Significance
Acute hemorrhagic conjunctivitis (AHC) is a painful and highly contagious infection of the eye, with reported incidence rates of up to 48%. No drugs or vaccines are available for treatment or prevention of AHC. Coxsackievirus A24 variant (CV-A24v) is the main etiological agent of AHC, being responsible for >10 million AHC cases worldwide during the last decades. We have identified the CV-A24v protein receptor and determined the high-resolution structure of the virus–receptor complex. Furthermore, we found that an adaptation which enhances binding to the receptor sialic acid may have contributed to the pathogenicity and pandemic nature of CV-A24v. These findings highlight the importance of sialic acid for viruses with ocular tropism, such as influenza A virus and several adenoviruses.
Keywords: conjunctivitis, coxsackievirus A24v, receptor, ICAM-1, sialic acid
Abstract
Acute hemorrhagic conjunctivitis (AHC) is a painful, contagious eye disease, with millions of cases in the last decades. Coxsackievirus A24 (CV-A24) was not originally associated with human disease, but in 1970 a pathogenic “variant” (CV-A24v) emerged, which is now the main cause of AHC. Initially, this variant circulated only in Southeast Asia, but it later spread worldwide, accounting for numerous AHC outbreaks and two pandemics. While both CV-A24 variant and nonvariant strains still circulate in humans, only variant strains cause AHC for reasons that are yet unknown. Since receptors are important determinants of viral tropism, we set out to map the CV-A24 receptor repertoire and establish whether changes in receptor preference have led to the increased pathogenicity and rapid spread of CV-A24v. Here, we identify ICAM-1 as an essential receptor for both AHC-causing and non-AHC strains. We provide a high-resolution cryo-EM structure of a virus–ICAM-1 complex, which revealed critical ICAM-1–binding residues. These data could help identify a possible conserved mode of receptor engagement among ICAM-1–binding enteroviruses and rhinoviruses. Moreover, we identify a single capsid substitution that has been adopted by all pandemic CV-A24v strains and we reveal that this adaptation enhances the capacity of CV-A24v to bind sialic acid. Our data elucidate the CV-A24v receptor repertoire and point to a role of enhanced receptor engagement in the adaptation to the eye, possibly enabling pandemic spread.
Acute hemorrhagic conjunctivitis (AHC) is characterized by a sudden onset of ocular pain, swelling, watering, and extensive subconjunctival hemorrhaging. Since AHC is highly contagious it spreads rapidly and can affect up to 48% of a population in a single outbreak (1, 2). Coxsackievirus A24 variant (CV-A24v), belonging to the genus Enterovirus within the family Picornaviridae, is the main etiological agent of AHC, being responsible for >10 million AHC cases worldwide during the last decades (2, 3). Coxsackievirus A24 (CV-A24) was first isolated in South Africa in 1951 (4) but was not associated with disease in humans. However, in 1970, a pathogenic variant of CV-A24 (CV-A24v) emerged that was responsible for a large AHC outbreak in Singapore (5, 6). Initially, this AHC-causing variant circulated only in Southeast Asia but later spread worldwide, suddenly causing numerous explosive AHC outbreaks and two pandemics, which started in 1985 and 2002 (7–9). Although both the CV-A24 variant and nonvariant strains are being detected in clinical samples worldwide, only strains belonging to the “variant” clade are associated with AHC. As yet, the molecular mechanism underlying this difference in pathogenicity is unknown. Also, it remains unclear what has led to the sudden onset of CV-A24v pandemics.
Receptor choice or the interplay between different receptors can be important determinants of viral tropism and pathogenicity. Most enteroviruses rely on an uncoating receptor that triggers destabilizing rearrangements of the capsid proteins, resulting in the formation of a capsid-mediated pore in the endosomal membrane through which the viral genome is delivered into the cytoplasm (10). In addition to uncoating receptors, many enteroviruses employ one or more attachment receptors that do not participate in uncoating but facilitate cell binding. Previous reports have shown that sialic acid (Sia) plays a role in infection of CV-A24v (11). Despite these findings, it remains unclear whether CV-A24v requires additional cell surface receptors and whether differences in receptor preference exist between AHC- and non-AHC-causing CV-A24 strains.
In this study we set out to identify a potential protein receptor and to investigate if possible changes in receptor requirements can explain the pathogenic and pandemic nature of AHC-causing CV-A24v strains. We identify ICAM-1 as an essential receptor for both AHC- and non-AHC–causing CV-A24 strains and provide a high-resolution cryo-EM structure of a virus in complex with ICAM-1, revealing in detail the interactions between CV-A24v and its receptor. Moreover, by combining structural data, phylogenetic analysis, and reverse genetics we identify a single amino acid substitution in the viral capsid that provides an improved Sia binding capacity and has been adopted exclusively by pandemic CV-A24 variant strains. These findings point toward a role of enhanced Sia binding in the adaptation of CV-A24v to the eye, which may have been a prerequisite for its pandemic potential.
