Significance
To circumvent our dependence on photoactivation in single-molecule microscopy, we devise a universal genetic system to precisely control copy number of fluorescently labeled molecules in a cell. Combined with photostable labels, this system enables long-term single-particle tracking of densely packed cellular organelles and proteins. This strategy expands spatiotemporal length scales of live-cell single-molecule measurements to quantitatively understand complex control of molecular dynamics in vivo.
Keywords: long-term single-molecule imaging, translational readthrough, stochastic labeling, synaptic vesicle transport, transcription factor dynamics
Abstract
Our ability to unambiguously image and track individual molecules in live cells is limited by packing of multiple copies of labeled molecules within the resolution limit. Here we devise a universal genetic strategy to precisely control copy number of fluorescently labeled molecules in a cell. This system has a dynamic range of ∼10,000-fold, enabling sparse labeling of proteins expressed at different abundance levels. Combined with photostable labels, this system extends the duration of automated single-molecule tracking by two orders of magnitude. We demonstrate long-term imaging of synaptic vesicle dynamics in cultured neurons as well as in intact zebrafish. We found axon initial segment utilizes a “waterfall” mechanism gating synaptic vesicle transport polarity by promoting anterograde transport processivity. Long-time observation also reveals that transcription factor hops between clustered binding sites in spatially restricted subnuclear regions, suggesting that topological structures in the nucleus shape local gene activities by a sequestering mechanism. This strategy thus greatly expands the spatiotemporal length scales of live-cell single-molecule measurements, enabling new experiments to quantitatively understand complex control of molecular dynamics in vivo.
Life is orchestrated by animate molecular processes in living cells. Decoding the dynamic behavior of single molecules enables us to investigate biology at this fundamental level that is often obscured in ensemble experiments. Sparse labeling is critical for live-cell single-molecule imaging (1, 2), because high copy numbers (103∼107) of protein molecules cooccupy the small volume in a cell (10∼20 µm in diameter). Thus, multiple protein molecules are packed within the light diffraction limit (200∼300 nm). For example, in a small area (10 × 10 µm) comparable to the size of a mammalian cell nucleus, single-molecule images begin to overlap with each other at low concentrations (>102∼103 copies), preventing precise localization (SI Appendix, Fig. S1). Accurate single-molecule tracking requires labeling at much lower densities (<102 copies) to link single-molecule positions across multiple frames with high fidelity (SI Appendix, Fig. S1B). Currently, the most reliable live-cell single-molecule imaging method is by stochastic switching of photoactivatable fluorescent proteins or dyes (e.g., sptPALM or dSTORM) (2–7). The stochastic photoactivation separates appearances of individual fluorophores temporally, allowing single-molecule imaging in densely labeled samples. However, under these conditions, only one fluorophore per molecule can be utilized for imaging. Thus, high laser powers (>1,000 W/cm2) potentially damaging to live specimens are applied to generate sufficient photon counts for localization (8). As a result, fluorescent molecules are quickly bleached, and single-molecule dynamics can only be probed on relatively short time scales.
Here we reason that if we can devise a genetic strategy to precisely control copy number of fluorescently labeled proteins in a cell, this would allow us to sparsely label any target protein with bright fluorescent tags [e.g., 3XGFP, 3XHaloTag, GFP11 (9), sfCherry11 (9), SunTag (10), and ArrayG (11)] for long-term single-molecule tracking. One unique advantage of this method is that single-molecule imaging across a large field of view can be realized by conventional microscopy without photoactivation. Currently, a common strategy to control labeling density is by using promoters with variable strengths to adjust mRNA transcript levels. However, there are no guidelines to engineer the strength of a promoter, because promoter strengths are cell type-dependent, and weak promoters are usually associated with high gene expression noise (12). These disadvantages make it challenging to fine tune protein copy numbers by transcriptional control.
Here, we demonstrate a general method to precisely adjust the copy number of fluorescently labeled proteins by translational readthrough (RT). This method enables sparse labeling of densely packed cellular organelles and molecules with bright fluorescent labels for long-term single-molecule imaging. Real-time imaging in neurons reveals a critical role of axon initial segment in enhancing synaptic vesicle anterograde transport. Long-term observation of transcription factor (TF; Sox2) binding dynamics shows that TF molecules translocate among clustered binding sites in the nucleus with confined local target search kinetics. Our studies thus provide evidence supporting a dynamic sequestering mechanism of topological structures in the nucleus.
Results and Discussion
Stochastic Protein Labeling with a Translational Readthrough Strategy.
