotranscriptionally folded RNA, molecules in the high FRET region before TPP injection were highly dynamic and showed a transition to a stable FRET state with an increased FRET value (Fig. 2C). This observation indicates that the high FRET population of the thermally annealed RNA in the presence of TPP is heterogeneous, comprising properly folded RNAs along with a significant proportion of misfolded RNAs. This observation is consistent with the view that the energy landscape for long RNAs is quite rugged (27, 28). With cotranscriptional folding, however, it is possible to reach higher levels of conformational homogeneity because it is sequential folding that determines the folding pathway.
Next, we used the D17 template to explore how cotranscriptional folding is affected by various elongation conditions. When we slowed transcription speed by using only 10 μM NTP instead of 200 μM, we observed a reduction in the high FRET population from 75% to 70% (Fig. 2G). This decrease may be small, but it is statistically significant (Fig. S2). On the other hand, the presence of TPP during elongation does not affect the folding populations (Fig. 2H), indicating that the aptamer folds into the off conformation independent of TPP.
Finally, we performed the experiment in Fig. 2C with other DNA templates. The high FRET population was highly dependent on the length of the DNA template downstream from the aptamer coding region (Fig. 2I and Fig. S3). Because the TPP riboswitch attains full TPP binding affinity only after aptamer synthesis is complete (24, 29), Fig. 2I allows us to accurately determine the RNA transcript length involved in folding at transcription stalling. By inspecting the FRET histograms and TPP binding kinetics, we concluded that the aptamer region is fully exposed on the D16–D19 templates (Fig. S4). In the subsequent discussions, each RNA transcript will be referred to as “R#,” e.g., R144, where # refers to the position number of the last nucleotide exposed outside RNAP during transcription stalling.
Cotranscriptional Folding of the Full TPP Riboswitch.
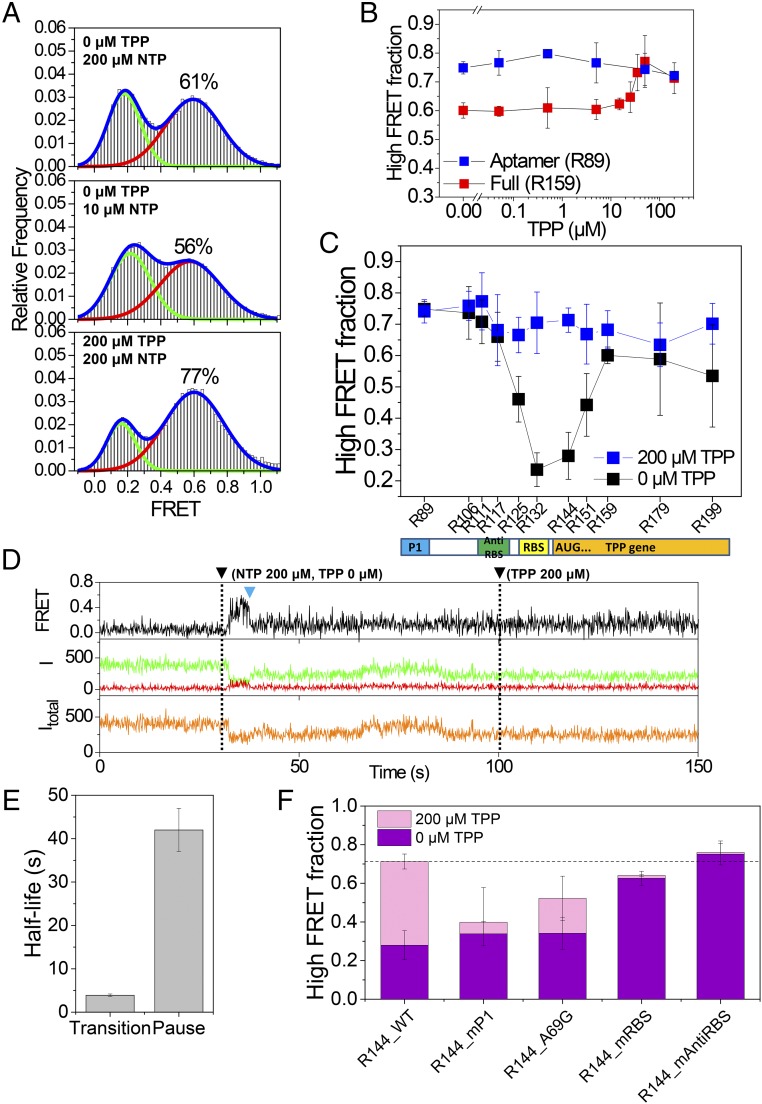
After establishing that the aptamer region folds independent of TPP, we studied the cotranscriptional folding of the full riboswitch (R159). Compared with the aptamer region, the high FRET population of the full riboswitch fell from 75% to 61% (Fig. 3A, Top), and this reduction was even more prominent when we reduced transcription speed (Fig. 3A, Middle). The addition of 200 μM TPP during transcription, however, restored the original high FRET population we observed with the aptamer region alone (Fig. 3A, Bottom). TPP’s ability to maintain the high FRET population depended on TPP concentration, with the midpoint falling around 30 μM (Fig. 3B and Fig. S5). This is almost 300-fold higher than the TPP Kd of the aptamer we observed in vitro (24, 30), but it is similar to the in vivo critical concentration that regulates thiamine production (31, 32). The fact that the TPP concentration that regulates riboswitch folding is much larger than the riboswitch Kd is consistent with the hypothesis that riboswitch folding is kinetically controlled (22).