Results
ICAM-1 Is an Essential CV-A24v Receptor.
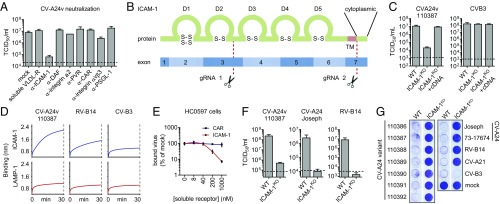
CV-A24v can bind Sia, but Sia was not found to initiate uncoating (12), suggesting that CV-A24v employs an additional, yet unknown, receptor. To identify this receptor, we performed neutralization assays with a CV-A24v clinical isolate in Sia-deficient HAP1 CMASKO cells (13), using soluble receptor domains (VLDL-R) or antibodies targeting various known enterovirus receptors (ICAM-1, DAF, integrin α2, PVR, CAR, integrin αvβ3, and PSGL-1) (Fig. 1A). The ability of these ligands to suppress receptor binding was confirmed using positive control viruses (Fig. S1A). Only the antibody against ICAM-1, a cell adhesion molecule consisting of five Ig-like domains (14) (Fig. 1B), blocked CV-A24v infection (Fig. 1A). To determine whether ICAM-1 is essential for CV-A24v, we knocked out ICAM-1 in various cell types by CRISPR/Cas9 genome editing (Fig. 1B). The ICAM-1 knockout (ICAM-1KO) genotype was verified by sequence analysis (Fig. S1B) and by assessing the susceptibility of ICAM-1KO cells to infection with ICAM-1–dependent coxsackievirus A21 (CV-A21) (15) and rhinovirus B14 (RV-B14) (16) and CAR-dependent coxsackievirus B3 (CV-B3) (17) (Fig. 1 F and G and Fig. S1 C–E). CV-A24v infection was reduced by ICAM-1 knockout in HAP1 CMASKO cells (Fig. S1C), WT HAP1 cells (Fig. S1D), and HeLa cells (Fig. 1C) and was rescued by transfecting ICAM1 cDNA (Fig. 1C). Biolayer interferometry (Fig. 1D) and CV-A24v neutralization with soluble ICAM-1 ectodomain in primary conjunctival cells (Fig. 1E) pointed to a direct interaction between CV-A24v and ICAM-1. Analysis of virus production in HeLa cells (Fig. 1F) and analysis of cytopathic effect induction in HeLa (Fig. 1G) or human corneal epithelial (HCE) cells (Fig. S1E) showed that several CV-A24v clinical isolates, the nonvariant prototype strain Joseph, and a nonvariant clinical isolate all require ICAM-1 for infection. Sialylation of ICAM-1 was not found to be essential for interaction with CV-A24v and CV-A24 Joseph, as shown by biolayer interferometry (Fig. S1F). Taken together, these data indicate that ICAM-1 functions as an essential receptor for AHC-causing variant strains as well as nonvariant strains.
Fig. 1.
ICAM-1 is an essential CV-A24 receptor. (A) Yields of infectious virus after a single replication cycle. Sia-deficient HAP1 CMASKO cells were treated with antibodies against enterovirus receptors (ICAM-1, DAF, integrin α2, PVR, CAR, integrin αvβ3, and PSGL-1) or soluble receptor (VLDL-R) and infected at 37 °C with CV-A24v 110387. (B) Schematic representation of ICAM-1 showing Ig-like domains 1–5 (D1–D5) and the two sites targeted by CRISPR/Cas9 (gRNA 1 and 2). Knockout was accomplished by disrupting a 1,489-bp region of ICAM1 encoding the transmembrane domain (TM), either by excision of this region or by introducing a frameshift mutation. (C) HeLa-R19 ICAM-1KO transfected with plasmid encoding ICAM-1 cDNA were exposed to virus and yields of infectious virus were measured after a single replication cycle. (D) Biolayer interferometry analysis of virus binding to either ICAM-1 or negative control receptor LAMP-1. (E) Relative levels of CV-A24v 110390 bound to primary conjunctival cells with expanded lifespan (HC0597), in the presence of ICAM-1 D1D5 or negative control receptor CAR D1. (F) HeLa-R19 cells were infected and yields of infectious virus were determined after a single replication cycle. Dashed lines (A, C, and F) represent virus input levels (T = 0). Error bars (A, C, E, and F) represent the mean ± SEM of three to four biological replicates. (G) HeLa-R19 cells were infected with seven CV-A24v clinical isolates, two CV-A24 nonvariant strains and ICAM-1-dependent (CV-A21, RV-14) or CAR-dependent (CV-B3) control viruses, followed by crystal violet staining of surviving cells.