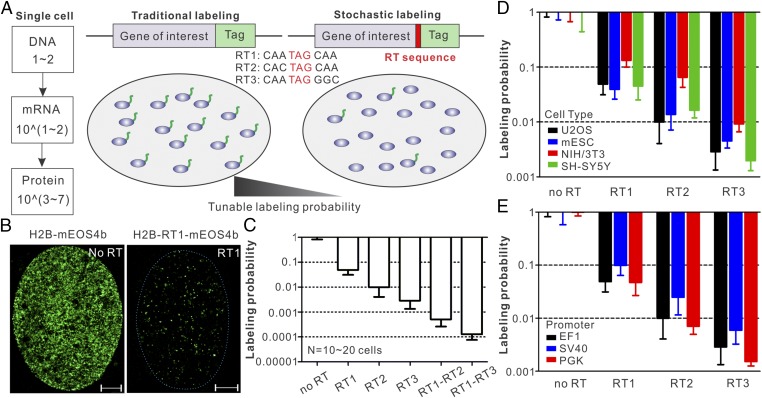
During protein translation, ribosome sometimes misincorporates an amino acid at the stop codon and continues to translate the downstream mRNA sequence. The RT mechanism is conserved from bacteria to mammals (13–15). Interestingly, the probability that a ribosome translates beyond the stop codon (the RT efficiency) is largely dictated by the sequence of the stop codon and its immediate flanking regions (14, 15). Simulation experiments suggest that, compared with using weak promoters, sparse labeling with a RT strategy would benefit from averaging effect at the translational level and give rise to much less labeling heterogeneity in single cells (SI Appendix, Fig. S2). To test this approach, we performed live-cell PALM experiments using cells expressing Histone H2B and mEOS4b separated by different RT sequences (Fig. 1 A and B). Although mRNAs were expressed at similar levels (SI Appendix, Fig. S3C), we observed dramatic reductions of mEOS4b labeling densities upon the RT sequence insertion (Fig. 1 B and C, SI Appendix, Fig. S3B, and Movie S1). We found that each RT sequence has a distinct labeling probability (RT1, ∼5%; RT2, ∼1.5%; RT3, ∼0.4%; Fig. 1C). Importantly, these probabilities are consistent across four cell types and three promoters (EF1, SV40, and PGK) that we tested (Fig. 1 D and E), suggesting that the RT system functions with similar efficiencies in different cell types and under the control of different promoters. We also discovered that RT sequences work synergistically in tandem, giving rise to even lower labeling probabilities (RT1–RT2, ∼0.06%; RT1–RT3, ∼0.01%; Fig. 1C). Therefore, the RT system can adjust labeling density linearly on the logarithmic scale with a large dynamic range (∼10,000-fold). If necessary, this range can be further extended by concatenation of RT sequences. In contrast, the internal ribosome entry site (IRES) only reduced the labeling density to ∼40% (SI Appendix, Fig. S3A), which is not suitable for sparse labeling of most proteins in a cell. It is important to note that past studies identified a repertoire of short sequences with RT efficiencies ranging from 0.1% up to ∼10% (16). These sequences can also be adapted to make fine adjustments to the labeling probability. Based on these data, we propose that the highly tunable RT system can be used to stochastically label protein in live cells for single-molecule imaging (Fig. 1A).
Fig. 1.
Sparse labeling with translational readthrough. (A) Schematics for precise labeling density control by translational readthrough. (Left) After transcription and translation, high copy numbers of functional protein molecules (103∼107) are typically generated in the cell. (Center) In the traditional labeling strategy, all molecules are labeled and high labeling densities make it challenging to observe single-molecule dynamics (SI Appendix, Fig. S1). (Right) By placing a short RT sequence between the target protein and the tag, we can accurately control the probability of protein fusion and sparsely label a faction of molecules for single-molecule imaging. (B) Representative single-cell PALM localization density map for EF1 promoter-driven H2B-mEOS4b in U2OS cells without (Left) and with (Right: RT1) readthrough (Movie S1). On average, 328 localizations per µm2 (no RT) and 17.2 localizations per µm2 (RT1) were detected within 4,500 frames. (Scale bar: 2 µm.) (C) Labeling probabilities for different RT sequences have a large dynamic range (from ∼5% down to ∼0.01%). EF1 promoter-driven H2B-mEOS4b without or with different RT sequences were transfected in U2OS cells, and live-cell PALM experiments were performed to estimate the readthrough efficiencies. Specifically for the probability calculation, H2B-mEOS4b localization density in each RT condition was normalized to the average density in no RT control. (D) The RT strategy is cell type-independent. EF1 promoter-driven H2B-mEOS4b without or with different RT sequences were transfected in different cell types, and live-cell PALM experiments were performed to estimate the readthrough efficiencies. (E) The RT strategy is promoter-independent. H2B-mEOS4b without or with different RT sequences driven by different promoters were transfected in U2OS cells, and live-cell PALM experiments were performed to estimate the readthrough efficiencies. Two-way ANOVA tests were performed for D and E, which indicate that the variations among different cell types or promoters are statistically nonsignificant. Error bars in C–E represent SD.
