Significance
Growing evidence indicates a correlation between colorectal cancer and intestinal dysbiosis or colonization by single bacterial species such as Streptococcus gallolyticus subsp. gallolyticus (SGG), yet a causality link remains to be established. To address this point experimentally, we colonized Apc+/− Notch-inducible mice with SGG. Apc+/− is a pioneer somatic mutation driving the occurrence of polyps. We did not observe significant changes in the occurrence or development of polyps in this model. However, we observed a strong SGG colonization in Apc+/− mice at the expense of resident enterococci. We related this ecological substitution to activation of a SGG-specific bacteriocin whose activity is induced in vivo by the detergent effect of an increased concentration of secondary bile acids in relation to oncogenic situation.
Keywords: APC/Notch, S. gallolyticus, S. bovis, colorectal cancer, bacteriocin
Abstract
Colonization by Streptococcus gallolyticus subsp. gallolyticus (SGG) is strongly associated with the occurrence of colorectal cancer (CRC). However, the factors leading to its successful colonization are unknown, and whether SGG influences the oncogenic process or benefits from the tumor-prone environment to prevail remains an open question. Here, we elucidate crucial steps that explain how CRC favors SGG colonization. By using mice genetically prone to CRC, we show that SGG colonization is 1,000-fold higher in tumor-bearing mice than in normal mice. This selective advantage occurs at the expense of resident intestinal enterococci. An SGG-specific locus encoding a bacteriocin (“gallocin”) is shown to kill enterococci in vitro. Importantly, bile acids strongly enhance this bacteriocin activity in vivo, leading to greater SGG colonization. Constitutive activation of the Wnt pathway, one of the earliest signaling alterations in CRC, and the decreased expression of the bile acid apical transporter gene Slc10A2, as an effect of the Apc founding mutation, may thereby sustain intestinal colonization by SGG. We conclude that CRC-specific conditions promote SGG colonization of the gut by replacing commensal enterococci in their niche.
Colorectal cancer (CRC) is the third most common cause of cancer mortality worldwide, with 1.4 million new cases and 694,000 deaths in 2012 (World Health Organization report, 2014). CRC is primarily a genetic disease that develops over many years via series of genetic changes (i.e., somatic mutations and epigenetic modifications) known as the adenoma–carcinoma sequence. The colon is a complex ecosystem characterized by the presence of rich and diverse microbiota, and CRC development has been consistently associated with a shift in microbiota composition (i.e., dysbiosis) (1–3). Although definitive identification of a CRC-specific core dysbiosis is still pending, overrepresentation of specific bacterial species in the microbiota of patients with CRC is now established (1–7). Among these, Streptococcus gallolyticus subsp. gallolyticus (SGG), formerly known as Streptococcus bovis biotype I, was one of the first bacteria clearly associated with CRC occurrence. SGG is responsible for endocarditis and bacteremia in elderly persons. It belongs to the S. bovis/equinus complex (SBSEC), encompassing a wide group of bacteria that, thanks to molecular taxonomy tools, can now be resolved into closely related species and subspecies such as the opportunistic pathogen S. gallolyticus subsp. pasteurianus (SGP) and the dairy starter S. gallolyticus subsp. macedonicus (SGM) (8). However, only SGG has been consistently linked to underlying CRC (9).
Detection of SGG in patient blood cultures has become an indication to perform colonoscopy. Indeed, 70% of patients with SGG invasive infection and undergoing colonoscopy are diagnosed with colonic neoplasia, compared with 32% of patients free of SGG but presenting symptoms or a family history of CRC (10). SGG presence is increased approximately 10-fold in tumoral colonic tissues compared with normal colonic tissues (5, 11). Similarly, 68% of patients with CRC are seropositive for SGG-specific IgG antibodies, compared with 17% in a sex- and age- matched control population (12). It is worth mentioning that SGG presence is more common in early-stage adenomas than in later-stage carcinomas (10, 12). Importantly, a significant association of exposure to SGG antigens and CRC risk was demonstrated in a large seroepidemiological study (13).
Despite accumulating association data, the mechanisms underlying SGG enrichment in CRC remain elusive, even though it was previously suggested that SGG benefits from the tumoral environment for its growth (14). It was also postulated that sites of neoplastic lesions might facilitate passage of SGG into the blood (15), but SGG infectious endocarditis could be detected several years before CRC diagnosis (16). Moreover, it has been shown that the relative risk of developing an invasive infection by SGG in the presence of a colon carcinoma is only 3–6%, whereas 60–75% of patients with SGG endocarditis simultaneously present a malignant colonic lesion that was not previously diagnosed (17).
High concentration of fecal secondary bile acids is a recognized risk factor for CRC, with patients exhibiting an elevated level of deoxycholic acid (DCA) and an increased lithocholic acid (LCA)/DCA ratio (18, 19). Importantly, a distinctive characteristic of SGG is its unusually high resistance to bile acids (20).
In this work, we demonstrate that Notch/APC mice, which are prone to develop low-grade adenoma in the colon, constitute a suitable experimental model in which to study the mechanism underlying SGG localization and enrichment in human CRC. We first showed enhanced colonization by SGG in Notch/APC compared with nontumoral Notch mice used as controls. Next, we demonstrated that Notch/APC mice displayed an increased level of excreted secondary bile acids, a tumor-associated condition favoring SGG colonization. We found a decreased transcriptional expression of Slc10A2, encoding a major ileal bile acid transporter, in Notch/APC mice. Finally, we identified a SGG-specific bacteriocin whose activity is enhanced by tumor microenvironment to create a colonization niche in which SGG outcompetes closely related bacterial species.
Results
Presence of Intestinal Polyps in the Notch/APC Murine Model Allows Gut Colonization and Persistence of SGG UCN34.