The Structure of CV-A24v in Complex with ICAM-1.
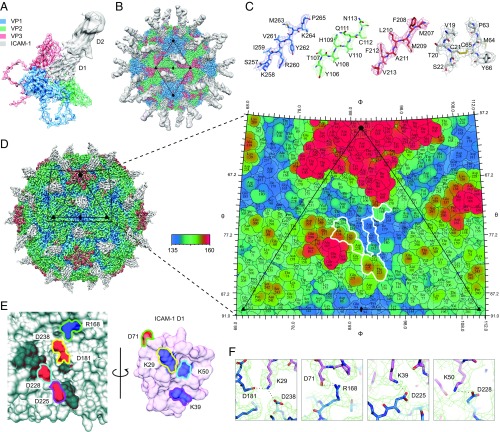
To elucidate the molecular interaction of CV-A24v with ICAM-1 we determined the structure of a complex between purified virions and a recombinant D1-D2 fragment of ICAM-1 (Fig. 2 A and B and Table S1). CV-A24v virions were immobilized on a lacey carbon support after which D1-D2 was added 30 s at 8 °C before blotting and plunge freezing. This rapid on-grid binding approach allowed us to capture the cryo-EM structure of CV-A24v with bound D1-D2, whereas longer incubation resulted in loss of particle integrity, probably due to receptor-mediated destabilization of the particle. The global resolution of the EM density is 3.9 Å (Fig. S2 A–D), the resolution of the viral capsid proteins is ∼3.6–3.8 Å, and that of the capsid-interfacing region of ICAM-1 D1 is ∼3.7 Å (Fig. S2 E–H), sufficient to resolve bulky side chains in the electron density map (Fig. 2C). The surface of an enterovirus particle is composed of 60 copies of the structural proteins VP1, VP2, and VP3 (Fig. 2A). A depression encircling each fivefold axis of symmetry forms the so-called canyon, which is the binding site for all uncoating receptors identified to date. The structure shows that the D1 domain binds in the canyon at the quasi-threefold axis of the capsid (Fig. 2B). In contrast to earlier studies, the interface between the capsid and ICAM-1 D1 is well defined in the final EM density map, enabling us to resolve secondary structure and the majority of side chains in this portion of the structure. This, in turn, allowed us to calculate an accurate footprint of the D1 component of the receptor on the virus surface, shown as a “roadmap” projection (Fig. 2D). The specificity in the interaction between ICAM-1 and CV-A24v is largely driven by electrostatic complementarity (Fig. 2E and Fig. S2I). Specifically, Lys29 (FG loop) of ICAM-1 interacts with VP3 Asp181 and VP1 Asp238 (Fig. 2F and Fig. S2J). In addition, Asp71 on the tip of the FG loop of ICAM-1 forms a salt bridge with VP1 Arg168. Finally, Lys50 (strand C) and Lys39 (strand D) of ICAM-1 form salt bridges with VP1 Asp228 and Asp225 (GH loop). Additional interactions include a hydrogen bonding network that tethers the DE loop of ICAM-1 D1 (Arg49, Asn47 and Pro45) to VP1 (Tyr230) and VP2 (Ala138, Lys139 and Thr140) (Fig. S2K). Many of these residues are conserved among other ICAM-1–binding viruses belonging to the enterovirus C species, suggesting a conserved mode of interaction with ICAM-1 (18). On the contrary, only VP1 Asp238 and VP3 Asp181 are present in ICAM-1–binding rhinoviruses.
Fig. 2.
The structure of CV-A24v in complex with ICAM-1. (A) Enlarged tilted view of density denoted by a black triangle in B, containing the corresponding atomic model. ICAM-1 D1 binds in the “canyon” located at the quasi-threefold axis. (B) The cryo-EM reconstruction of CV-A24v in complex with ICAM-1 (D1D2) viewed down the icosahedral twofold axis. VP1-3 (3.6σ), D1 (1σ), and D2 (0.6σ). (C) Typical example of the cryo-EM electron density of VP1-3 and D1 domain. (D) Radially colored isosurface representation of CV-A24v in complex with D1 (gray) viewed down the icosahedral twofold axis (Left). The stereographic projection of the viral surface (Right), where the polar angles θ and φ represent latitude and longitude, respectively. Amino acids which interact with ICAM-1 D1 are located at the floor and wall of the canyon (blue) and are circled in white. The radial coloring key is shown in angstroms. (E) Surface representation of the EM-derived atomic model for the quasi-threefold axis of CV-A24v (gray) and ICAM-1 D1 (pink), with residues forming salt bridges labeled and colored in respect to their interacting partners. (Left) The surface of CV-A24v. (Right) The surface of ICAM-1 D1 that interacts with CV-A24v. Residues forming salt bridges are colored according to charge (red is negative, blue is positive) and capsid residues that hydrogen-bond with ICAM-1 are dark gray. (F) Overview of electrostatic interactions shown in E between ICAM-1 (pink) and CV-A24v (blue) with potential hydrogen bonds denoted as dotted black lines and EM density as a green mesh.
