Significance
One of the outstanding questions in understanding how new species form is how reproductive isolation arises. In particular, the relative roles of gene flow and natural selection in creating two separate species remains open for debate. Here we show within the four continuously speciating lineages of a poplar that local genomic differentiation of populations is not associated with either rate of recent gene flow or time of species divergence. By contrast, we found that these genomic islands of divergence most likely came about by selective processes—sorting of ancient genetic polymorphisms and the incidental hitchhiking of linked variations. These findings substantially enhance our understanding of genomic changes in speciation.
Keywords: speciation, paraphyletic, genome divergence, natural selection, gene flow
Abstract
How genome divergence eventually leads to speciation is a topic of prime evolutionary interest. Genomic islands of elevated divergence are frequently reported between diverging lineages, and their size is expected to increase with time and gene flow under the speciation-with-gene-flow model. However, such islands can also result from divergent sorting of ancient polymorphisms, recent ecological selection regardless of gene flow, and/or recurrent background selection and selective sweeps in low-recombination regions. It is challenging to disentangle these nonexclusive alternatives, but here we attempt to do this in an analysis of what drove genomic divergence between four lineages comprising a species complex of desert poplar trees. Within this complex we found that two morphologically delimited species, Populus euphratica and Populus pruinosa, were paraphyletic while the four lineages exhibited contrasting levels of gene flow and divergence times, providing a good system for testing hypotheses on the origin of divergence islands. We show that the size and number of genomic islands that distinguish lineages are not associated with either rate of recent gene flow or time of divergence. Instead, they are most likely derived from divergent sorting of ancient polymorphisms and divergence hitchhiking. We found that highly diverged genes under lineage-specific selection and putatively involved in ecological and morphological divergence occur both within and outside these islands. Our results highlight the need to incorporate demography, absolute divergence measurement, and gene flow rate to explain the formation of genomic islands and to identify potential genomic regions involved in speciation.
Understanding how and why genomes diverge during speciation is fundamental to an understanding of how species evolve (1). With the advance of high-throughput sequencing technologies, considerable progress has been made in documenting the genomic landscape of divergence between recently evolved species (2–23). Most genomic speciation studies have revealed a highly heterogeneous pattern of genomic divergence, with peaks of elevated differentiation scattered across the genome. This “mosaic” pattern of elevated genomic divergence has been hypothesized to be associated with the presence of genomic regions that are resistant to gene flow (1, 23), most likely because they contain barrier variants related to local adaptation and/or reproductive isolation (24). Under this speciation-with-gene-flow model, the size and number of these regions, generally called genomic islands (1, 2, 25), are predicted to expand due to divergence hitchhiking. Moreover, both are expected to grow with divergence time and greater gene flow between incipient lineages (26, 27).
However, genomic islands with elevated divergence (usually quantified by Wright’s fixation index, FST) may also occur in the absence of recent gene flow and can be created by sorting of ancient divergent haplotypes, recent selection at ecologically relevant loci regardless of gene flow and recombination rate, and/or recurrent background selection and selective sweeps in regions of low recombination (12, 21, 28–31). These alternative deterministic and stochastic processes are not mutually exclusive and are therefore difficult to disentangle. Dxy, an absolute measure of genetic divergence between incipient lineages, is less affected than FST by genomic reductions in genetic diversity (12). Thus, Dxy is expected to be elevated in regions with reduced gene flow but unchanged or decreased in regions under the effect of recurrent background selection or selective sweeps (21). This is because recurrent background selection or selective sweeps tend to reduce genetic diversity in the ancestral population, which leads to reduced Dxy due to decreased time to the most recent common ancestor between individuals representing the two descendent lineages (1, 23). Such a scenario should be more apparent in genomic regions of low recombination (12, 15, 20, 32), resulting in a general positive correlation between genetic diversity and recombination rate (33). Nonetheless, divergent sorting of ancient divergent haplotypes will also elevate Dxy and cause island-like patterns of genomic divergence to originate between diverging lineages as in the speciation-with-gene-flow model (21, 34). Thus, analyses of Dxy will only discriminate between some of the alternative factors that cause genomic islands of divergence identified by FST (12, 21, 23, 29, 35, 36). These problems can be alleviated by (i) reconstructing the past demography of the lineages concerned and (ii) comparing lineages with contrasted gene flow and times of divergence along the speciation continuum (22, 23). This is especially critical for plants in which interspecific gene flow occurs more frequently than in animals (1).