To understand the molecular mechanisms underlying enhanced SGG colon colonization in tumor conditions, we compared the colonization efficiency of our SGG model strain, UCN34, in healthy and tumor-bearing mice. Apc+/1638N mice (henceforth referred to as APC mice) are heterozygous for a loss-of-function allele of Apc (adenomatous polyposis coli), a key negative regulator of β-catenin/Wnt pathway mutated in more than 80% of human colonic tumors. APC mice spontaneously develop intestinal polyps following loss of the second Apc allele, leading to constitutive activation of the Wnt signaling pathway. As these mice develop polyps mainly in the small intestine and not in the colon, we decided to use Notch/APC mice resulting from the crossing of APC mice with previously described Vil-Cre/Nic mice (21), henceforth referred to as Notch mice. In addition to the constitutive Apc+/1638N mutation, Notch/APC mice contain a second genetic modification allowing tamoxifen-inducible activation of the Notch receptor in intestinal epithelial cells (21). Notch activation in Notch/APC mice leads to a decrease in the latency period for the development of low-grade adenomas compared with APC mice and, more importantly, to the appearance of large numbers of dysplastic lesions in the colon (21). As Notch mice do not develop any intestinal polyps following tamoxifen injection, the development of colonic polyps in Notch/APC mice results from the synergy between Notch and Wnt pathways in intestinal epithelium (21).
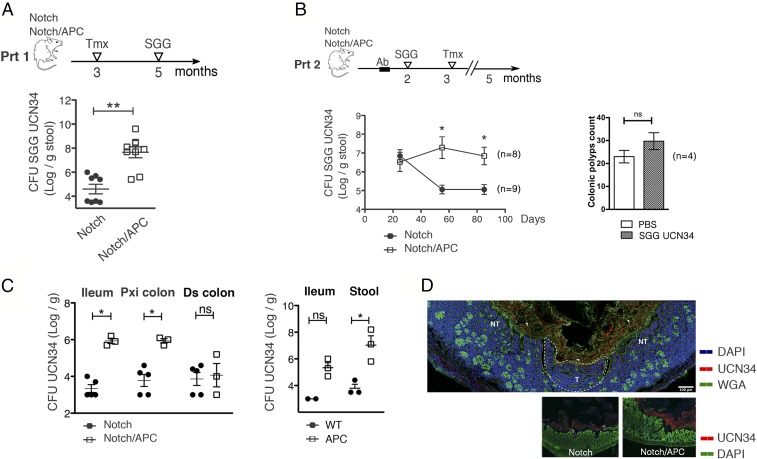
Two months after Notch receptor activation in Notch/APC mice, as many as 30 colonic polyps per mouse were detected, mostly located in the proximal colon. We then monitored the capacity of SGG strain UCN34 to colonize the mouse gut 7 d after oral gavage by plating the stools on selective agar plates (protocol 1). By using tamoxifen-injected Notch mice as control littermates, we found that the UCN34 colonization capacity was 1,000 fold higher in Notch/APC tumor-bearing mice compared with healthy control Notch mice (Fig. 1A). It is important to note that, in tamoxifen-induced Notch/APC mice, SGG was able to colonize the intestinal tract without any antibiotic treatment.
Fig. 1.
Presence of colonic polyps enhances gut colonization by SGG UCN34. (A) cfu counts of SGG UCN34 in the stool of tamoxifen (Tmx)-injected Notch and Notch/APC mice. SGG was inoculated by oral gavage 2 mo following tamoxifen injection, and cfu counts were enumerated 7 d later (Prt 1). (B) cfu counts of SGG UCN34 in the stool of Notch and Notch/APC mice at different time points following its oral inoculation. SGG was administered 1 mo before tamoxifen injection following a 1-wk antibiotic (Ab) treatment. Macroscopic adenoma count in the colon of SGG colonized vs. PBS solution-treated Notch/APC mice at the time of euthanasia, 2 mo following tamoxifen injection (mean ± SEM; **P ≤ 0.01 and *P ≤ 0.05, t test). (C) Mean ± SEM cfu counts of SGG UCN34 in the ileum and proximal (Pxi) and distal (Ds) colon of Notch, Notch/APC, WT, and APC mice 3 mo after oral inoculation (Prt 2; *P ≤ 0.05, t test). (D) Immunofluorescence staining of SGG UCN34 in Notch/APC colonic tissues 3 mo after its inoculation (Prt 2) shows tumoral (marked as “T”) and adjacent nontumoral (NT) areas. Bacteria (in red) were stained with a specific rabbit anti-UCN34 polyclonal antibody and anti-rabbit IgG coupled to Alexa Fluor 568, respectively. Mucus was visualized with wheat germ agglutinin (WGA) lectin coupled to Alexa Fluor 488 (green), and nuclei were labeled with DAPI (cyan blue). ns, not significant. (Scale bar, 100 µm.)
We then aimed at determining the impact of intestinal tumors on the persistence of gut colonization by SGG in the Notch/APC model. However, tumor-bearing Notch/APC mice exhibited a limited life expectancy, with almost all mice dying within 3 mo following Notch receptor activation. Thus, the experimental setup was slightly modified to introduce SGG UCN34 in the mouse 1 mo before Notch activation as previously described in C57BL/6 WT mice (22). To reach detectable colonization in these pretumoral conditions, mice were pretreated with antibiotic agents for 1 wk, allowing the study of SGG persistence over several months (protocol 2). In this setting, SGG UCN34 persisted at a constant level for more than 3 mo in tumor-bearing Notch/APC mice, whereas SGG carriage decreased significantly in control Notch mice 2 mo after colonization (Fig. 1B). Macroscopic inspection of adenomas at the time of animal euthanasia did not reveal any increase (in number and/or size) in tumor formation promoted by SGG UCN34 (Fig. 1B). A preferential colonization by SGG UCN34 was found in the ileum and proximal colon of Notch/APC mice, but not in the distal colon (Fig. 1C).
To evaluate the contribution of Apc+/1638N mutation alone in SGG persistence, the same experiment (i.e., protocol 2) was performed in APC mice vs. WT control mice. Importantly, SGG colonization capacity was found to be 1,000 fold higher in APC mice compared with WT control in ileum and stools (Fig. 1C). Thus, Apc+/1638N mutation is sufficient to promote SGG intestinal colonization.
Next, immunohistochemical analyses were carried out by using a specific polyclonal antibody raised against SGG UCN34 to visualize bacterial adhesion on intestinal tissues. SGG UCN34 was found entrapped in the mucus layer overlaying the epithelium in Notch/APC mice, whereas SGG was barely detectable in control Notch mice (Fig. 1D). A similar localization was reported in WT C57BL/6 mice, with even lower numbers of SGG UCN34 (22). Of note, we did not observe any preferential attachment of SGG to the tumor sites, and bacterial density appeared similar in the lumen regardless of the presence of a tumor in underlying mucosal tissue (Fig. 1D).