Sia Is an Attachment Receptor Supporting ICAM-1–Mediated CV-A24v Infection.
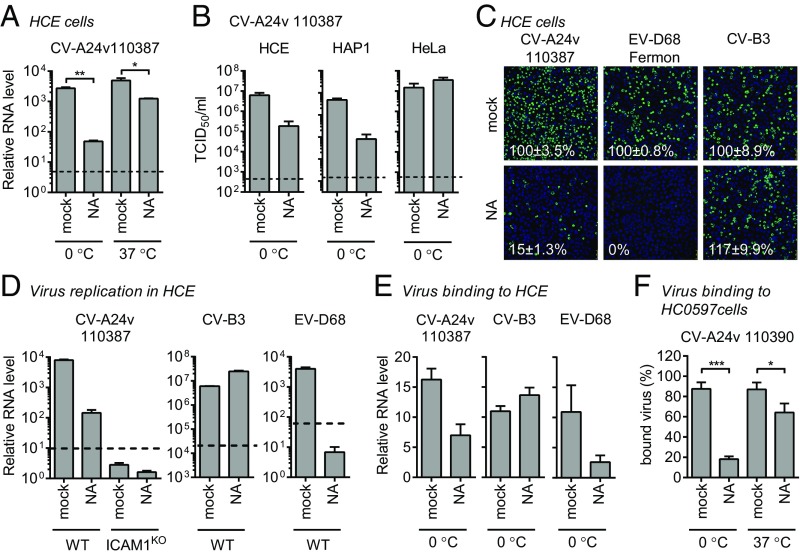
In addition to their uncoating receptor, several enteroviruses exploit additional receptors to enhance cell attachment. Since previous studies identified a role of Sia for CV-A24v (11, 12), we set out to investigate how Sia and ICAM-1 cooperate during CV-A24v infection. Removal of cell-surface Sia by CMAS knockout or neuraminidase (NA) treatment revealed that Sia supports CV-A24v infection (Fig. 3A and Fig. S3A). The role of Sia was more prominent when infection was performed on ice, compared with infection at 37 °C (Fig. 3A). This may be due to stabilization of the interaction at low temperature, as was previously described for several other Sia-binding viruses (19, 20). NA treatment of several cell types revealed that Sia supports CV-A24v infection of HCE and HAP1 cells, but not of HeLa cells (Fig. 3B). This difference might result from variability in the expression levels of ICAM-1/Sia or in the types of Sia expressed on cells. Sia removal from HCE cells (Fig. 3 C and D and Fig. S3B) or HAP1 cells (Fig. S3C) incompletely inhibited CV-A24v infection, whereas knockout of ICAM-1 completely blocked CV-A24v infection (Fig. 3D). Analysis of virus binding to HCE cells (Fig. 3E) and primary conjunctival cells (Fig. 3F) showed that Sia plays a role in cell attachment of CV-A24v. Together, these results show that, while ICAM-1 is the main CV-A24v receptor, Sia functions as an auxiliary attachment receptor.
Fig. 3.
Sia is an attachment receptor supporting ICAM-1–mediated CV-A24v infection. (A) HCE cells were treated with a mixture of A. urefaciens and Vibrio cholerae NA (1:30), infected with CV-A24v 110387 on ice (0 °C) or at 37 °C, followed by qPCR analysis of viral RNA levels after a single replication cycle. (B) NA-treated cells were infected with CV-A24v 110387 and yields of infectious virus were determined after a single replication cycle. (C) NA-treated HCE cells were infected on ice with CV-A24v, Sia-dependent enterovirus D68 (EV-D68), and Sia-independent CV-B3, followed by staining of dsRNA (green) and nuclei (blue). Shown are representative confocal micrographs. Percentage values denote mean ± SEM of three technical replicates, normalized to mock. (D and E) HCE cells were treated with a mixture of A. urefaciens and V. cholerae NA (1:30) and infected on ice. After incubation with virus, viral RNA levels were determined by qPCR either directly (E) or after a single replication cycle (D). (F) Levels of radioactively labeled CV-A24v 110390 bound to NA-treated (V. cholerae 10 mU/mL) primary conjunctival cells with expanded lifespan (HC0597) on ice (0 °C) or at 37 °C. Dashed lines (A, B, and D) represent virus input levels (T = 0). Error bars represent the mean ± SEM of three (A, B, D, and E) or eight (F) biological replicates. P values (A and F) were calculated by an unpaired two-sided t test; *P < 0.05; **P < 0.01; ***P < 0.001.