Populus euphratica Oliv. (Pe) and Populus pruinosa Schrenk (Pp) are two sister species in section Turanga (37–39), both of which are well adapted to the extreme desert environment (SI Appendix, Fig. S1). The natural range of P. euphratica extends discontinuously from Morocco in North Africa through the Middle East to central and western Asia, whereas P. pruinosa is restricted to western China and adjacent regions (40) (Fig. 1A). Despite their close relationship, these two species are morphologically well differentiated and grow in distinctly different desert habitats. Leaves of P. euphratica are glabrous and vary in shape from linear or lanceolate on juvenile plants and basal twigs of mature plants, to rounded, ovate, or obovate on older twigs of mature plants. In contrast, leaves of P. pruinosa are always ovate or kidney-shaped with thick hairs (SI Appendix, Fig. S2). Although their flowering periods partially overlap, P. euphratica flowers earlier than P. pruinosa in western China (41). It is also evident that where they co-occur, P. euphratica occupies sites with deep underground water, whereas P. pruinosa occurs where underground water is close to the surface, near ancient or extant rivers. Both species form clones by vegetative propagation, mainly through root suckers (38, 40). Since asexual reproduction leads to more extensive linkage disequilibrium, it can play a part in speciation and the maintenance of species. A previous study of nuclear sequence variation in samples from western China has shown that these two species diverged during the Pleistocene in the presence of strong gene flow (42). Therefore, P. euphratica and P. pruinosa provide an excellent system for evaluating how various evolutionary forces could have shaped patterns of genomic divergence during speciation, especially for tree species.
Fig. 1.
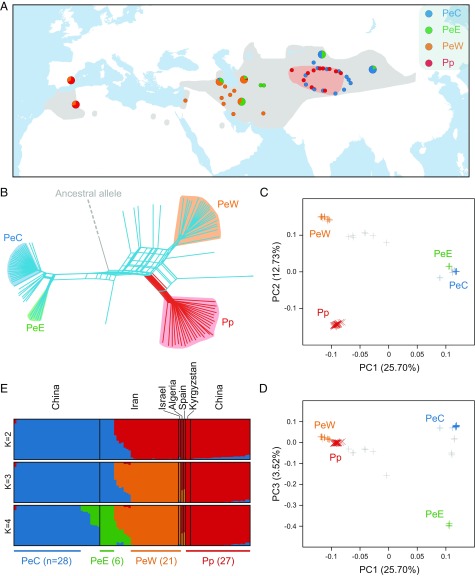
Phylogenetic and population genetic analyses of P. euphratica and P. pruinosa. (A) The biogeographic regions of whole-genome–sequenced individuals for P. euphratica (three lineages identified here: PeC, PeE, and PeW) and P. pruinosa (a single lineage identified here: Pp). The colored areas indicate biogeographic distribution of P. euphratica (gray) and P. pruinosa (pink), and the frequencies of each lineage in potential hybrid and/or backcross populations are shown using larger pie charts. (B) Phylogenetic network inferred using the Neighbor-Net method based on genome-wide SNPs. The ancestral state was identified by genotyping the segregating sites with 10 additional poplar species (see SI Appendix for detailed information). (C and D) PCA plots of SNP data for P. euphratica (+) and P. pruinosa (x). (E) Population structure bar plots. Each vertical bar represents a single individual, and the height of each color represents the probability of assignment to that cluster. The number of individuals in each lineage is also shown.
We resequenced the genomes of both species across their biogeographical distributions to examine genomic patterns of divergence, and in particular to infer the relative roles played by gene flow, divergence time, lineage sorting, recombination, and selection during the speciation process. We resolved a paraphyletic pattern of divergence between the two species with four lineages originating during two continuous phases of divergence. Furthermore, we found that divergent sorting of ancient polymorphisms and divergence hitchhiking contributed mainly to the heterogeneous patterns of genomic divergence between the lineages detected. Levels of gene flow and time since divergence were found not to affect the general formation, size, and number of genomic islands that distinguish lineages. Our study not only provides insights into the speciation histories and the corresponding genomic changes of the two desert poplars but also identifies important lineage-specific genes under positive selection that could be responsible for the ecological, physiological, and morphological diversification between these two species.
Results
Genome Resequencing and Genetic Relatedness.
We resequenced the genomes of 122 P. euphratica and 34 P. pruinosa individuals spanning their worldwide geographic distributions (mean sequencing coverage of 11.36×; Fig. 1A and SI Appendix, Table S1). Sequences were first aligned to the reference genome of P. euphratica (43), and population genetic statistics based on the site frequency spectrum were then estimated using methods that accounted for uncertainty in the assignment of genotypes (44, 45). Additional tools (46, 47) were used to increase the stringency of variant calls when required. In total, 4,285,372 and 2,460,093 high-quality SNPs were identified in P. euphratica and P. pruinosa, respectively, of which 1,688,107 were shared between the two species (39.4% and 68.6% of the total SNPs in each species, respectively).
Given that poplar trees are known to form clones by vegetative propagation, mainly through root suckers (38, 40), we estimated the genetic relatedness between individuals on the basis of pairwise comparisons of SNP data. We found extensive clonal growth in almost all populations, with P. euphratica populations in Spain, Algeria, and Israel most extreme in this regard (SI Appendix, Fig. S3). The latter likely reflects strong founder effects by a few or even a single individual, followed by extensive clonal propagation. Therefore, we discarded individuals that were more related than third-degree relationships, retaining a total of 99 individuals (SI Appendix, Fig. S3) for subsequent population genetic analyses.