Long-Term Observation of Synaptic Vesicle Moving Dynamics.
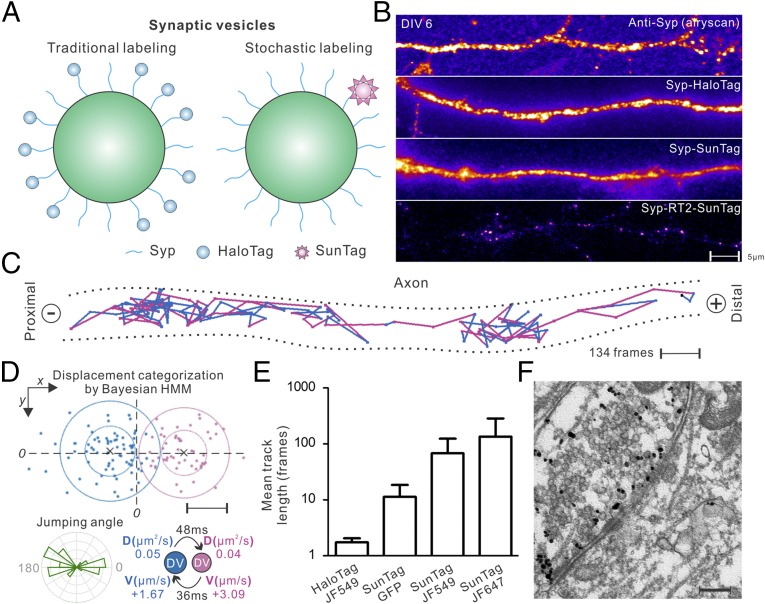
We first sought to apply this stochastic labeling strategy to an important question in neuroscience—how neuronal trafficking polarity is dynamically established. Specifically, synaptic vesicle precursors (SVPs) packaged at the soma must be specifically targeted to presynaptic regions for release and recycle as synaptic vesicles (SVs) (17). Airyscan superresolution imaging (Fig. 2B and SI Appendix, Fig. S4A) and EM studies (18) reveal that high copy numbers of SVPs/SVs are densely packed below the diffraction limit in mammalian neurons, particularly in boutons and presynaptic regions. Consistent with these results, when we labeled a classical synaptic vesicle marker, Synaptophysin (Syp), according to traditional strategies at C terminus (19, 20) with weak expression systems (e.g., CMVd3 and SunTag v1), individual SVPs/SVs cannot be distinguished from each other (Fig. 2 A and B and SI Appendix, Fig. S5). Next, we implemented the RT strategy to sparsely label Syp with the SunTag (Fig. 2A and SI Appendix, Figs. S4B and S14A)—a fluorescence signal amplification system capable of directing 24 copies of fluorescence protein to the target (10). After optimizing the RT sequence under the control of neuron-specific human synapsin1 (hSyn1) promoter, we can directly observe movements of bright, sparsely spaced SVPs/SVs in different compartments of the cell (Fig. 2B and Movie S2). The advantages for this strategy include the following: (i) there are ∼30 copies of Syp in each SVP/SV (21, 22) and on average only one copy of Syp is labeled (Fig. 2A). Thus, we expect that the large size (>1.4 MDa) of the SunTag would not significantly affect the SVP/SV function. Indeed, both immunofluorescence staining and high-resolution EM experiments demonstrated that SVPs/SVs labeled with the SunTag were correctly targeted to presynaptic regions (Fig. 2F and SI Appendix, Fig. S6). (ii) Due to sparse labeling, the movements of single SVPs/SVs covering large distances in the cell can be unambiguously imaged and tracked with high accuracy (Fig. 2C and Movies S3 and S7). Conversely, individual SVPs/SVs could not be resolved without the RT sparse labeling (Fig. 2B), and diffraction-limited spots in the images showed much lower diffusion rates (SI Appendix, Fig. S5E) and likely contain multiple SVPs/SVs. These results suggest that, because of dense labeling, the traditional tracking-in-bulk kymograph method might not be able to capture individual SVP/SV diffusion events in live cells. (iii) Because of the signal amplification from the SunTag, ∼20-fold less laser powers (∼50 W/cm2) can be used compared with the sptPALM or dSTORM method for fast single-molecule imaging (50 Hz). Thus, imaging induced phototoxicity is greatly reduced without trading off imaging speed. (iv) By incorporating HaloTag and bright Janelia Fluor (JF) dyes (5) into the SunTag system, average trajectory lengths showed ∼100-fold improvement over the traditional single HaloTag strategy (Fig. 2E and Movie S2). To further validate the applicability of this method, we sparsely labeled SVPs/SVs in live zebrafish using the Syp-SunTag RT system (SI Appendix, Fig. S7A). We can observe dynamic transport of single SVPs along neurites in vivo (SI Appendix, Fig. S7 B and C and Movies S5 and S6).