Differential Sia Dependency Between AHC-Causing Strains and Non-AHC Strains.
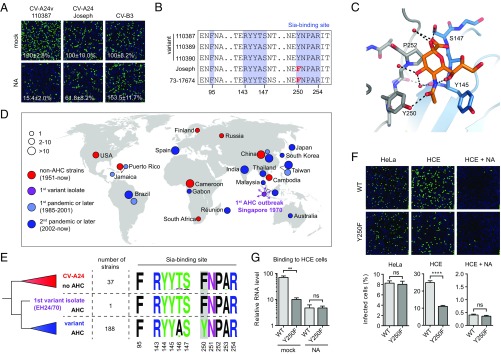
Next, we tested whether AHC strains and non-AHC strains differ in their dependency on cell surface Sia, either by treating HCE cells with NA to remove Sia (Fig. 4A and Fig. S4A) or with NaIO3 to oxidize carbohydrates (Fig. S4B). The AHC strains were more reliant on Sia than a non-AHC clinical isolate and the non-AHC prototype strain Joseph. Comparison of VP1 residues that constitute the Sia-binding site (12) showed that only one residue present in all AHC strains (VP1 Tyr250) was absent in the non-AHC strains (VP1 Phe250) (Fig. 4B). Although a Sia-binding site lacking this residue might be functional, the presence of Tyr250 could increase the affinity by hydrogen bonding with the 5-N-acyl carbonyl group in Sia (Fig. 4C). A similar mode of Sia stabilization is seen in other viral lectins, such as coronavirus hemagglutinin-esterases (20) and influenza NAs (19) (Fig. S4C).
Fig. 4.
Pandemic CV-A24v strains have acquired a capsid residue that enhances Sia binding. (A) NA-treated HCE cells were infected on ice, followed by staining of infected cells with a dsRNA antibody (green) and nuclei with DAPI (blue). Percentage values denote mean ± SEM of three technical replicates, normalized to mock. (B) Amino acid sequence alignment of tested CV-A24 strains, with the residue unique for nonvariant strains highlighted in red. (C) Binding site for Sia (orange) in CV-A24v 110390 (PDB ID code 4Q4X), showing that the 5-N-acyl group of Sia is stabilized by two hydrogen bonds (dashed lines). Two adjacent VP1 proteins are colored gray and blue, respectively. Red spheres represent water molecules. Oxygen and nitrogen atoms are colored red and blue, respectively. (D) Geographic origins of non-AHC- (red) and AHC-causing (purple, light blue, blue) CV-A24 strains of which complete VP1 sequences are available, with circle sizes proportional to the number of isolates. (E) Phylogenetic tree of CV-A24 isolates, showing on the right side the frequencies of amino acids in the Sia-binding site among the indicated number of strains. Frequency plots were generated with WebLogo. Residues at VP1 position 250 are highlighted in gray. (F) HCE cells were treated with NA and infected on ice with CV-A24v 110390 or the VP1 Y250F mutant, followed by staining of dsRNA (green) and nuclei (blue). Quantifications shown at the bottom denote mean ± SEM of four technical and two biological replicates. Images shown in A and F are representative confocal micrographs. (G) HCE cells were treated with a mixture of A. urefaciens and V. cholerae NA (1:30) and incubated with virus on ice, followed by qPCR analysis of bound virus. Error bars represent the mean ± SEM of three biological replicates. P values (F and G) were calculated by an unpaired two-sided t test; not significant (ns), P ≥ 0.05; **P < 0.01; ****P < 0.0001.
Pandemic CV-A24v Strains Have Acquired a Capsid Residue That Enhances Sia Binding.