Population Structure.
To examine genetic relationships between individuals, we constructed a phylogenetic network based on nuclear genome polymorphisms. This revealed an evolutionary history distinctly different from the phenotype-based classification (Fig. 1B). P. euphratica comprises three lineages (PeE, PeW, and PeC) and is paraphyletic with P. pruinosa; that is, P. pruinosa individuals belong to a single lineage (Pp) sister to one of the three P. euphratica lineages, PeW. The other two lineages, PeC and PeE, are closely related. PeW is mainly distributed in western Iran and extends to Israel, while PeE occurs mainly in eastern Iran. Both PeC and Pp mainly occur in western China and adjacent regions (Fig. 1A). This paraphyletic pattern was also confirmed by our maximum-likelihood phylogenetic analyses based on complete chloroplast sequences (SI Appendix, Fig. S4), although, in this case, a few P. pruinosa individuals grouped with the lineage PeC, which may reflect recent gene flow between P. pruinosa and PeC in western China. The results of a principal components analysis (PCA) of SNP genotypes across all individuals clearly reflected a history of lineage divergence (Fig. 1 C and D). The first principal component (PC1; variance explained = 25.70%; Tracy–Widom test, P = 1.99e-14) separated PeC and PeE from the two other lineages, while the second one (PC2; variance explained = 12.73%; Tracy–Widom test, P = 7.94e-44) separated PeW from Pp. Lineages PeC and PeE were clearly differentiated by PC3 (variance explained = 3.52%; Tracy–Widom test, P = 1.87e-134), confirming them as genetically distinct. The PCA also showed some individuals with intermediate positions among PeW, PeE, and PeC, indicating population admixture and recent hybridization and backcrossing between these lineages. Population structure analyses using NGSadmix (48) (Fig. 1E) further indicated that potential hybrids and backcrosses (with >10% of the genome from another lineage) between PeW and PeE are distributed in central Iran, while those between PeC and PeE occur in western China (Fig. 1A). These admixed individuals were excluded from downstream analyses (SI Appendix, Table S2). Moreover, this analysis showed that individuals from Spain and Algeria are hybrids between PeW and Pp. Because such hybrids occur disjunctively from the current distributions of both PeW and Pp lineages, it is feasible they were introduced as clones to these areas by humans. Alternatively, both lineages may have been much more widely distributed in the past, with the hybrids in Spain and Algeria representing remnants of their previous co-occurrence and contact in these areas.
The genetic diversity (π) of each lineage ranged from 5.07 to 8.30 × 10−3 (SI Appendix, Table S2). Lineage Pp exhibited the highest genetic diversity and showed much more extensive genome-wide linkage disequilibrium (LD) than PeC and PeW (SI Appendix, Fig. S5). This could be partly explained by a more recent bottleneck in Pp and strong asymmetric genetic introgression from PeC into Pp as previously suggested (42). Pairwise genome-wide averages of genetic divergence (FST) between lineages ranged from 0.19 to 0.36, indicating increased divergence times among lineages. This pattern was also evident from the mean pairwise nucleotide difference in interlineage comparisons (Dxy) (SI Appendix, Table S2).
Demographic History.
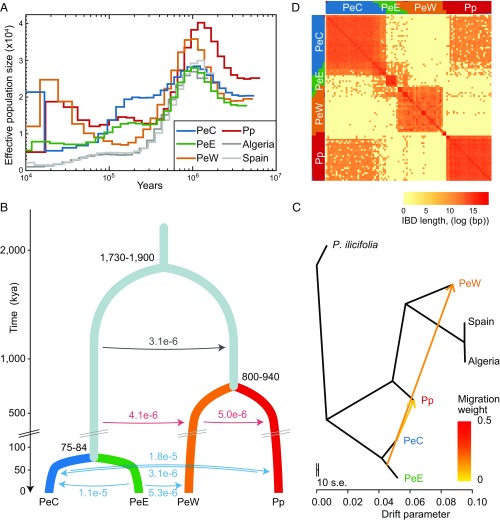
To investigate the evolutionary history of the four lineages, we first examined the historical changes in effective population size (Ne) for each lineage using the Pairwise Sequentially Markovian Coalescent method (49). This showed that all populations/lineages experienced a period of dramatic decline in Ne starting ∼1 Mya (Fig. 2A and SI Appendix, Fig. S6). Algerian and Spanish populations of P. euphratica exhibited the smallest historical Ne, consistent with a clonal origin. The lineage PeW underwent a more severe bottleneck than lineages PeE and Pp ∼50–300 kya, while PeC maintained a relatively large Ne between 100 and 300 kya followed by a sharp decrease. To evaluate alternative divergence models (SI Appendix, Fig. S7), we further analyzed pairwise joint site frequency spectra using a composite likelihood approach as implemented in fastsimcoal2 (50). The best-supported model (Fig. 2B and SI Appendix, Figs. S8 and S9 and Tables S3 and S4) suggests that the common ancestor of PeC and PeE split from the common ancestor of PeW and Pp at least 1.86 Mya [95% highest posterior density (HPD) = 1.73–1.90 Mya], while PeW and Pp diverged 0.88 Mya (95% HPD = 0.80–0.94 Mya) and PeC and PeE diverged more recently, ∼80 kya (95% HPD = 75–84 kya). In addition, our simulations suggest that the two continued phases of divergence gave rise to the four lineages and these were accompanied by different rates of gene flow. Gene flow was highest from PeC to Pp (>PeE to PeC > PeE to PeW > Pp to PeC), as also suggested by TreeMix analyses (51) (Fig. 2C and SI Appendix, Fig. S10). Moreover, the much more extensively shared identical-by-descent haplotypes of PeC and Pp indicate that gene flow occurred recently between these two lineages (Fig. 2D). Thus, the contrasting patterns and rates of gene flow between lineages provide a good model for examining the potential effect of gene flow on patterns of genomic divergence and speciation.