Fig. 3.
Cotranscriptional folding of the full TPP riboswitch. (A) FRET histograms of R159 under three different elongation conditions. FRET histograms were obtained in the presence of 200 μM TPP. (B) Fraction of the high FRET population of the aptamer (R89) and the full riboswitch (R159) with varying TPP concentrations during elongation. Error bars represent SD from three experiments, each with more than 50 molecules. (C) Fraction of the high FRET population of the riboswitch with varying lengths. Error bars represent SD from three experiments, each with more than 50 molecules. (D) Representative time trace that shows a transition (blue triangle) from the off conformation to the on conformation for R144 RNA. Fluorescence intensities of a donor and an acceptor are colored in green and red, respectively, while the corresponding FRET appears in black. The total intensity (Itotal) is shown in orange. NTP (200 μM) and TPP (200 μM) were injected at 30 s and 100 s, respectively (dashed lines). (E) Half-life of the off state of R144 and that of the transcriptional pausing of E. coli RNAP at the translation start codon region. (F) Fraction of the high FRET population of the riboswitch mutants. Error bars represent SD from three experiments, each with more than 50 molecules.
Effects of Transcriptional Pausing on TPP Riboswitch Folding.
The data in Fig. 3 A and B clearly show that the final folding products of the full TPP riboswitch are influenced by the presence of TPP during transcription, and the amount of the effect depended on transcription speed. Based on these observations, we speculated that if transcription speed affects the final folding products of the TPP riboswitch, transcription pausing may have a more significant effect on riboswitch folding (33). Using a gel assay, we found that long transcription pauses in the range of tens of seconds occur near the translation start codon in transcription by E. coli RNAP, but not phage T7 RNAP (Fig. S6). The pausing sites we observed with E. coli RNAP are consistent with previous reports (33, 34). To emulate the effect of a transcriptional pause, we studied the folding of transcripts of varying lengths. We observed a large TPP effect when the RBS and translation start codons were exposed at transcription stalling (Fig. 3C and Fig. S7), proving the importance of transcriptional pausing in folding regulation. Single-molecule FRET time traces of R144 in the absence of TPP show that the huge decrease of the off conformation is due to the fact that the transition from the off conformation to the on conformation occurs during stalling (Fig. 3D and Fig. S8). The off-to-on transition occurred with a half-life of 3.9 s, which is more than 10-fold smaller than the half-life of the transcriptional pausing (Fig. 3E and Fig. S9). We did not observe any such transition in the presence of 200 μM TPP; the on or off state was maintained during 150 s of observation time (Fig. S10). This is unsurprising because the binding lifetime of TPP to the riboswitch was measured as 71 s (24).