After its emergence in 1970 in Singapore the pathogenic CV-A24 variant initially circulated only in Southeast Asia but started to spread globally in 1985, causing numerous AHC outbreaks and two pandemics. Concurrently, CV-A24 strains that do not belong to the variant clade and do not cause AHC continued to circulate worldwide and are sporadically found in stool samples. To understand whether the presence of Tyr250 correlates with the ability of CV-A24v to cause AHC, we compared the Sia-binding sites between variant and nonvariant strains. To classify these strains, we constructed a phylogenetic tree of all available complete CV-A24 VP1 sequences (Fig. S4D), originating from isolates collected worldwide during seven decades, including the first AHC outbreak in 1970 and the two subsequent pandemics that started in 1985 and 2002 (Fig. 4D). This tree showed that of the 226 strains 189 belong to the variant clade, which is delineated by the earliest AHC-causing strain EH24/70 that was isolated in Singapore in 1970 (Fig. 4E). A comparison of amino acid frequencies in the Sia-binding site revealed that most nonvariant strains, as well as the first AHC strain EH24/70, have a phenylalanine at VP1 position 250, whereas all variant strains isolated since the first AHC pandemic possess VP1 Tyr250 (Fig. 4E). Such a genetic change could result from a population bottleneck that variant strains may have undergone when invading a new target tissue. However, the absence of this residue in the “ancestral” strain EH24/70 suggests that Tyr250 was not present during the initial tropism switch but was adopted by its pandemic descendants in consequence of positive selection in the eye. The selective pressure to maintain this residue might be due to a more prominent role of Sia for infection in the ocular tissue to which AHC strains have adapted. To establish whether Tyr250 promotes Sia usage we constructed an infectious cDNA clone of CV-A24v strain 110390 and mutated Tyr250 to Phe. This mutant and the WT virus were equally infectious in HeLa cells, in which Sia does not play a role. In HCE cells, however, WT CV-A24v was more infectious than the Tyr250Phe mutant (Fig. 4F), and this difference was abolished by NA treatment. Consistently, the WT virus bound better to Sia-expressing HCE cells than the Tyr250Phe mutant (Fig. 4G), indicating that Tyr250 increases the capacity of CV-A24v to bind Sia. Together, these data demonstrate that the tropism switch of CV-A24 toward the eye was followed by the acquisition of a residue that enhances Sia-binding, an adaptation which coincides with the emergence of pandemic CV-A24v.
Discussion
This study provides insights into the receptor requirements of CV-A24v and reveals that a subtle change in the engagement of a secondary attachment receptor, Sia, may have promoted the adaptation of a pathogen to a new niche. Although VP1 Tyr250 is not a prerequisite for Sia binding (Fig. 4F), our results show that the presence of this residue can enhance Sia binding. Possibly together with other mutations outside the Sia-binding site, the adoption of an enhanced Sia-binding capacity by pandemic AHC-causing strains may have been a crucial evolutionary step in becoming a well-adapted ocular virus that can rapidly spread worldwide and cause AHC pandemics. Sia is also a receptor for several other ocular viruses such as adenovirus type 37, enterovirus D70, and influenza A virus (21–23), suggesting that Sia is important particularly for infection of the eye. Improved Sia-binding properties may promote CV-A24v attachment to and infection of conjunctival cells, thereby increasing the viral load in the eye and enhancing transmission. Since CV-A24v is believed to be transmitted via hand-to-eye contact, the fate of CV-A24v after the initial disposition into the eye might be largely determined by its capacity to attach to the conjunctival tissue before it is cleared by the continuous draining of tear film through the nasolacrimal duct. A weak and dynamic interaction with highly sialylated mucus proteins might facilitate this initial attachment and rapidly secure virions in the mucus layer, without eliminating virus mobility.
While the emergence of pandemic CV-A24v may have been promoted by the acquisition of VP1 Tyr250, it remains unknown what has caused the initial adaptation of CV-A24 to the eye. The genetic change underlying this tropism switch may be a combination of multiple synergistic mutations, which could reside in the capsid-coding region, nonstructural genes, or noncoding regulatory elements in the viral genome. For CV-A24v, it has been reported that the emergence of the AHC-causing CV-A24v variant was accompanied by a loss of amino acid diversity (Table S2), probably as a result of a population bottleneck (24). Consequently, numerous genetic differences exist between variant and nonvariant strains. Such changes in amino acid diversity also occurred at several CV-A24 capsid sites that interact with ICAM-1 (Table S3), some of which may have caused subtle changes in the engagement of ICAM-1 that facilitated ocular tropism. Thus, the identification of ICAM-1–binding residues in CV-A24v may help future studies investigating whether changes in ICAM-1 binding have contributed to the earliest manifestation of AHC.
Materials and Methods
Extended methods and information about cell lines, viruses (including construction of CV-A24v 110390 infectious cDNA clones), chemicals, and reagents are described in SI Materials and Methods.
EM.
Cryo-EM grids were prepared by applying 3 µL of purified CV-A24v (10 mg/mL) to 400-mesh lacey grids coated in a 3-nm carbon film (Agar Scientific). The sample was left to adsorb for 30 s before most of the sample was blotted away manually. On-grid binding of the receptor was performed by applying 3 µL of ICAM-1 D1-D2 (9.85 mg/mL) to the preblotted, virus-containing grid and leaving it for 30 s before blotting and freezing using a Leica EM GP plunge-freeze (Leica Microsystems) device. The Leica EM chamber temperature was set to 8 °C with 80% relative humidity and liquid ethane was used for sample vitrification. Grids were glow-discharged for 30 s before application of the samples. CV-A24v – ICAM-1 D1-D2 data were collected on an FEI Titan Krios (Astbury Biostructure Laboratory, University of Leeds) transmission electron microscope at 300 kV, using a total electron dose of 60 e-/Å2 and a magnification of 75,000×. The final calibrated object sampling was 1.065 Å per pixel. A total of 2,643 exposures were recorded using the EPU automated acquisition software on a FEI Falcon III direct electron detector operating in linear mode. Each exposure movie had a total exposure of 1 s and contained 40 frames. Image processing, model building, and refinement are described in SI Materials and Methods.