Fig. 2.
Inferred demographic history for P. euphratica (PeC, PeE, and PeW) and P. pruinosa (Pp). (A) Changes in effective population size (Ne) through time inferred by the Pairwise Sequentially Markovian Coalescent model. (B) Schematic of demographic scenario modeled using fastsimcoal2. Split times (kya), population size, and migration rates correspond to 95% CIs obtained from this model are shown in SI Appendix, Fig. S8 and Table S4. Estimates of gene flow between populations are given in the migration fraction per generation. (C) The maximum likelihood tree inferred using TreeMix with two allowed migration events. P. ilicifolia was assigned as an outgroup. The scale bar shows 10 times the average SE of the entries in the sample covariance matrix, and migration edges are depicted as arrows colored by migration weight. (D) Estimated haplotype sharing between individuals. Heat-map colors represent the total length of IBD blocks for each pairwise comparison. IBD, identical-by-descent.
Genomic Islands of Divergence Between Lineages.
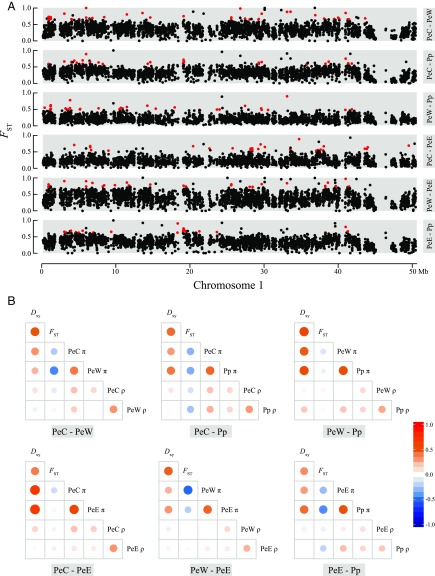
The shape of the FST distribution varied between population pairs (Kolmogorov–Smirnov test, P < 2.2e-16 for all pairwise comparisons) (SI Appendix, Fig. S11). Consistent with a divergence scenario that includes a burst of recent gene flow, the FST values between PeC and Pp showed an l-shaped distribution with only 9,546 fixed differences (SI Appendix, Table S2). This contrasts with comparisons between PeC and PeW, which exhibit similar divergence times but reduced introgression with a greater proportion of highly divergent loci (75,467 fixed differences, accounting for 1.99% of the total number of polymorphisms in these two lineages). We next surveyed the genomic landscape of divergence using a sliding window approach (Fig. 3A and SI Appendix, Fig. S12). As previously found in other species (9, 15, 16), genetic divergence along the genome was highly heterogeneous, irrespective of the level of genome-wide differentiation and geographical separation. In addition, coalescent simulations using msms (52) under the best-fitting demographic model showed that the distributions of simulated polymorphisms and divergence were generally in agreement with the observed patterns across the genome (SI Appendix, Fig. S13). This indicates that much of the genomic heterogeneity in divergence can simply be attributed to the effects of demographic processes and genetic drift (31, 53).
Fig. 3.
The heterogeneity of genomic divergence in P. euphratica (PeC, PeE, and PeW) and P. pruinosa (Pp). (A) Pairwise genetic divergence (FST) in 10-kb sliding windows along an example chromosome (chromosome 1) for all comparisons (see SI Appendix, Fig. S12, for all chromosomes). Genomic islands of divergence are shown in red. (B) Correlation of 10-kb window-based estimates of genetic divergence (FST), absolute divergence (Dxy), nucleotide diversity (π), and population-scaled recombination rate (ρ) for all six pairwise comparisons. Red circles indicate a positive correlation, while blue ones indicate a negative correlation. The color intensity and circle size are proportional to Spearman’s correlation coefficient.