Finally, we tested the effects of riboswitch mutations on the folding of R144. The TPP riboswitch has several sites that are highly conserved across species (23). Our TPP riboswitch regulation model predicts that the sequence of the aptamer region plays a functional role during ligand recognition, whereas the sequence of the expression platform plays a role during the off-to-on structural transition. As expected, we found a functional dichotomy between the aptamer and the expression platform. Mutations in the aptamer region (R144_P1 and R144_A69G) reduced the off-state population, indicating a corresponding reduction in ligand binding (Fig. 3F and Fig. S11). Still, we did observe significant regulatory effects of TPP in those mutants (Fig. 3F and Fig. S11). Mutations in the expression platform (R144_mRBS and R144_mAntiRBS), on the other hand, led to the near-complete disappearance of the regulatory effects of TPP, while leaving the off-state population relatively unchanged (Fig. 3F and Fig. S11).
Discussion
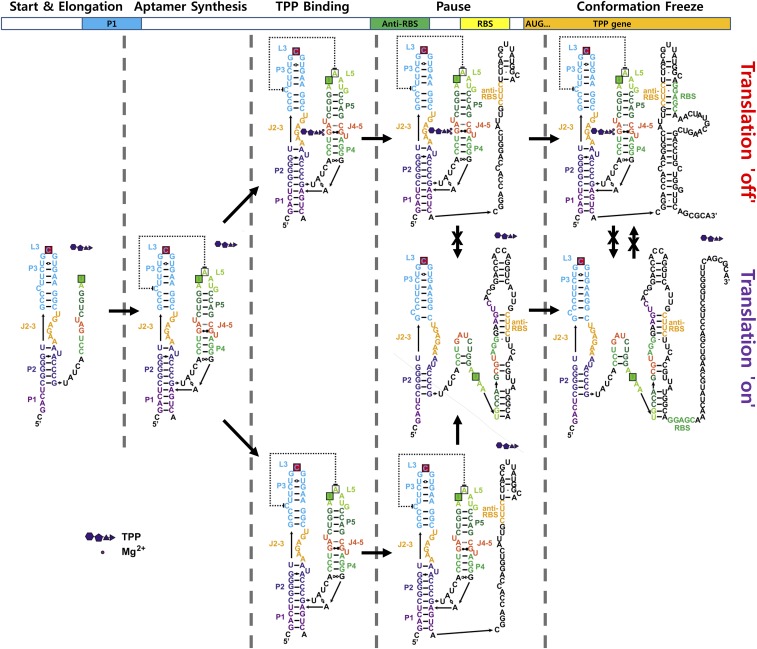
Our single-molecule cotranscriptional folding assay provided the following observations. First, the aptamer folds into the off conformation independent of TPP (Fig. 2H), and TPP efficiently binds the aptamer only after the aptamer region is fully exposed (Fig. 2I). Second, the off conformation switches to the on conformation during transcription from the aptamer region to RBS (Fig. 3A), and the transition is enhanced by the presence of the transcriptional pause that occurs as RNAP nears the translation start codon (Fig. 3 C and D and Figs. S8 and S9). Third, the off-to-on transition that occurs during transcription and the transcriptional pause is inhibited in the presence of TPP (Fig. 3 A and C and Fig. S10). Finally, the regulatory effect of TPP mostly disappears after the transcription of the RBS (Fig. 3C). Based on these observations, we propose a regulation mechanism for the TPP riboswitch (Fig. 4) that is consistent with mutational studies in Fig. 3F. In this model, TPP has a brief time window for translation regulation that begins with aptamer synthesis and ends after the transcription of the RBS. Once this window has passed, the transition between the off and on states becomes more difficult, effectively blocking TPP’s ability to regulate the riboswitch.
Fig. 4.
Model for cotranscriptional folding and regulation of the TPP riboswitch.