Colonization and Persistence of SGG in Tumor-Bearing Mice Is Mediated by a Specific Bacteriocin That Overrides Commensal Gut Enterococci.
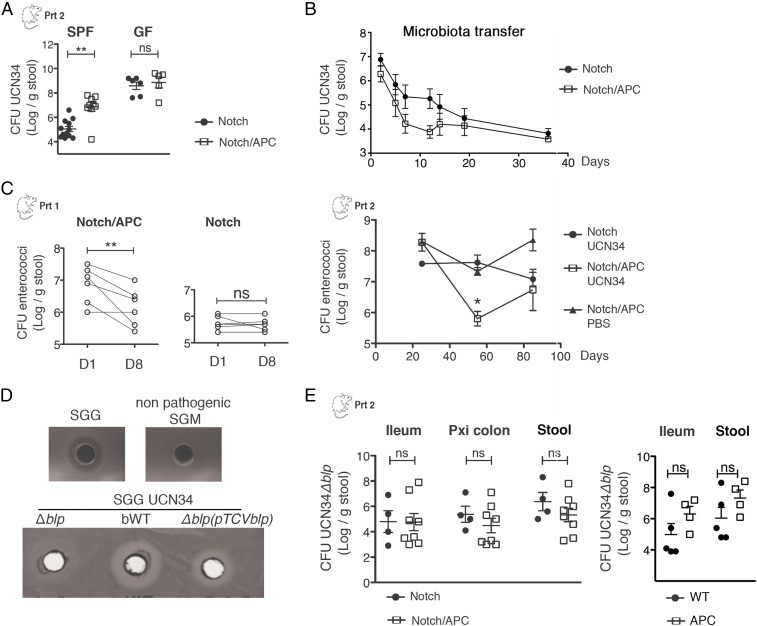
To test whether the tumor alone was sufficient to mediate enhanced colonization by SGG, persistence of UCN34 was quantified in specific pathogen-free (SPF) and germfree (GF) Notch and Notch/APC mice. Although GF Notch/APC mice developed fewer tumors than their SPF counterparts, these mice still harbored tens of colonic adenomas following tamoxifen injection. We confirmed that UCN34 more efficiently colonized SPF Notch/APC mice compared with SPF Notch mice. However, in the absence of an endogenous gut microbiota, no difference could be seen in the amounts of SGG UCN34 recovered from the stools of tumor-bearing and healthy mice (Fig. 2A). Moreover, fecal microbiota transfer from Notch/APC mice to GF WT mice did not enhance SGG UCN34 colonization compared with the transfer of control Notch mice microbiota (Fig. 2B), indicating that it is not tumor-induced microbiota dysbiosis per se that influences SGG colonization.
Fig. 2.
Involvement of a specific bacteriocin in the persistence of SGG in tumor-bearing mice. (A) Mean ± SEM cfu counts of SGG UCN34 in the stool of SPF and GF Notch and Notch/APC mice 3 mo after SGG oral inoculation (**P ≤ 0.01, t test). (B) cfu counts of SGG UCN34 in the stool of GF mice colonized with microbiota from Notch/APC or Notch mice (n = 5). (C) cfu counts of enterococci in the stool of Notch and Notch/APC mice at different time points following SGG UCN34 oral inoculation (D0). Enterococci cfu counts in the stool of Notch/APC mice receiving PBS solution instead of SGG are also depicted (mean ± SEM; **P ≤ 0.01, paired t test). (D) Agar diffusion assay showing the capacity of SGM and different strains of SGG [UCN34, Δblp, bWT, Δblp(pTCVblp)] to inhibit the growth of commensal E. faecalis. Δblp, bacteriocin-deficient mutant; Δblp(pTCVblp), complemented Δblp mutant; bWT, back to WT strain. (E) Mean ± SEM cfu counts of the Δblp mutant in the ileum, proximal colon, and stools of Notch, Notch/APC, WT, and APC mice 3 mo after its oral inoculation. ns, not significant.
To monitor SGG colonization in SPF mice, we used selective entero agar plates (also called Slanetz and Bartley) to discriminate between SGG (pink) and enterococci (purple), the latter being a prominent member of murine microbiota (Fig. S1A). Interestingly, the increase of SGG UCN34 in Notch/APC mice was concomitant with a decrease in the fecal carriage of commensal enterococci in both experimental settings (protocols 1 and 2; Fig. 1). Indeed, total numbers of enterococci significantly decreased after SGG inoculation in tumor-bearing Notch/APC mice compared with control Notch mice (Fig. 2C). This result suggests that SGG UCN34 is able to outcompete commensal enterococci in tumor-bearing mice.