To further understand the evolutionary forces shaping the heterogeneous landscape of genomic divergence, we detected outlier windows for each pair of lineages. These were defined as those falling within the top 1% of the empirical FST distribution that exhibited significant (false discovery rate < 0.01) divergence compared with a null distribution obtained using a permutation approach (16). Across all comparisons, a total of 1,869 extreme 10-kb outlier windows were identified (SI Appendix, Table S5). A few of these outlier windows were shared between pairwise comparisons (SI Appendix, Table S6). After examining their genomic distribution and overlap relationships, we combined adjacent outlier windows and established that these genomic islands of divergence were rather small and scattered across the genome (Fig. 3A and SI Appendix, Figs. S12 and S14). In addition, these islands were characterized by strongly reduced levels of nucleotide diversity (π), skewed allele frequency spectra toward an excess of low-frequency variants (i.e., more negative Tajima’s D), elevated levels of LD, and significantly higher proportions of positively selected genes [PSGs; 108 PSGs of 1,537 genes in these regions; χ2 test, χ2 (1) = 210.62, P < 2.2e-16], indicating that linked selection acted on these regions (SI Appendix, Fig. S15 and Tables S5 and S7).
We further examined what factors contributed to the formation of genomic islands based on values of Dxy (12, 21). We compared the level of Dxy in genomic islands to that of genome background in each pair of lineages. We recovered significantly elevated Dxy in genomic islands for all pairwise comparisons (SI Appendix, Table S7), regardless of rate of gene flow between lineages under comparison. This observation suggests that haplotypes that include genomic islands may have become genetically isolated before the rest of the genomes of the lineage pairs under comparison. No significant difference was observed in the number and size of genomic islands among comparisons for pairwise lineages with contrasting amounts of gene flow and divergence times [number of genomic islands: χ2 test, χ2 (5) = 5.35, P = 0.375; size of genomic islands: nonparametric Kruskal–Wallis rank sum test, χ2 (5) = 5.53, P = 0.355]. We additionally examined changes of recombination rates and found that the population-scaled recombination rate (ρ = 4Nec) was significantly reduced in genomic islands compared with the rest of the genome for all paired comparisons (SI Appendix, Table S7). However, the correlation between recombination rates ρ and absolute divergence Dxy across the whole genome, although significant, was very low (close to zero; Fig. 3B and SI Appendix, Figs. S16 and S17).
Genes Under Positive Selection.
Despite being closely related, P. euphratica and P. pruinosa differ in ecology, physiology, and morphology (41, 42). To explore the genetic basis of this differentiation, we conducted HKA (Hudson-Kreitman-Aguadé) (54) and PBS (Population Branch Statistic) (55) tests to identify genes with high interlineage divergence under recent natural selection following their paraphyletic divergence. Among 28,148 genes analyzed (SI Appendix, SI Text), a total of 451 were identified under positive selection (188, 204, and 74 in lineages PeC, PeW and Pp, respectively; SI Appendix, Table S8). In addition, 15 of these PSGs were shared between any two lineages, a proportion that is significantly higher than expected by chance (for simulation, we randomly sampled 188, 204, and 74 out of 28,148 genes without replacement for lineages PeC, PeW, and Pp, respectively, and a maximum of 12 overlaps between any two lineages were observed after 100,000 replications). This suggests the presence of diverged haplotypes in their ancestral population possibly followed by strong divergent selection. Importantly, only 108 PSGs were located inside identified genomic islands, while the rest were distributed outside them. Gene ontology enrichment analyses of all PSGs yielded several candidate genes associated with organ development, local adaptation, and reproductive isolation (SI Appendix, Table S9). Among these genes, four (GIS, MYB66, CPC, and RSL2) are associated with trichome differentiation and branching, whereas eight (TCP13, LCR, RDR6, CRF5, YUC4, TRN2, NAC083, and IAA24) are involved in regulation of leaf development and morphology (SI Appendix, Table S10). GIS encodes a putative C2H2 transcription factor that plays a key role in trichome initiation and branching in Arabidopsis thaliana. In addition, GIS loss-of-function mutations exhibit a premature decrease in trichome production on successive leaves, stem internodes, and branches, while overexpression has an opposite effect and causes significant delay in flowering compared with wild-type (56). TCP13 is a member of the CINCINNA-like TCP (TEOSINTE BRANCHED1, CYCLOIDEA, PCF) group of transcription factors, which play a pivotal role in signaling pathways that generate different leaf forms via negative regulation of the expression of boundary-specific genes (57, 58). LCR (LEAF CURLING RESPONSIVENESS) encodes an F-box protein (SKP1-Cullin/CDC53-F-box) involved in the regulation of leaf curling-related morphology. Overexpression of LCR leads not only to abnormal leaf development but also to a salt-tolerant phenotype of A. thaliana in an abscisic acid-dependent manner (59, 60). Moreover, we found that several genes subjected to selection encoded proteins associated with reproductive function, including flowering time (e.g., EFM, HLP1, CIB1, and GAS41), anther development (RPK2), pollen tube growth and guidance (GLR5, PIP5K4, and DOK1), and the development of male and female gametophytes (ARID1, PIN8, DHAD, and RH36) (SI Appendix, Table S10). These reproductive proteins have diverged rapidly across lineages and emerged as candidates involved in postzygotic isolation between lineages.