Fig. 2.
Automated long-term tracking of individual synaptic vesicle precursors in live neurons. (A) SVP/SV labeling either by tagging Syp with the HaloTag (Left) or sparsely with the SunTag via RT sparse labeling (Right). (B) High packing densities prevent imaging single SVP/SV particles with traditional labeling methods in days in vitro (DIV) 6-cultured hippocampal neurons [from top to bottom, anti-Syp staining (Airyscan); HaloTag fusion (TIRF); SunTag without RT sparse labeling (TIRF)]. When sparsely labeled with the SunTag (Bottom, TIRF) using RT2, individual and sparsely spaced SVPs/SVs (scFv-HaloTag-JF646) can be observed and imaged. (Scale bar: 5 µm.) (C) One representative trajectory tracking SVP movements in axon: steps are color-coded according to transport modes classified by the HMM-Bayes algorithm shown in D. The + and “−” signs denote anterograde and retrograde transport directions, respectively. (Scale bar: 200 nm.) See Movie S3 for raw imaging data. (D) Displacements between frames for the trajectory shown in C can be classified into two main categories with one transport component pointing to the + direction and the other to the − direction. (Lower Left) Rose histogram for jumping angles for trajectory shown in C. (Lower Right) Transport velocities (V), diffusion coefficients (D), and mean lifetimes for the two states defined in Upper. (Scale bar: 100 nm.) (E) Track length comparison for different labeling strategies. Data in lane 1 were acquired using the dSTORM strategy with high laser powers (>1,000 W/cm2). Data in lanes 2–4 were acquired with relatively low laser powers (∼50 W/cm2) with sparse SunTag labeling. Error bars represent SD. (F) Immuno-EM images showing SunTag-labeled SVPs/SVs (RT1) were targeted to presynaptic regions in DIV 12 hippocampal neurons. (Scale bar: 200 nm.)
Enhanced Anterograde Transport of SVP in Axon Initial Segment.
It is worth noting that SVP/SV diffusion dynamics has been imaged before with traditional GFP labeling (20, 23) or extracellular loading of antibody–dye conjugates (24–26). However, because of dense and selective labeling of recycling SVs by antibodies, these methods do not allow long-term, fast tracking of SVP transport events in different neuronal compartments. Because reliable classification of transitions between active transport and random diffusion requires continuous analysis of displacements in long trajectories (27, 28), we applied a recently developed hidden Markov model (HMM)-Bayes algorithm to analyze our SVP/SV tracking data (27). This program effectively separates SVP/SV diffusion, anterograde, and retrograde transport processes (Figs. 2 C and D and 3B and SI Appendix, Fig. S7C) and provides detailed kinetic information, such as diffusion coefficients, transport velocities, and lifetimes for each state. Hence, long-term dynamics of individual SVPs/SVs can be imaged and quantitatively analyzed in live mammalian neurons with high spatiotemporal resolution.
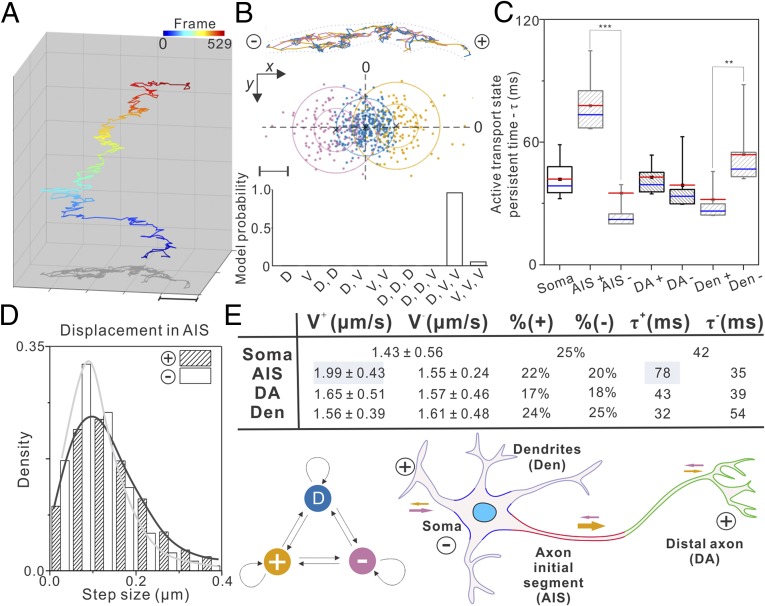
Fig. 3.