To test whether SGG UCN34 could inhibit the growth of commensal enterococci in vitro, killing assays were performed by using a simple agar spot test. Briefly, the “target” bacteria, Enterococcus faecalis, were flooded on the surface of agar plates and the “predator” bacteria, SGG UCN34, were spotted in holes punched in agar. After growth of target bacteria at 37 °C, a clear zone of inhibition became visible around SGG spots, suggesting the production of a diffusible antibacterial molecule by SGG UCN34 (Fig. 2D). Interestingly, the majority of the SGG clinical isolates from our collection were similarly able to inhibit the growth of E. faecalis, whereas the colonizing isolates identified as SGP or the nonpathogenic SGM lacked this activity (Fig. S1B). The inhibitory activity of SGG UCN34 was demonstrated on six different E. faecalis strains, including murine autochthonous and human isolates (Fig. S1C). Interestingly, only one isolate of Enterococcus faecium was found to be sensitive to this bacteriocin, whereas Enterococcus hirae isolate was found resistant (Fig. S1C). In silico search for genes encoding annotated bacteriocins in the genome of SGG UCN34 provided three hits: gallo_2020, gallo_2021, and gallo_2203. BLAST searches revealed that gallo_2203 was also present in the genome of the nonpathogenic SGM strain ACA-DC 198, whereas gallo_2020 and gallo_2021 were specific and highly conserved in many SGG strains. In silico analysis revealed that gallo_2021 and gallo_2020 encode small hydrophobic proteins of 83 and 65 aa, respectively, that belong to the class II bacteriocins (Pfam 10439) showing a characteristic double-glycine leader peptide. These bacteriocins are usually constituted of two genes encoding short peptides, named alpha and beta, that fold into α-helical structures and insert into target bacterial membranes to alter their permeability. First, we verified at DNA and RNA levels that gallo_2021 and gallo_2020 were present and transcribed in SGG UCN34 but not in our nonpathogenic SGM strain CIP105683T (Fig. S2). Transcription of gallo_2020 correlated with inhibitory activity of SGG bacteriocin against E. faecalis (Fig. S2). To test the role of these genes in the inhibitory effect of SGG against enterococci isolates, we constructed a targeted deletion mutant of both gallo_2021 and gallo_2020, named ∆blp, in strain UCN34 (Fig. S2). This mutant appeared unable to inhibit the growth of E. faecalis strains in vitro by using the agar spot assay, and this activity was restored by complementation with the deleted genes (Fig. 2D). These results demonstrated that UCN34 gallo_2021 and gallo_2020—which we propose to rename blpA and blpB (for bacteriocin-like peptide), respectively—encode bacteriocins active against E. faecalis (Fig. S2). In addition to E. faecalis, a number of closely related Gram-positive bacteria such as SGM or Streptococcus thermophilus were found susceptible to this bacteriocin, which we propose to name gallocin, whereas no effect was detected on Gram-negative bacteria such as Escherichia coli or Bacteroides thetaiotaomicron (Fig. S1C).
Next, we tested the ability of this mutant UCN34∆blp to colonize the gastrointestinal tract of Notch/APC vs. Notch mice. Colonization levels of UCN34∆blp in the ileum, proximal colon, or stool were similar in tumor-bearing Notch/APC mice compared with control Notch mice (Fig. 2E). Similarly, the 3-log increase of SGG UCN34 colonization in APC vs. WT mice was reduced to a 1-log increase by using only the isogenic UCN34∆blp mutant (Fig. 2E). These results indicate that the specific bacteriocin activity encoded in the blpAB region is critical for the enhanced SGG colonization in the genetic context predisposing to CRC (Apc mutation).
SGG-Specific Bacteriocin Activity Is Enhanced by Gut Bile Acids.
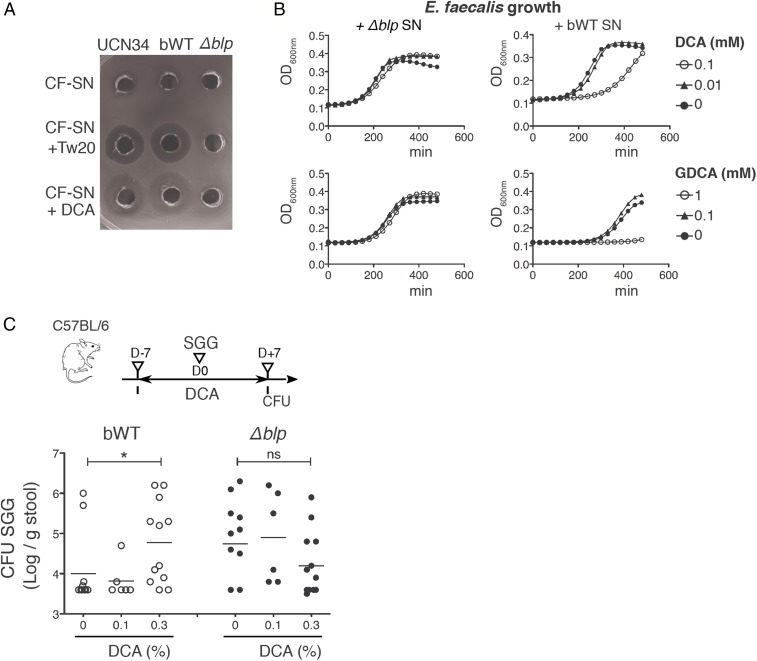
Class II bacteriocins consist of two different peptides that need to fold into α-helical structures to acquire the capacity to insert into target membranes and exert their antimicrobial activity. Helical structuring is favored when the peptides are exposed to a more hydrophobic or membrane-like environment, i.e., solutions containing micelles or liposomes (23). Consistently, addition of detergents at greater than micelle concentrations was required to reveal SGG antimicrobial activity targeting enterococci in the culture supernatant as previously described (24). By using our spot agar diffusion assay, we found that cell-free sterile-filtered supernatant of strain UCN34 was poorly inhibiting enterococci growth unless a subinhibitory dose of Tween 20 (1%) was added (Fig. 3A). This effect was not specific to Tween 20 and could be obtained by using other detergents such as Tween 60, Nonidet P-40, Triton X-100, or SDS (Fig. S1D). Bile acids, which are natural detergents found in the host intestine that solubilize diet fats, may account for enhancement of the gallocin activity in vivo. Primary bile acids are synthesized in the liver from cholesterol. They are secreted in the gut lumen in a conjugated form before being transformed by the intestinal microbiota into unconjugated and secondary bile acids. As gallocin activity was preferentially expressed in tumor conditions, we hypothesized that microbiota-dependent secondary bile acids such as DCA and LCA, the levels of which have been linked to colon cancer development in animal models and in humans, could explain the prevalence of SGG in Notch/APC mice but not in control Notch mice.
Fig. 3.
Bile acid deoxycholate enhances SGG bacteriocin activity in vitro and in vivo. (A) Inhibitory potential of cell-free (CF) sterile-filtered supernatant (SN) of different SGG strains (UCN34, bWT, Δblp mutant) against E. faecalis growth. Supernatants from overnight cultures were used alone or in combination with Tween 20 (Tw20) or DCA. (B) Growth curves of E. faecalis in TH broth containing 50% of SGG UCN34 sterile filtered supernatant (SN) from the bWT or Δblp mutant strain and supplemented with different concentrations of DCA or GDCA. (C) Mean ± SEM cfu counts of SGG UCN34 or the Δblp mutant in the stools 7 d after oral inoculation in C57BL/6 mice treated with different doses of DCA for 14 d (0.1% or 0.3%, wt/wt ratio; *P ≤ 0.05, t test). ns, not significant.