Discussion
Our population genomic analyses of P. euphratica within its vast but fragmented range across Eurasia and North Africa, and additional analyses of P. euphratica and P. pruinosa within the sympatric area in west China, together suggest a scenario of paraphyletic divergence of these two morphologically delimited species. Four lineages were identified: PeW, PeE, and PeC representing P. euphratica, and Pp representing P. pruinosa (Fig. 1). Coalescent-based simulations indicate that these lineages diverged during the Pleistocene. Although the three P. euphratica lineages show limited morphological differentiation, the possibility of incipient speciation occurring within this species is supported by the finding that divergence between PeW and P. pruinosa occurred later than between their ancestral clade and the ancestral clade of the other two lineages, PeE and PeC. While P. euphratica is genetically divided into three lineages, there is little congruence between the current geographical distributions and the genetic distances between them, suggesting a complex expansion and colonization history of the total complex, with some populations possibly established or affected by humans (e.g., Spanish populations and those from North Africa and Central Asia).
Few speciation studies have explicitly reconstructed the demographic history and estimated the amount of gene flow between diverging lineages (23, 24). In this study, we assessed the timing and amount of gene flow and changes in population size of the recovered lineages, revealing pairs of lineages with either large or limited amounts of gene flow occurring between them. In particular, a relatively high level of recent gene flow was shown to have occurred between PeC and Pp. The level of divergence between pairs of lineages was highly heterogeneous along the genome, which could be largely explained by historical demographic processes and genetic drift as suggested by our simulations (SI Appendix, Fig. S13). In addition, we identified numerous small genomic islands (quantified by FST) scattered across the genome. Dxy was elevated in these islands (SI Appendix, Table S7), which is consistent with a model in which these islands were derived from divergent sorting of ancient divergent haplotypes (21). An alternative explanation is the speciation-with-gene-flow model in which restricted gene flow at genomic islands would be expected to increase both absolute and relative measures of genomic divergence (12). Under this alternative hypothesis, genomic islands are predicted to be more pronounced with increased gene flow and divergence time (26, 27). However, our findings indicate that Dxy was of the same magnitude across genomic islands, and no significant difference in size and number of genomic islands was observed among interlineage comparisons with contrasting levels of gene flow and divergence times. Therefore, our observations suggest that recent gene flow is unlikely to be a major factor in generating genomic islands of divergence in these desert poplars.
Several selection signals, including elevated levels of LD and reduced levels of genetic diversity and Tajima’s D, were observed across genomic islands. In addition, we observed a strong association between islands of divergence and low recombination rates. Such an association suggests the possible occurrence of divergence hitchhiking, or background selection and/or recurrent selective sweeps within regions of low recombination (12, 21), both of which tend to reduce genetic diversity and increase FST. If genomic islands are mainly produced by low recombination rate and linked selection, levels of Dxy within them will not be elevated (12, 21), which is inconsistent with our observations (SI Appendix, Table S7). In conjunction with the covariation of population genomic parameters (Fig. 3B and SI Appendix, Figs. S15–S17), genomic islands of divergence observed here are best explained by divergent sorting of ancient polymorphisms predating lineage divergence (12, 15, 20, 21, 61). The link between low recombination rates and high Dxy of genomic islands most likely resulted from “divergence hitchhiking” because divergent sorting of anciently diverged haplotypes tends to reduce gene flow in surrounding linked regions (62).
Interestingly, we found that genes under positive selection and exhibiting high interlineage divergence were detected both within and outside genomic islands (SI Appendix, Table S8). These genes are putatively functionally related to organ development, local adaptation, and reproductive isolation, which may underlie ecological and morphological divergence between lineages (SI Appendix, Table S10). Several genes showed high divergence and selection signatures between P. pruinosa and the P. euphratica lineages. Two of these, TCP13 and IAA24, are involved in the regulation of leaf development and morphology, of which a major difference exists between P. euphratica and P. pruinosa. Similarly, the fixation of different alleles for the gene GIS could be a cause of dense trichome development on leaves of P. pruinosa, while high allelic divergence and fixation of the gene GAS41 in P. pruinosa may contribute to its later flowering relative to PeC (40, 41). In addition, numerous single-nucleotide differences outside these islands or undetectable here may have also contributed to dramatic differentiation between lineages (or species) through an accumulation of small individual effects underlying polygenic barriers (63). What drove cryptic differentiation within P. euphratica and between it and P. pruinosa remains to be investigated in the future.