Axon initial segment gates synaptic vesicle precursor transport polarity. Long-term single-particle imaging experiments were performed in different neuronal compartments: soma, dendrite (Den), AIS, and DA based on the localization of the AIS marker Ankyrin G. The SVP trajectories were obtained with single-molecule localization and tracking analysis. After the rotation correction of the trajectories (SI Appendix, Fig. S9), the HMM-Bayes algorithm could be applied to analyze SVP dynamics. (A) A representative SVP trajectory consists of 529 frames showing dynamic transitions between diffusion and active transport states. See Movie S7 for the raw imaging data. The frame number is color-coded as indicated by the color bar above. (Scale bar: 1 µm.) (B, Top) Individual steps in the trajectory showed in A are color-coded by the three hidden states classified in the Middle. (B, Middle) Displacements between frames for the trajectory shown in A can be classified by the HMM-Bayes algorithm into one diffusion state (blue) and two transport states with opposing directions: anterograde (+, yellow) and retrograde (−, pink). (B, Bottom) The probabilities of competing models for correctly describing the hidden states in the trajectory. D, diffusion; V, active transport. (Scale bar: 100 nm.) (C) The persistent lifetime for each active transport state in the indicated compartment. The + and − signs denote anterograde and retrograde transport ,respectively (number of cells analyzed >10; number of trajectories analyzed >300). In the box chart, top and bottom error bars represent 95% and 5% percentile, respectively; box represents the range from 25th to 75th percentile; center blue line represents the median; center red line and the small square represent mean value. ***P < 0.001; *P < 0.05. (D) The step size distribution for anterograde (+) and retrograde (−) transport processes in the AIS. (E, Upper) statistic summary for anterograde (+) and retrograde (−) transport processes in different compartments of the neuron. V, transport velocity; %, time fraction; τ, state lifetime. Velocities shown in mean ± SD. (E, Lower Left) HMM diagram describing the dynamic behavior of SVP in the cell. (E, Lower Right) Schematics explaining the abbreviations for different functional compartments of the neuron and the directions of active transport. The size of arrows indicates the degree of cargo transport processivity.
With the imaging and analyzing methods established, we sought to understand the moving and transport dynamics of SVP/SV in different neuronal compartments. First, we divided each neuron into four distinct functional regions: dendrites (Den), soma, axon initial segment (AIS), and distal axon (DA) (Fig. 3E), based on the localization of Ankyrin G—an AIS-specific scaffold protein (SI Appendix, Fig. S8). Then, we performed long-term single-particle tracking of SVP/SVs in these compartments. After the tracking of single SVP/SVs, the trajectories were rotated to align along the x axis with anterograde transport pointing to the “+” direction (SI Appendix, Figs. S9 and S10). Then, we applied HMM-Bayes algorithm to analyze SVP/SV dynamics in each functional region. The results reveal several kinetic features that explain the steady-state SVP/SV distributions and target search mechanisms in neurons: (i) we found that SVPs/SVs display compartment-specific search kinetics by dynamically partitioning between short-lived (30∼80 ms) anterograde and retrograde transport movements interspersed by relatively long-lived slow diffusion states (100∼200 ms; 0.06∼0.09 μm2/s; Fig. 3 B, C, and E and SI Appendix, Fig. S11C). (ii) In the AIS, the processivity of the anterograde transport is substantially enhanced, with anterograde transport lifetime and average movement velocity significantly increased (Fig. 3 C–E). At the same time, the slow diffusion state has a substantially shortened lifetime in the AIS (SI Appendix, Fig. S11C). These observations suggest that specialized functional components in the AIS promote SVP anterograde transport while limiting transient binding and retrograde backtrack movements, in analogy to how a waterfall facilitates unidirectional water flows. (iii) Interestingly, in dendrites and distal axons, the strengths of anterograde and retrograde transport are relatively balanced (Fig. 3 C and E and SI Appendix, Fig. S11 A and B), suggesting that SVPs unbiasedly sample these regions, dynamically searching for binding sites. The slow diffusion fraction is significantly higher in distal axons than in dendrites (SI Appendix, Fig. S11C), consistent with selective recruitment of SVPs/SVs to boutons and presynaptic regions. Previous studies show that the AIS consists of specifically arranged nanoscale cytoskeleton architectures (29), and the AIS serves as a passive filter controlling the selective entry of axonal cargos (30). Data from our high-resolution kinetics analysis are complementary to these observations, highlighting a crucial role of the AIS in actively enhancing the axonal transport polarity by promoting the anterograde transport processivity (Fig. 3E). It is worth noting that our results independently converge with a recent study revealing that microtubule-associated protein 2 (MAP2) promotes the processivity of fast cargo transport in the AIS by selectively inhibiting a slow motor protein KIF5 (31).
Imaging Long-Lived Transcription Factor Binding Dynamics.