By using our spot agar diffusion assay, we first showed that DCA used at 10 mM (∼0.4%) was able to replace Tween 20 (1%) to detect gallocin activity (Fig. 3A). Similar effect was observed with all bile acids that were tested: LCA, chenodeoxycholic acid (CDCA), cholic acid (CA), taurodeoxycholic acid, and glycodeoxycholic acid (GDCA). To compare the contribution of the various bile acids, we set up a quantitative test whereby E. faecalis growth in broth was monitored in the presence of UCN34 sterile-filtered supernatant supplemented with various concentrations of bile acids. As negative control, we used the sterile-filtered supernatant of our isogenic UCN34∆blp mutant. In this assay, we noticed that DCA was 10 times more efficient at synergizing with gallocin than its conjugated form GDCA (Fig. 3B). Moreover, LCA, another secondary bile acid known for its toxicity, appeared 10 times more efficient than DCA.
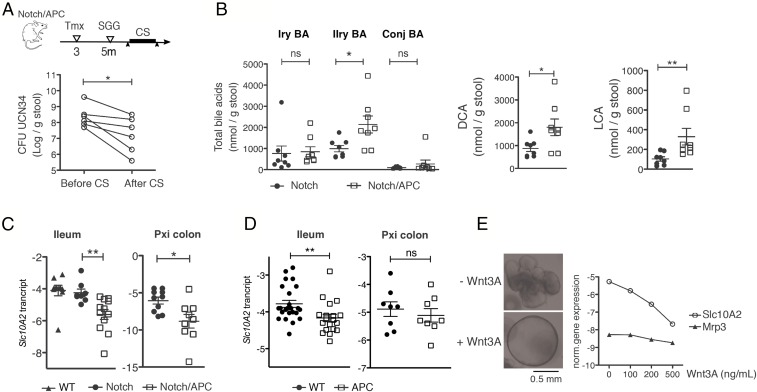
We next tested whether an increase in secondary bile acids in our murine model could facilitate SGG UCN34 colonization by enhancing gallocin activity. UCN34 colonization efficiency was assessed in C57BL/6J WT mice fed two different doses (0.1% and 0.3%) of DCA for 1 wk. This short-term DCA treatment did not induce significant weight loss in mice. DCA administration at the highest dose enhanced gut colonization level by SGG UCN34. In contrast, intestinal colonization of the ∆blp mutant was not significantly different among the different groups of mice (Fig. 3C). To further evaluate the role of bile acids in the colonization capacity of SGG UCN34 in tumor-bearing mice, we used cholestyramine (CS), a bile acid chelator, to reduce the availability of bile acids in the gut. Notch/APC mice colonized with SGG UCN34 received a 1-wk treatment with 2% CS, and stools were plated before and after treatment. Comparison of SGG colonization levels, although variable from mouse to mouse, showed a consistent reduction in colonization efficiency, confirming that bile acids are playing an important role in SGG UCN34 colonization in Notch/APC mice (Fig. 4A).
Fig. 4.
Increased excretion of secondary bile acids in Notch/APC mice. (A) Mean ± SEM cfu counts of SGG UCN34 in the stool of Notch/APC mice 1 d before the beginning and 1 d after the end of a 10-d CS oral treatment (*P ≤ 0.05, paired t test). (B) Mean ± SEM fecal concentration of primary (Iry), secondary (IIry), and conjugated (Conj) bile acids (BA), DCA, and LCA in Notch and Notch/APC mice 2 mo after Notch receptor activation (**P ≤ 0.01, t test). (C) Slc10A2 gene relative expression level in the ileum and proximal colon of WT, Notch, and Notch/APC mice 2 mo after Notch receptor activation (mean ± SEM; **P ≤ 0.01, t test). (D) Slc10A2 gene expression level in the ileum and proximal colon of 3-mo-old WT and APC mice. (E) Slc10A2 and Mrp3 gene expression level in small intestinal organoids produced from C57BL/6 mice and cultivated for 20 h in the presence of different concentrations of Wnt3A (0, 100, 200, and 500 ng/mL). ns, not significant.
Increased Bile Acid Excretion in Notch/APC Mice Is Associated with Decreased Slc10A2 Bile Acid Transporter Gene Expression.
To characterize tumor effects on intestinal bile acid profiles in the Notch/APC model, selection of four primary (α-muricholic acid, β-muricholic acid, cholic acid, CDCA), three secondary (DCA, LCA, ursodeoxycholic acid), and 11 conjugated bile acids were quantified by MS in the stools of tumor-bearing Notch/APC and control Notch mice (Fig. 4B). Although we did not observe any significant difference in the quantity of primary and conjugated bile acids, we found a significant increase in secondary bile acids (Fig. 4B). DCA and LCA were both significantly enriched in the stools of tumor-bearing Notch/APC mice compared with control Notch mice (Fig. 4B).
Intestinal epithelial cells play a major role in regulating bile acid levels in the gut lumen. Indeed, the majority of ileal bile acids are recycled back to the liver following active reabsorption across the epithelium by apical and basolateral transporters. We thus hypothesized that epithelial signaling alteration in CRC-predisposed Notch/APC mice may disrupt bile acid reabsorption, resulting in their increased excretion. We compared expression levels of a panel of genes involved in epithelial bile acid transport in Notch/APC and Notch mice. Whereas we did not observe any difference in the expression level of genes encoding basolateral transporters (Osta, Ostb) and intracellular bile acid sensors (Fabp6, Fgf15, Fxr; Fig. S3), we could demonstrate a significant down-regulation in expression of the Slc10A2 gene, encoding an apical bile acid transporter in Notch/APC mice compared with Notch mice, in the ileum and proximal colon (Fig. 4C). In contrast, the expression levels of genes encoding bile acid transport (i.e., Ntcp, Oatp1, Bsep, Mrp3), synthesis (i.e., Cyp7a1, Cyp8b1, Cyp27a1, Akr1d1), intracellular sensing (i.e., Shp, Fxr), and conjugation (i.e., Baat, Bacs, Hnf4a1, Taut, Csd) in the liver were similar between Notch and Notch/APC mice (Fig. S4).