Infectivity Assays.
Cells were incubated with virus for 1 h at 37 °C or, where indicated, on ice. After virus binding, cells were washed once with PBS (ice-cold PBS was used after virus binding on ice). Cells were supplied with fresh medium and incubated at 37 °C for a total of 8 h, which represents a single replication cycle. In virus production assays cells were subjected to three freeze/thaw cycles and virus titers were determined by end-point dilution. Crystal violet staining was performed 4 d postinfection. In neutralization assays, cells were pretreated with antisera or soluble receptors for 45 min at 37 °C. Unless otherwise indicated, NA treatments were performed by incubating cells with a 1:30 dilution of Arthrobacter urefaciens NA in serum-free medium for 1 h at 37 °C. Treatments with periodate, which oxidizes vicinal diols in glycans, was performed by incubation of cells with 5 mM NaIO3 in PBS for 1 h on ice.
Immunofluorescence Assays.
Cells were fixed by submersion in a 4% paraformaldehyde solution for 15 min. Fixed cells were stained with 1:2,000 diluted mouse monoclonal anti-dsRNA (J2; English and Scientific Consulting). Cells were examined by confocal microscopy (Leica SPE-II) and the number of infected cells was quantified with ImageJ.
Virus Binding Assays.
HCE cells were successively incubated with NA for 1 h at 37 °C, 1% BSA in PBS for 10 min on ice, virus for 1 h on ice, and washed three times with ice-cold PBS. RNA was isolated from cells using the NucleoSpin RNA Isolation Kit (740955.250; Macherey-Nagel). cDNA was generated using the TaqMan Reverse Transcription Reagents kit (N8080234; Applied Biosystems). Quantitative PCR was performed using the Roche Lightcycler 480 SYBR Green I Master kit (04 887 352 001; Roche). For binding assays with HC0597 cells, CV-A24v was 35S-labeled as described previously (11), followed by treatment with soluble proteins for 1 h at 37 °C. Adherent HC0597 cells were detached (with PBS–0.05% EDTA; Merck) and recovered in growth medium at 37 °C. After 1 h, 100,000 cells per sample were washed and diluted in 50 µL binding buffer [HC0597 media + 0.5% BSA (Roche) + Hepes, pH 7.4 (EuroClone)] before the addition of 5,000 35S-labeled CVA24v virions per cell. Cells were washed and the radioactivity of the cells was measured using a Wallac 1409 scintillation counter (PerkinElmer).
Biolayer Interferometry.
Biolayer interferometry was performed with the Octet Red 384 (ForteBio) using biotinylated ICAM-1 and LAMP-1. Streptavidin sensors were loaded with a 2 µg/μL solution of each protein for 15 min. Association of viruses to the sensor was performed for 60 min.
Phylogenetic Analysis.
All available CV-A24(v) sequences spanning the entire VP1 protein were downloaded from GenBank (January 1, 2017) and aligned using the ClustalW (1.6) algorithm implemented in MEGA version 7, followed by manual refinement. A phylogenetic tree of VP1 sequences was constructed using the neighbor-joining method and the maximum composite likelihood method implemented in MEGA version 7. The phylogenetic tree was visualized using Figtree version 1.4.3.
Supplementary Material
Acknowledgments
We thank Dieter Blaas for providing ICAM-1–specific antiserum (supersup). This work was supported by Netherlands Organization for Scientific Research Grant NWO-VICI-91812628 (to F.J.M.v.K.), by German Research Foundation Grant SFB685 (to T.S. and G.Z.), and Wellcome Trust PhD Studentship support 102572/B/13/Z (to D.L.H.). All EM was performed in the Astbury Biostructure Laboratory, which was funded by the University of Leeds and the Wellcome Trust (108466/Z/15/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM reconstruction and atomic model have been deposited in the Electron Microscopy Data Bank (entry no. 3880) and Protein Data Bank (PDB ID code 6EIT), respectively. VP1 sequences have been deposited in the GenBank database (accession nos. MG272373, MG272374, and MG272375).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713284115/-/DCSupplemental.