We next sought to apply this method to imaging an entirely different molecular phenomenon—the binding of TFs to chromatin. One highly debated area in this field is how long individual TFs dwell on chromatin. Two popular single-molecule residence time measurement methods were developed recently: one is based on time-lapse imaging (4, 32) and the other relies on motion blur by using a long acquisition time of 500 ms (1, 3). However, technical problems associated with dense labeling and photobleaching prevent these methods from reliably measuring the long-lived chromatin binding events at the single-molecule level. To address this problem, we devised an N-terminal RT stochastic labeling strategy, in which the RT sequence is placed upstream of the 3XHaloTag-TF sequence (Fig. 4A), and therefore protein overexpression is minimized.
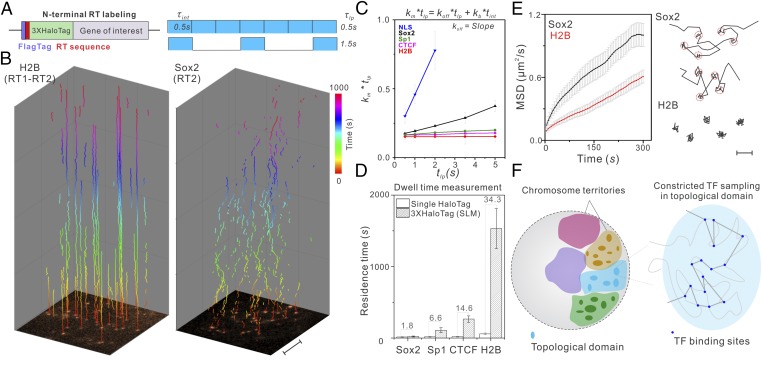
Fig. 4.
Long-time observation of TF–chromatin binding dynamics in the nucleus. (A, Left) The RT sequence is placed between a FLAG tag and a 3XHaloTag to achieve sparse labeling of transcription factors in live cells. (A, Right) Due to the low labeling density, relatively long dark times between imaging frames can be introduced to probe long-lived binding events without tracking contaminations (see SI Appendix, Fig. S12A for details). Imaging integration time () is 500 ms; lapse time (), time between exposure starts can be varied. (B) With sparse labeling, single molecules are visible from the first frame shown at the bottom and can be faithfully tracked up to 1,000 s for H2B molecules (Left) with a lapse time of 5 s. Conversely, Sox2 binding is more dynamic (Right). In each trajectory, the time component displayed in the z direction is color-coded as indicated by the color bar. Imaging was performed using the Nikon TiE microscope (HILO illumination). (Scale bar: 5 µm.) (C) By using a varied lapse-time imaging method as previously described (4), the true chromatin dissociation rate (Koff) of a TF can be calculated using the slope of the linear regression of as a function of . This calculation renders dissociation rate (Koff) independent of photobleaching rate (Kb). See SI Appendix, Eqs. S1–S8 for calculation details. Kb is estimated as according to SI Appendix, Eq. S8. Tracking data from 8–10 cells were used in this analysis. Average number of trajectories for each data point: ∼1,850. H2B, SP1, CTCF, and NLS imaging experiments were performed in U2OS cells. Sox2 experiments were performed in mouse ES cells. (D) Dwell-time measurement comparison using the traditional dense single HaloTag labeling strategy and the 3XHaloTag RT labeling strategy. The number above each bar reflects the fold of dwell-time differences. (E, Left) MSD plots show that, even when we selectively image less-mobile fractions using long lapse times (5 s) and a long acquisition time of 500 ms, Sox2 molecules are more mobile than H2B [number of trajectories: 1,692 (H2B); 1,049 (Sox2)]. Error bars in C–E represent SD. (E, Right) A close examination of Sox2 and H2B trajectories shows distinct binding behaviors. Specifically, Sox2 hops between adjacent stable binding sites (highlighted by 50-nm red circles). In contrast, H2B molecules remain relatively static at fixed locations. (Scale bar: 100 nm.) (F) Long-term tracking suggests that Sox2 molecules translocate among binding sites in spatially restricted regions inside the nucleus, supporting a sequestering mechanism of topological chromosome domains in regulating local gene activities.