Furthermore, we found that, in APC mice, Slc10A2 was significantly down-regulated in the ileum, but not in the proximal colon, compared with WT mice, even before the appearance of visible polyps (age 2 mo; Fig. 4D). These results indicate that Apc mutation, which is an early event in CRC development, leads to the transcriptional down-regulation of Slc10A2 expression in the intestinal epithelium.
To decipher the mechanisms underlying Slc10a2 down-regulation in APC mice, we used the organoid model of primary intestinal epithelial cell culture, which constitutes a pure epithelial system recapitulating all cell types, including stem cells, proliferating progenitors, and differentiated cells from different lineages (i.e., goblet cell, enterocytes, enteroendocrine cells, Paneth cell, stem cells) (25). We produced small intestinal organoids from WT C57BL/6 mice and exposed them to increasing doses of Wnt3A for 20 h to mimic constitutive activation of the Wnt pathway that supports polyp development in Apc+/− mice. Strong induction of Wnt signaling in small intestinal organoids is known to impair epithelial differentiation (26) and to promote the formation of cystic structures, reminiscent of tumoral organoids (Fig. 4E) (27). In this context, we found that Wnt activation resulted in Slc10A2 gene expression down-regulation, whereas the expression of Mrp3, encoding another epithelial membrane transporter, was unchanged (Fig. 4E). Bile acid malabsorption and subsequent SGG colonization may thus constitute very early biomarkers of CRC development.
Discussion
SGG colonization of the gut is strongly associated with the presence of colonic neoplasia. Whether SGG plays an etiological role precipitating the occurrence of polyps and tumors or acquires a strong colonization property in response to a favorable ecological environment created by the tumor itself or preexisting conditions remains unknown. It is worth mentioning that, even though asymptomatic carriage of SGG is estimated to be between 5% and 15% in the literature, mostly using enrichment on selective media and culture techniques, a recent study indicated a much higher carriage rate (∼60%) by using quantitative real-time PCR (28).
Sporadic CRC remains a genetic disease that results from the occurrence of successive somatic mutations, Apc being one of the first, driving precancerous polyp development in humans. It is found in more than 80% of human sporadic colon cancers and is also responsible for the familial adenomatous polyposis (FAP) syndrome, one of the main forms of hereditary colon cancer. Apc loss leads to the constitutive activation of Wnt pathway, one of the earliest signaling alterations in CRC, which in turn induces cell proliferation and rapid loss of epithelial differentiation (29) that can be recapitulated in intestinal organoids (26). Of note, Apc mutation in APCmin/+ mice is associated with intestinal dysbiosis even before the appearance of any visible polyps (30).
We first demonstrated greater SGG gut colonization in mice bearing the CRC-predisposing loss-of-function mutation in the Apc gene. Although Apc mutation alone is sufficient to lead to few polyps in the small intestine, development of an increased number of adenomas preferentially located in the proximal colon requires the synergistic activation of Wnt and Notch pathways in intestinal epithelium as shown previously in Notch/APC mice (27). By using this double transgenic model, we confirmed significantly greater colonization of SGG UCN34 in tumor-bearing mice compared with control Notch mice. Strikingly, SGG UCN34 was able to colonize the intestinal gut of Notch/APC mice at a high level without any antibiotic treatment (Fig. 1A). Histological analyses showed greater colonization of ileum and proximal colon on the entire mucosa and not preferentially at the tumor sites. As shown previously (22), SGG UCN34 was found entrapped in the mucus layer (Fig. 1D). Although we did not observe any obvious difference in the thickness of the mucus layer between tumor-bearing Notch/APC and control Notch mice, we have not compared the mucus composition between these two genotypes and thus cannot rule out the possibility that minor differences in mucin composition may also contribute to differential colonization. Next, we found that SGG-enhanced colonization in tumor conditions was occurring at the expense of another closely related bacterium, E. faecalis, present in the murine gut microbiota. Further studies led to the identification of a bacteriocin, here named gallocin, specifically produced and secreted by SGG, whose activity is strongly enhanced in CRC-associated conditions that kill E. faecalis, one of the first bacterial colonizers of the naïve mammalian gut microbiota, allowing successful SGG colonization in the murine intestinal tract.
Gallocin is encoded by two genes, blpA (gallo_2021) and blpB (gallo_2020), which are absent from closely related bacteria belonging to the SBSEC. Gallocin is a class II bacteriocin, and members of this family are widespread in lactic acid bacteria, including streptococci. These molecules are usually directed against closely related bacteria competing within the same environment. The genetic locus encoding gallocin in SGG UCN34 is complex and shares many similarities with the blp genomic region of Streptococcus pneumoniae encoding several ORFs encoding small bacteriocin peptides, interspersed with immunity genes and membrane transporters (Fig. S2). BlpI and BlpJ are the closest orthologs of BlpA and BlpB, but their expression and role in S. pneumoniae have not been studied to our knowledge. Only pneumococcal blpMN-encoded bacteriocin has been shown to be involved in intraspecies competition and important for nasopharynx colonization (31).
Importantly, we found that gallocin activity was strongly enhanced in a bile acid-enriched environment, a broadly recognized risk factor for CRC. Patients with CRC show elevated DCA level and LCA/DCA ratio in their stools (18, 19). Furthermore, chronic long-term administration of DCA in animal models promotes cancer development (32, 33). DCA has particularly been shown to promote CRC development in Apc(min/+) mice by enhancing Wnt signaling (32). Bile acids’ positive effect on gallocin activity is likely a result of their detergent-like properties, as shown in vitro by using various nonionic and ionic detergents (Fig. S1). In addition, detergents are known activators of these bacteriocins, as they provide a membrane-like scaffold allowing proper α-helical folding of the two interacting hydrophobic peptides (23, 34–36).
Elevated levels of intestinal bile acids, which means increased risk of CRC, can result from environmental factors such as high-fat diet (37) and/or inherited host genetic factors. Increased fecal bile acid has been detected in patients with FAP in whom multiple adenomas develop (38). Polymorphisms in the SLC10A2 gene, encoding an apical bile acid transporter, have been linked to CRC risk (39). This is consistent with the observation that inactivation of this gene in mice enhanced CRC development (40). Here, we identified a link to one of the earliest signaling alterations in CRC development, i.e., Wnt pathway activation and elevated levels of bile acids. We showed that constitutive activation of the Wnt pathway in intestinal epithelial cells resulted in down-regulation of the expression of Slc10A2 gene encoding a major apical bile acid transporter.