Transcriptional pausing near the translation start codon is a frequently observed phenomenon (35, 36). In the case of the TPP riboswitch, the functional role of this transcriptional pausing is clear. By inspecting the secondary structures of the TPP riboswitch, it became clear that when the riboswitch is elongated and exposed downstream of the translation start codon, the on conformation becomes more favorable than the off conformation (). Thus, transcriptional pauses at this position may be selected by evolution to provide RNAs adequate time to switch to the on conformation.
In the model shown in Fig. 1A, TPP induces the structural transition of the TPP riboswitch from the on conformation to the off conformation. Our data, in contrast, show that the TPP riboswitch folds into the off conformation in the absence of TPP, and that TPP works as a kind of “safety pin” that inhibits the structural transition from the off state to the on state during transcriptional pausing. It will be interesting to see how general this mechanism proves to be across other riboswitches.
Riboswitches regulating transcription termination are thought to be kinetically controlled, whereas riboswitches regulating translation initiation are thought to be thermodynamically controlled (22, 37). Our work clearly shows that the E. coli thiM riboswitch, which regulates translation initiation, is kinetically controlled. This suggests kinetic control mechanisms are more widespread than expected. It is also evident that an RNA molecule’s folding should be kinetically controlled if the regulation of that RNA should be accomplished as soon as RNA is synthesized. Even nascent RNAs with a time gap between their synthesis and the achievement of their functional regulation may not have enough time to reach a point of thermodynamic equilibrium, in terms of conformation, before they function. As RNAs become longer, this tendency becomes even more evident. In these cases, the kinetic control of RNA folding makes more sense.
In general, the energy landscapes for RNA folding are quite rugged. Sequential folding and transcription kinetics can therefore play significant roles in RNA folding. We successfully established a single-molecule FRET assay to study cotranscriptional influences on RNA folding. Compared with assays based on optical trapping (17, 38), our single-molecule FRET assay has a few important merits: it is more efficient, can distinguish different folding conformations, and can report ligand binding/dissociation without unfolding the RNA.
Host polymerases also have significant influence over RNA folding. Although the influence of RNAP over RNA folding has primarily been attributed to transcriptional pausing, transcription speed is also an important factor. Compared with E. coli RNAP, T7 RNAP rarely exhibits any transcriptional pausing. Our single-molecule FRET assay can take advantage of this absence of transcriptional pausing with T7 RNAP. We can take advantage of the absence of intrinsic pause sites, effectively emulating transcriptional pausing by forcing pauses at specific positions; the presence of pause sites, as with E. coli RNAP, would make it hard to control and study the effects of transcriptional pausing. T7 RNAP transcribes faster than E. coli RNAP. Our assay also allowed us to study the effects of transcription speed by controlling NTP concentration.
In summary, our assay can be generally used to study the cotranscriptional folding of bacterial or eukaryotic RNAs. It allows us to perform well-controlled studies of the cotranscriptional effects of the two main physical mechanisms: transcriptional pausing and speed. Although our assay provides a simple means to directly study the physical mechanisms of cotranscriptional folding, there are also several other factors in vivo that affect transcription speed, transcriptional pausing, and conformational rearrangement of RNAs. To study these factors, more complicated cotranscriptional folding assays should be developed.
Methods
Synthetic Aptamer Preparation.
Detailed procedures are described in SI Methods.
EC Reconstitution.
The seed RNA (800 nM) was incubated in a tube with a template DNA strand (200 nM) and T7 RNAP (40 nM; New England Biolabs) for 50 min at 37 °C. A donor-labeled UTP (Cy3-UTP, 100 μM; GE Healthcare) and a nontemplate DNA strand (400 nM) were added to the tube and incubated for 20 min before the single-molecule experiment. More detailed procedures are provided in SI Methods.
Single-Molecule FRET Experiments.
After streptavidin (0.2 mg/mL; Invitrogen) coating of polymer-coated quartz slides (39) for 5 min, the EC (400 pM) was immobilized on the surface and imaged using a homemade wide-field total internal reflection fluorescence microscope equipped with a green laser (532 nm, Compass215M; Coherent) and a red laser (640 nm, Cube640-100C; Coherent). All experiments were performed at 37 °C with 50-ms exposure times in an alternating laser excitation mode (40, 41). Various NTP and TPP concentrations were used for transcription elongation, but high FRET fractions for the RNA transcripts were determined in the presence of 200 μM TPP. More details are provided in SI Methods.