After optimizing the RT sequence for each TF under the control of an EF1 promoter, we can visualize single molecules from the first frame without a high laser-power prebleaching step (Fig. 4B and SI Appendix, Fig. S12B). This is important for two reasons: (i) because of the sparse labeling, on average only one labeled molecule exists in one diffraction limited area. Therefore, long dark times (0∼4.5 s) between imaging frames can be safely introduced without the risk of tracking errors where different nearby molecules across frames are linked as one (SI Appendix, Fig. S12A); and (ii) our measurements do not rely on a single fluorophore switched back from the triplet state (dSTORM). Without high laser-power photobleaching, more than one copy of JF549 dye molecules that are not predamaged ensure long-term observation of single-molecule dynamics (Fig. 4B, SI Appendix, Figs. S12 B, D, and E and S14B, and Movie S8). When we combined time-lapse imaging (, 0.5∼5 s) to minimize photobleaching and motion blur (500 ms exposure time) to selectively capture stable binding events (Fig. 4A), we were able to extend the temporal length scale of single-molecule dwell time measurement by ∼2 orders of magnitude into the 10- to ∼20-min range (Fig. 4 B–C and D and Movie S8), which is more consistent with the FRAP measurements on the residence times of binding-dominated factors such as H2B (33). Single-molecule imaging is more suitable to dissect subpopulation associated binding dynamics for diffusion dominant TFs, and indeed average residence times for different classes of chromatin binding proteins (Sox2, 21.3 ± 7.8 s; Sp1, 109 ± 35.6 s; CTCF, 265 ± 45.7 s; and H2B, 1,532 ± 278.4 s) measured by this approach are well separated in the temporal domain (Fig. 4D), consistent with their distinct roles in gene regulation. These results are in contrast to 12.1 ± 4.5 s (Sox2), 16.5 ± 6.4 s (Sp1), 18.1 ± 3.7 s (CTCF), and 44.6 ± 12.6 s (H2B) measured by the strategy described in previous reports (3, 34).
One long-standing hypothesis in gene regulation is that topological chromosome domains might compartmentalize local gene activities by physically sequestering gene regulatory machinery. This notion was derived from extensive Hi-C (chromosome conformation capture coupled with high-throughput sequencing) experiments (35, 36) and has yet to be substantiated by live cell data. Previously, it was shown that the stable binding sites of a pluripotency factor, Sox2, are spatially clustered in living ES cells (37), suggesting the existence of high-order organization of cis-regulatory elements in the nucleus. However, due to technical problems associated with dense labeling and fast photobleaching, it is challenging to perform long-term tracking experiments to investigate whether Sox2 dynamics is altered in these binding site clusters. Here, long-time observation of slow diffusion events reveals distinctive dynamic differences between Sox2 and H2B (a chromatin marker) at the single-molecule level (Fig. 4B). Specifically, mean square displacement (MSD) plots suggest that Sox2 molecules are more mobile than H2B even when we selectively imaged slow diffusion events by motion blur and long lapse times (2, 5 s) (Fig. 4E and SI Appendix, Fig. S13A). A detailed analysis of individual trajectories revealed that Sox2 dynamically hops and interacts locally in the nucleus (Fig. 4 B and E, SI Appendix, Fig. S13, and Movies S8 and S9). In contrast, H2B molecules are generally more static, bound to fixed positions (Fig. 4 B and E and SI Appendix, Fig. S13A). However, we note that H2B diffusion dynamics that we observed likely encompass movements of whole chromosomes as well as movements of nucleosomes along the DNA or association/dissociation of H2B molecules from chromatin. Importantly, Sox2 molecules in the same field of view displayed both stable binding and hopping behaviors (SI Appendix, Fig. S13B and Movie S9), excluding the possibility that the hopping is an artifact of cell movement or imaging platform drift. These results suggest that topological structures in the nucleus kinetically facilitate Sox2 molecules to explore locally, confining target search and gene regulatory activities (see SI Appendix, Fig. S13A and Eqs. S12 and S13 for details of the confined motion analysis). Our studies thus provide evidence supporting a dynamic sequestering mechanism of topological structures in the nucleus of a live cell (Fig. 4F).
In summary, here we demonstrate a simple genetic handle to adjust protein labeling density in live cells. Pairing with bright fluorescent labels, this method enables long-term single-particle tracking of densely packed cellular organelles and proteins. We note that the C-terminal RT strategy is suitable for sparse labeling membrane proteins, of which the N-terminal tag may affect their trafficking. The C-terminal RT strategy is best combined with CRISPR/Cas9 knock-in to achieve sparse labeling from the endogenous locus, whereas the N-terminal RT strategy could achieve sparse labeling and at the same time minimize overexpression in transgenic expression conditions. The beauty of this method lies in its simplicity. Only conventional microscopy is required for long-term single-molecule imaging. However, this strategy can be readily combined with advanced imaging modalities (8), such as lattice light-sheet microscopy (38), adaptive optics microscopy (39), and the MINFLUX method (40) to study in vivo kinetics and control logics of molecular systems with extended imaging depth at higher spatiotemporal resolution.