In conclusion, Wnt pathway activation and increased secondary bile acids are both part of a vicious circle supporting CRC development (26). This study shows that this loop creates an optimal environment for SGG colonization. Overall, our results demonstrate that the tumor environment allows SGG to prevail and colonize the host intestinal tract. In that sense, SGG is not a bona fide pathogen that is intrinsically able to cause disease in a healthy host, but rather an opportunist pathobiont benefiting from a favorable ecological niche offered by the intestinal oncogenic environment, which eventually promotes its translocation and systemic dissemination. However, a very recent study demonstrated that SGG strain TX2005 can promote colorectal tumor development through increase of epithelial cell proliferation (41). Finally, it is worth mentioning that combined detection of increased levels of luminal bile acids and of SGG may constitute useful diagnostic tools for the detection of CRC at early stages.
Materials and Methods
Mouse Experiments.
All animal experiments were carried out under approval by the Use Committee of Institut Pasteur and by the French Ministry of Agriculture (committee protocol no. 2013–0030). Notch/APC mice have been previously described (21). They were obtained from Sylvie Robine (Institut Curie, Paris, France) and bred under specific opportunistic pathogen-free conditions at Institut Pasteur. GF Notch and Notch/APC mice were produced at Institut Pasteur facility by sterile C-section and kept in sterile isolators. Six-week-old C57BL/6J RJ mice were bought from Janvier Labs.
UCN34 Colonization Experiments in Notch/APC Mice.
Notch and Notch/APC mice were genotyped and put in separate cages right after weaning (age 4 wk). Three-month-old Notch and Notch/APC mice received i.p. injection of tamoxifen (50 μg/g animal body weight; MP Biomedicals) every day for five consecutive days. For SGG UCN34 colonization experiments, bacteria were grown overnight in Todd–Hewitt (TH) broth at 37 °C and subcultured in fresh medium until exponential phase (OD600 0.4–0.6). Bacteria were then washed once in PBS solution, and each mouse received 1010 bacteria in 200 μL PBS solution by oral gavage using a straight feeding cannula (Bioseb N-020). For long-term persistence experiments, 2-mo-old Notch/APC mice were first treated with a broad-spectrum antibiotic mixture including vancomycin (50 μg/g), neomycin (100 μg/g), metronidazole (100 μg/g), amphotericin B (1 μg/g), and ampicillin (1 g/L) for 8 d as previously described (42). Antibiotic-treated mice were then fed SGG for three consecutive days. Bacteria colonization was determined by cfu counts. Briefly freshly collected stools were weighed and homogenized by using a Precellys homogenizer (Bertine) for 2 × 15 s at a frequency of 5,000 rpm. Following serial dilutions, samples were plated on Enterococcus agar-selective media (BD Difco) for counting of SGG colonies, exhibiting a specific pink color on these plates as described previously (5).
Microbiota Transfer Experiment.
Stool from Notch and Notch/APC mice was freshly collected, homogenized in PBS solution (100 mg/mL), and passed through a 100-μm filter. GF mice were fed 300 μL of the stool suspension once. Two days later, recipient mice were fed 1010 cfu of S. gallolyticus strain UCN34 (WT).
DNA Extraction from Intestinal Content and Microbiota Composition Analysis.
Intestinal contents were collected from each group of mice and immediately stored at −80 °C. Total DNA was extracted from samples by using the PowerFecal DNA isolation kit (MoBio 12830). PCR inhibition was tested with DNA dilutions by using the TaqMan exogenous internal positive control (Applied Biosytems). Conditions for quantitative PCR amplifications were as follows: one cycle at 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 40 cycles at 60 °C for 1 min. The ABI PRISM 7000 sequence detection system and 7000 system software (version 1.2.3; Applied Biosytems) were used. Total bacteria were detected by using the TaqMan universal PCR system. E. coli, Enterococcus, and Lactobacillus/Leuconostoc were detected by using the SYBR Green PCR system. Probes and primers used are shown in Table 1. Standard curves generated from 10-fold serial dilutions of DNA samples of specific strains were used for quantification.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
| UCN34∆blp mutant | |
| delta-5′_EcoRI | GGAATTCTCTCAAATGAGCCAG |
| delta-5′ | TGCAGCTTCATCTGTCATTACATCA |
| delta- 3′ | GATGAAGCTGCAATGATCTGTGTGG |
| delta 3′_BamHI | GGATCCCGTCAGAAAACTAGTGTTAGCTTC |
| For complementation | |
| Blp forward-BamH1 | CGGATCCATCTGTCAGTTGCAACGAC |
| Blp reverse-Sph1 | CGCATGCGCCATAAAAATATGAATCTG |
| Gene expression (quantitative RT- PCR) in mice | |
| Akr1d1 forward | TGCACACCACCAAATATCCCT |
| Akr1d1 reverse | CTTCACTGCCACATAGGTCTTC |
| Baat forward | GTCCTCCCTTGGATAGCCTGA |
| Baat reverse | CCGGATGCGGCTTTCCTTTA |
| Bacs forward | ACCCTGGATCAGCTCCTGGAT |
| Bacs reverse | GTTCTCAGCTAGCAGCTTGG |
| Bsep forward | CTGCCAAGGATGCTAATGCA |
| Bsep reverse | CGATGGCTACCCTTTGCTTCT |
| Csd forward | CCAGGACGTGTTTGGGATTGT |
| Csd reverse | ACCAGTCTTGACACTGTAGTGA |