In conclusion, our results revealed lineage divergence within P. euphratica and a paraphyletic relationship of this species with P. pruinosa. Genomic islands of divergence that distinguish the four lineages of this poplar complex most likely derived from divergent sorting of ancient polymorphisms and divergence hitchhiking but not heterogeneous gene flow. Our findings highlight the need to integrate information on demographic history, gene flow, and absolute genomic divergence to distinguish evolutionary processes that generate genomic islands and identify genomic regions potentially involved in speciation.
Materials and Methods
Silica gel-dried leaves were collected from western China, Kyrgyzstan, Iran, Israel, Spain, and Algeria (SI Appendix, SI Text). Total genomic DNA was extracted using a standard cetyltrimethylammonium bromide protocol and sequenced on the Illumina HiSeq 2000 and 2500 platforms. After data-quality control, reads were mapped to the reference genome using BWA-MEM. Population genetic structure was conducted using the Neighbor-net method, PCA, and admixture estimation. Population genetic statistics π, FST, Dxy, and Tajima’s D were estimated based on the site frequency spectrum inferred by analyses of next generation sequencing data. Detailed information on materials and methods with associated references are available in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Lakhdar Khelifi and Abdelkader Morsli (Laboratoire des Ressources Génétiques et Biotechnologies, Ecole Nationale Supérieure Agronomique, Algiers) for helping collect materials from Algeria. Financial support was provided by National Natural Science Foundation of China (91731301, 31590821, 31500502, and 31561123001), National Key Research and Development Program of China (2016YFD0600101), National High Technology Research and Development Program of China (863 Program, No. 2013AA100605), Youth Science and Technology Innovation Team of Province (2014TD003), National Key Project for Basic Research (2012CB114504), Ministry of Science and Technology of the People’s Republic of China (2010DFA34610), International Collaboration 111 Projects of China, and the 985 and 211 Projects of Sichuan University.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (BioProject PRJNA380894 and PRJNA380891).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713288114/-/DCSupplemental.
References
- 1.Seehausen O, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15:176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- 2.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harr B. Genomic islands of differentiation between house mouse subspecies. Genome Res. 2006;16:730–737. doi: 10.1101/gr.5045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawniczak MK, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 6.Nadeau NJ, et al. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos Trans R Soc Lond B Biol Sci. 2012;367:343–353. doi: 10.1098/rstb.2011.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin SH, et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–1828. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renaut S, et al. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat Commun. 2013;4:1827. doi: 10.1038/ncomms2833. [DOI] [PubMed] [Google Scholar]
- 9.Brawand D, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carneiro M, et al. The genomic architecture of population divergence between subspecies of the European rabbit. PLoS Genet. 2014;10:e1003519. doi: 10.1371/journal.pgen.1003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarkson CS, et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 2014;5:4248. doi: 10.1038/ncomms5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- 13.Poelstra JW, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344:1410–1414. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- 14.Soria-Carrasco V, et al. Stick insect genomes reveal natural selection’s role in parallel speciation. Science. 2014;344:738–742. doi: 10.1126/science.1252136. [DOI] [PubMed] [Google Scholar]
- 15.Burri R, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 2015;25:1656–1665. doi: 10.1101/gr.196485.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feulner PG, et al. Genomics of divergence along a continuum of parapatric population differentiation. PLoS Genet. 2015;11:e1004966. doi: 10.1371/journal.pgen.1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malinsky M, et al. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350:1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foote AD, et al. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat Commun. 2016;7:11693. doi: 10.1038/ncomms11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marques DA, et al. Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genet. 2016;12:e1005887. doi: 10.1371/journal.pgen.1005887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Street NR, Scofield DG, Ingvarsson PK. Variation in linked selection and recombination drive genomic divergence during allopatric speciation of European and American aspens. Mol Biol Evol. 2016;33:1754–1767. doi: 10.1093/molbev/msw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han F, et al. Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin’s finches. Genome Res. 2017;27:1004–1015. doi: 10.1101/gr.212522.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riesch R, et al. Transitions between phases of genomic differentiation during stick-insect speciation. Nat Ecol Evol. 2017;1:82. doi: 10.1038/s41559-017-0082. [DOI] [PubMed] [Google Scholar]