Methods
To make the RT constructs, the RT sequences (RT1: CAATAGCAATTA, RT2: CACTAGCAATTA; RT3: CAATAGGGCTTA, RT1-GGTTCA-RT2, RT1-GGTTCA-RT3) are inserted into the desired region. For live-cell PALM experiments, we put different RT sequences between H2B and mEOS4b-HA under the control of EF1, SV40, or human PGK promoters. For SV/SVP labeling with C-terminal RT strategy, we put RT1 or RT2 between Syp and SunTag under the control of hSyn1 promoter. For TF labeling with N-terminal RT strategy, we put the RT sequences between flag or 6×flag and 3×HaloTag-TF under the control of EF1 promoter. Detailed methods are available in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank M. Radcliff, S. Moorehead, and C. Morkunas for assistance; M. G. Paez-Segala in the Looger laboratory (Janelia Research Campus, HHMI) for the mEOS4b construct; and Y. Liang and R. Turcotte in the Ji laboratory and R. Chhetri in the Keller laboratory (Janelia Research Campus, HHMI) for helping us test imaging conditions. This work is funded solely by the HHMI.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713895115/-/DCSupplemental.
References
- 1.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gebhardt JC, et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimm JB, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 7.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Lavis LD, Betzig E. Imaging live-cell dynamics and structure at the single-molecule level. Mol Cell. 2015;58:644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 9.Kamiyama D, et al. Versatile protein tagging in cells with split fluorescent protein. Nat Commun. 2016;7:11046. doi: 10.1038/ncomms11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh R, Draper W, Franklin JM, Shi Q, Liphardt J. A fluorogenic nanobody array tag for prolonged single molecule imaging in live cells. bioRxiv. 2017 doi: 10.1101/111690. [DOI] [Google Scholar]
- 12.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertram G, Innes S, Minella O, Richardson J, Stansfield I. Endless possibilities: Translation termination and stop codon recognition. Microbiology. 2001;147:255–269. doi: 10.1099/00221287-147-2-255. [DOI] [PubMed] [Google Scholar]
- 14.Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes–The impact of RNA sequence. RNA Biol. 2015;12:950–958. doi: 10.1080/15476286.2015.1068497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassan M, Rousset JP. UAG readthrough in mammalian cells: Effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol Biol. 2001;2:3. doi: 10.1186/1471-2199-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schueren F, Thoms S. Functional translational readthrough: A systems biology perspective. PLoS Genet. 2016;12:e1006196. doi: 10.1371/journal.pgen.1006196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 18.Gray EG. Axo-somatic and axo-dendritic synapses of the cerebral cortex: An electron microscope study. J Anat. 1959;93:420–433. [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer MP, Smith SJ. Evidence from in vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. J Neurosci. 2006;26:3604–3614. doi: 10.1523/JNEUROSCI.0223-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakata T, Terada S, Hirokawa N. Visualization of the dynamics of synaptic vesicle and plasma membrane proteins in living axons. J Cell Biol. 1998;140:659–674. doi: 10.1083/jcb.140.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Wilhelm BG, et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- 23.Maeder CI, San-Miguel A, Wu EY, Lu H, Shen K. In vivo neuron-wide analysis of synaptic vesicle precursor trafficking. Traffic. 2014;15:273–291. doi: 10.1111/tra.12142. [DOI] [PubMed] [Google Scholar]
- 24.Westphal V, et al. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 25.Kraszewski K, et al. Synaptic vesicle dynamics in living cultured hippocampal neurons visualized with CY3-conjugated antibodies directed against the lumenal domain of synaptotagmin. J Neurosci. 1995;15:4328–4342. doi: 10.1523/JNEUROSCI.15-06-04328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joensuu M, et al. Subdiffractional tracking of internalized molecules reveals heterogeneous motion states of synaptic vesicles. J Cell Biol. 2016;215:277–292. doi: 10.1083/jcb.201604001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monnier N, et al. Inferring transient particle transport dynamics in live cells. Nat Methods. 2015;12:838–840. doi: 10.1038/nmeth.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouzigues C, Dahan M. Transient directed motions of GABA(A) receptors in growth cones detected by a speed correlation index. Biophys J. 2007;92:654–660. doi: 10.1529/biophysj.106.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song AH, et al. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136:1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Gumy LF, et al. MAP2 defines a pre-axonal filtering zone to regulate KIF1- versus KIF5-dependent cargo transport in sensory neurons. Neuron. 2017;94:347–362.e7. doi: 10.1016/j.neuron.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 32.Normanno D, et al. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat Commun. 2015;6:7357. doi: 10.1038/ncomms8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura H, Cook PR. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swinstead EE, et al. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell. 2016;165:593–605. doi: 10.1016/j.cell.2016.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pombo A, Dillon N. Three-dimensional genome architecture: Players and mechanisms. Nat Rev Mol Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z, et al. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife. 2014;3:e04236. doi: 10.7554/eLife.04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen BC, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji N, Milkie DE, Betzig E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat Methods. 2010;7:141–147. doi: 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- 40.Balzarotti F, et al. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.