References
- 1.Ghazali O, et al. An outbreak of acute haemorrhagic conjunctivitis in Melaka, Malaysia. Singapore Med J. 2003;44:511–516. [PubMed] [Google Scholar]
- 2.Kurokawa M, Rai SK, Ono K, Gurung R, Ruit S. Viral investigation of acute hemorrhagic conjunctivitis outbreak (2003) in Nepal using molecular methods. Southeast Asian J Trop Med Public Health. 2006;37:904–910. [PubMed] [Google Scholar]
- 3.Oh MD, et al. Acute hemorrhagic conjunctivitis caused by coxsackievirus A24 variant, South Korea, 2002. Emerg Infect Dis. 2003;9:1010–1012. doi: 10.3201/eid0908.030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Measroch V, Gear J, Faerber GI. Studies in poliomyelitis; the isolation of a coxsackie-like virus from the faeces of apparently healthy bantu infants. S Afr Med J. 1951;25:421–424. [PubMed] [Google Scholar]
- 5.Kono R, Sasagawa A, Ishii K, Sugiura S, Ochi M. Pandemic of new type of conjunctivitis. Lancet. 1972;1:1191–1194. doi: 10.1016/s0140-6736(72)90921-x. [DOI] [PubMed] [Google Scholar]
- 6.Mirkovic RR, Schmidt NJ, Yin-Murphy M, Melnick JL. Enterovirus etiology of the 1970 Singapore epidemic of acute conjunctivitis. Intervirology. 1974;4:119–127. doi: 10.1159/000149850. [DOI] [PubMed] [Google Scholar]
- 7.Miyamura K, et al. The first epidemic of acute hemorrhagic conjunctivitis due to a coxsackievirus A24 variant in Okinawa, Japan, in 1985-1986. Jpn J Med Sci Biol. 1988;41:159–174. doi: 10.7883/yoken1952.41.159. [DOI] [PubMed] [Google Scholar]
- 8.Ishiko H, et al. Phylogenetic analysis of a coxsackievirus A24 variant: The most recent worldwide pandemic was caused by progenies of a virus prevalent around 1981. Virology. 1992;187:748–759. doi: 10.1016/0042-6822(92)90477-7. [DOI] [PubMed] [Google Scholar]
- 9.Lévêque N, Huguet P, Norder H, Chomel J-J. Les Enterovirus responsables de conjonctivite aiguë hémorragique. Med Mal Infect. 2010;40:212–218. doi: 10.1016/j.medmal.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Rossmann MG, He Y, Kuhn RJ. Picornavirus-receptor interactions. Trends Microbiol. 2002;10:324–331. doi: 10.1016/s0966-842x(02)02383-1. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson EC, Jamshidi F, Johansson SMC, Oberste MS, Arnberg N. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J Virol. 2008;82:3061–3068. doi: 10.1128/JVI.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zocher G, et al. A sialic acid binding site in a human picornavirus. PLoS Pathog. 2014;10:e1004401. doi: 10.1371/journal.ppat.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baggen J, et al. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc Natl Acad Sci USA. 2016;113:1399–1404, and correction (2016) 113:E1588. doi: 10.1073/pnas.1524498113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Stolpe A, van der Saag PT. Intercellular adhesion molecule-1. J Mol Med (Berl) 1996;74:13–33. doi: 10.1007/BF00202069. [DOI] [PubMed] [Google Scholar]
- 15.Shafren DR, Dorahy DJ, Greive SJ, Burns GF, Barry RD. Mouse cells expressing human intercellular adhesion molecule-1 are susceptible to infection by coxsackievirus A21. J Virol. 1997;71:785–789. doi: 10.1128/jvi.71.1.785-789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greve JM, et al. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 17.Bergelson JM, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 18.Xiao C, et al. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure. 2005;13:1019–1033. doi: 10.1016/j.str.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Varghese JN, et al. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc Natl Acad Sci USA. 1997;94:11808–11812. doi: 10.1073/pnas.94.22.11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakkers MJG, et al. Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation. Proc Natl Acad Sci USA. 2016;113:E3111–E3119. doi: 10.1073/pnas.1519881113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnberg N, Edlund K, Kidd AH, Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander DA, Dimock K. Sialic acid functions in enterovirus 70 binding and infection. J Virol. 2002;76:11265–11272. doi: 10.1128/JVI.76.22.11265-11272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belser JA, Rota PA, Tumpey TM. Ocular tropism of respiratory viruses. Microbiol Mol Biol Rev. 2013;77:144–156. doi: 10.1128/MMBR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smura T, et al. The evolution of Vp1 gene in enterovirus C species sub-group that contains types CVA-21, CVA-24, EV-C95, EV-C96 and EV-C99. PLoS One. 2014;9:e93737. doi: 10.1371/journal.pone.0093737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.