- 23.Wolf JB, Ellegren H. Making sense of genomic islands of differentiation in light of speciation. Nat Rev Genet. 2017;18:87–100. doi: 10.1038/nrg.2016.133. [DOI] [PubMed] [Google Scholar]
- 24.Ravinet M, et al. Interpreting the genomic landscape of speciation: A road map for finding barriers to gene flow. J Evol Biol. 2017;30:1450–1477. doi: 10.1111/jeb.13047. [DOI] [PubMed] [Google Scholar]
- 25.Wu C-I. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. [Google Scholar]
- 26.Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- 27.Feder JL, Nosil P. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution. 2010;64:1729–1747. doi: 10.1111/j.1558-5646.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- 28.Turner TL, Hahn MW. Genomic islands of speciation or genomic islands and speciation? Mol Ecol. 2010;19:848–850. doi: 10.1111/j.1365-294X.2010.04532.x. [DOI] [PubMed] [Google Scholar]
- 29.Noor MA, Bennett SM. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity (Edinb) 2009;103:439–444. doi: 10.1038/hdy.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renaut S, Owens GL, Rieseberg LH. Shared selective pressure and local genomic landscape lead to repeatable patterns of genomic divergence in sunflowers. Mol Ecol. 2014;23:311–324. doi: 10.1111/mec.12600. [DOI] [PubMed] [Google Scholar]
- 31.Campagna L, Gronau I, Silveira LF, Siepel A, Lovette IJ. Distinguishing noise from signal in patterns of genomic divergence in a highly polymorphic avian radiation. Mol Ecol. 2015;24:4238–4251. doi: 10.1111/mec.13314. [DOI] [PubMed] [Google Scholar]
- 32.Irwin DE, Alcaide M, Delmore KE, Irwin JH, Owens GL. Recurrent selection explains parallel evolution of genomic regions of high relative but low absolute differentiation in a ring species. Mol Ecol. 2016;25:4488–4507. doi: 10.1111/mec.13792. [DOI] [PubMed] [Google Scholar]
- 33.Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 34.Sicard A, et al. Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nat Commun. 2015;6:7960. doi: 10.1038/ncomms8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Mol Ecol. 2016;25:2337–2360. doi: 10.1111/mec.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeaman S, Aeschbacher S, Bürger R. The evolution of genomic islands by increased establishment probability of linked alleles. Mol Ecol. 2016;25:2542–2558. doi: 10.1111/mec.13611. [DOI] [PubMed] [Google Scholar]
- 37.Eckenwalder JE. Systematics and evolution of Populus. In: Stettler RF, Bradshaw HD, Heilman PE, Hinckler TM, editors. Biology of Populus and Its Implications for Management and Conservation. Canadian Government Publishing; Ottawa: 1996. pp. 7–32. [Google Scholar]
- 38.Isebrands JG, Richardson J. Poplars and Willows: Trees for Society and the Environment. CAB International; Wallingford, UK: 2014. [Google Scholar]
- 39.Liu X, Wang Z, Shao W, Ye Z, Zhang J. Phylogenetic and taxonomic status analyses of the Abaso section from multiple nuclear genes and plastid fragments reveal new insights into the North America origin of Populus (Salicaceae) Front Plant Sci. 2017;7:2022. doi: 10.3389/fpls.2016.02022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickmann DI, Kuzovkina J. Poplars and willows of the world, with emphasis on silviculturally important species. In: Isebrands JG, Richardson J, editors. Poplars and Willows: Trees for Society and the Environment. CABI; Wallingford, UK: 2014. pp. 8–91. [Google Scholar]
- 41.Wang S, Chen B, Li H. Euphrates Poplar Forest. China Environmental Science Press; Beijing: 1996. [Google Scholar]
- 42.Wang J, et al. Speciation of two desert poplar species triggered by Pleistocene climatic oscillations. Heredity (Edinb) 2014;112:156–164. doi: 10.1038/hdy.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma T, et al. Genomic insights into salt adaptation in a desert poplar. Nat Commun. 2013;4:2797, and erratum (2014) 5:3454. doi: 10.1038/ncomms3797. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS One. 2012;7:e37558. doi: 10.1371/journal.pone.0037558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browning BL, Browning SR. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skotte L, Korneliussen TS, Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ewing G, Hermisson J. MSMS: A coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics. 2010;26:2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: From genotyping to genome typing. Nat Rev Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- 54.Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gan Y, et al. Glabrous inflorescence stems modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell. 2006;18:1383–1395. doi: 10.1105/tpc.106.041533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell. 2010;22:3574–3588. doi: 10.1105/tpc.110.075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song JB, Huang SQ, Dalmay T, Yang ZM. Regulation of leaf morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol. 2012;53:1283–1294. doi: 10.1093/pcp/pcs080. [DOI] [PubMed] [Google Scholar]
- 60.Song JB, et al. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 2013;13:210. doi: 10.1186/1471-2229-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vijay N, et al. Evolution of heterogeneous genome differentiation across multiple contact zones in a crow species complex. Nat Commun. 2016;7:13195. doi: 10.1038/ncomms13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Via S. Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow. Philos Trans R Soc Lond B Biol Sci. 2012;367:451–460. doi: 10.1098/rstb.2011.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.