The Promoter-Initiated Pause Assay with E. coli and Phage T7 RNAP.
A linear DNA template (50 nM) containing the T7A1 promoter followed by the TPP riboswitch was incubated with 50 nM E. coli RNAP holoenzyme for 10 min at 37 °C in transcription buffer. Halted ECs (+20) were formed by adding 150 μM ApU dinucleotide (Trilink), 20 μM GTP and CTP, 1 μM ATP, and 10 μCi [α-32P] ATP (3,000 Ci/mmol). Transcription was then allowed to resume by addition of all four NTPs (50 μM) and heparin (100 μg/mL). In vitro transcription assays using T7 RNAP were performed similarly. More details are provided in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by a Creative Research Initiative Program (Grant 2009-0081562 to S.H.) and by the National Research Council of Science and Technology (Grant DRC-14-2-KRISS to C.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712983115/-/DCSupplemental.
References
- 1.Badelt S, Hammer S, Flamm C, Hofacker IL. 2015. Thermodynamic and kinetic folding of riboswitches. Methods in Enzymology, Computational Methods for Understanding Riboswitches, eds Chen S-J, Burke-Aguero DH (Academic, Cambridge, MA), pp 193–213. [DOI] [PubMed]
- 2.Draper DE. RNA folding: Thermodynamic and molecular descriptions of the roles of ions. Biophys J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein JA, Khodursky AB, Lin P-H, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 6.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Lau MW, Ferré-D’Amaré AR. Ribozymes and riboswitches: Modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell R, et al. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thirumalai D, Hyeon C. RNA and protein folding: Common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 10.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 11.Miranda-Ríos J. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure. 2007;15:259–265. doi: 10.1016/j.str.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 14.Rentmeister A, Mayer G, Kuhn N, Famulok M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007;35:3713–3722. doi: 10.1093/nar/gkm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. The ligand-free state of the TPP riboswitch: A partially folded RNA structure. J Mol Biol. 2010;396:153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anthony PC, Perez CF, García-García C, Block SM. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci USA. 2012;109:1485–1489. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird NJ, Ferré-D’Amaré AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haller A, Altman RB, Soulière MF, Blanchard SC, Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci USA. 2013;110:4188–4193. doi: 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steen K-A, Malhotra A, Weeks KM. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease. J Am Chem Soc. 2010;132:9940–9943. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 23.Kulshina N, Edwards TE, Ferré-D’Amaré AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhm H, Hohng S. Ligand recognition mechanism of thiamine pyrophosphate riboswitch aptamer. Bull Korean Chem Soc. 2017;38:1465–1473. [Google Scholar]
- 25.Huang J, Sousa R. T7 RNA polymerase elongation complex structure and movement. J Mol Biol. 2000;303:347–358. doi: 10.1006/jmbi.2000.4150. [DOI] [PubMed] [Google Scholar]
- 26.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 29.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 31.Kawasi T, Iwashima A, Nose Y. Regulation of thiamine biosynthesis in Escherichia coli. J Biochem. 1969;65:407–416. doi: 10.1093/oxfordjournals.jbchem.a129028. [DOI] [PubMed] [Google Scholar]
- 32.Song Q, Singleton CK. Mitochondria from cultured cells derived from normal and thiamine-responsive megaloblastic anemia individuals efficiently import thiamine diphosphate. BMC Biochem. 2002;3:8. doi: 10.1186/1471-2091-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauvier A, et al. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat Commun. 2017;8:13892. doi: 10.1038/ncomms13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong TN, Pan T. RNA folding during transcription: Protocols and studies. Methods Enzymol. 2009;468:167–193. doi: 10.1016/S0076-6879(09)68009-5. [DOI] [PubMed] [Google Scholar]
- 35.Larson MH, et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vvedenskaya IO, et al. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemay J-F, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S, Lee J, Hohng S. Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS One. 2010;5:e12270. doi: 10.1371/journal.pone.0012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.