| Cyp27a1 forward | GCCTCACCTATGGGATCTTCA |
| Cyp27a1 reverse | TCAAAGCCTGACGCAGATG |
| Cyp7a1 forward | GGGATTGCTGTGGTAGTGAGC |
| Cyp7a1 reverse | GGTATGGAATCAACCCGTTGTC |
| Cyp8b1 forward | GGCTGGCTTCCTGAGCTTATT |
| Cyp8b1 reverse | ACTTCCTGAACAGCTCATCGG |
| Fabp6 forward | CTTCCAGGAGACGTGATTGAAA |
| Fabp6 reverse | CCTCCGAAGTCTGGTGATAGTTG |
| Fgf15 forward | ACGTCCTTGATGGCAATCG |
| Fgf15 reverse | GAGGACCAAAACGAACGAAATT |
| Fxr forward | GCTTGATGTGCTACAAAAGCTG |
| Fxr reverse | CGTGGTGATGGTTGAATGTCC |
| Gapdh forward | TGTGCAGTGCCAGCCTC |
| Gapdh reverse | ATGAAGGGGTCGTTGATGGC |
| Hnf4a1 forward | AAATGTGCAGGTGTTGACCA |
| Hnf4a1 reverse | CACGCTCCTCCTGAAGAATC |
| Mrp3 forward | TCCCACTTTTCGGAGACAGTAAC |
| Mrp3 reverse | ACTGAGGACCTTGAAGTCTTGGA |
| Ntcp forward | ATGACCACCTGCTCCAGCTT |
| Ntcp reverse | GCCTTTGTAGGGCACCTTGT |
| Oatp1 forward | GTGCATACCTAGCCAAATCACT |
| Oatp1 reverse | CCAGGCCCATAACCACACATC |
| Osta forward | AGGCAGGACTCATATCAAACTTG |
| Osta reverse | TGAGGGCTATGTCCACTGGG |
| Ostb forward | AGATGCGGCTCCTTGGAATTA |
| Ostb reverse | TGGCTGCTTCTTTCGATTTCTG |
| Shp forward | CGATCCTCTTCAACCCAGATG |
| Shp reverse | AGGGCTCCAAGACTTCACACA |
| Slc10a2 forward | TGGGTTTCTTCCTGGCTAGACT |
| Slc10a2 reverse | TGTTCTGCATTCCAGTTTCCAA |
| Taut forward | GCACACGGCCTGAAGATGA |
| Taut reverse | ATTTTTGTAGCAGAGGTACGGG |
| Primers and probes used for bacteria quantification in mouse ileal content | |
| All bacteria | CGG TGA ATA CGT TCC CGG |
| TAC GGC TAC CTT GTT ACG ACT T | |
| 6FAM-CTT GTA CAC ACC GCC CGT C | |
| Lactobacillus/Leuconostoc | AGC AGT AGG GAA TCT TCC A |
| CGC CAC TGG TGT TCY TCC ATA TA | |
| E. coli | CAT GCC GCG TGT ATG AAG AA |
| CGG GTA ACG TCA ATG AGC AAA | |
| Enterococcus | CCC TTA TTG TTA GTT GCC ATC ATT |
| ACT CGT TGT ACT TCC CAT TGT | |
Construction of a Bacteriocin-Deficient Mutant UCN34 ∆blp.
SGG strains were grown at 37 °C in TH broth in standing filled flasks. The UCN34∆blp-deficient mutant was constructed as previously described (43) by using splicing-by-overlap extension PCR. The resulting recombinant vector pG1-∆blp was introduced directly in SGG UCN34 by electroporation. The deleted region covers the two ORFs encoding the bacteriocin (gallo_2021–gallo_2020). The deletion mutant was identified by PCR and confirmed by sequence analysis on chromosomal DNA. The primers used are listed in Table 1. We used the back-to-WT strain as a control in all experiments.
Mouse Treatment with Deoxycholate and CS.
Eight-week-old C57BL/6 mice were given a deoxycholate-enriched diet. Sodium deoxycholate (0.1% or 0.3% weight ratio; Sigma) was combined with minced chow and left for free access in the cage. Fresh deoxycholate was added daily for 12 consecutive days. At day 5, UCN34∆blp mutant or the parental UCN34 strain were inoculated by oral gavage. Colonization efficiency was monitored 7 d after inoculation by using an Enterococcus-selective agar plate. For the CS experiment, tamoxifen-injected 5-mo-old Notch/APC mice were colonized with SGG UCN34. Ten days later, 2% (weight ratio) CS (Sigma) was combined daily with minced chow and left freely for mice to eat for 7 d before monitoring of UCN34 colonization level on selective agar plates.
Immunohistochemistry for SGG UCN34 Detection in Intestinal Lumen.
Intestinal tissues were recovered, and full rolls were placed in PBS solution/PFA 4% baths for 24 h before paraffin embedding following routine procedures. Sections (10 μm) were permeabilized with 0.1% Triton X-100 for 30 min and blocked for 5 min with Ultra V block (Thermo Scientific). Samples were then incubated for 1 h in PBS solution/10% Ultra V block with rabbit anti-UCN34 (1:200). Secondary Alexa Fluor 568-conjugated goat anti-rabbit antibody (1:200; Life Technologies) and WGA coupled to Alexa Fluor 488 (1:200; Life Technologies) in PBS solution/10% Ultra V block were added, and samples were incubated for 45 min at room temperature. Following 3 min DAPI incubation (1:1,000), slides were mounted in Prolong Gold Antifade reagent and imaged by using an Opterra swept-field confocal microscope (Bruker).
Bacteriocin Activity Assay.
For well diffusion assays, one colony of Enterococcus was put in 10 mL of saline solution, and 5 mL of this suspension was poured on a TH plate. After removal of excess liquid, the plate was dried for 5 min. Using sterile tips, 5-mm-diameter wells were dug into the agar. Each well was then filled with 50–100 μL of the tested solutions: overnight bulk culture or sterile filtered supernatant supplemented or not with different detergents (Tween 20, Tween 60, Nonidet P-40, Triton X-100, SDS) or with deoxycholate (12 mM). The plate was then kept at 37 °C overnight, and inhibition zones were measured around the wells. For liquid-phase bacteriocin activity test, 2 106 E. faecalis bacteria were inoculated in 96-well plates in 200 μL of TH broth containing 50% freshly prepared sterile filtered UCN34 overnight supernatant (∆blp mutant or WT). Different concentrations of bile acids were added to the wells, and bacteria growth was monitored by OD600 measurement during a period of 8 h at 37 °C with an Infinite 200 PRO plate reader (Tecan). DCA, LCA, and GDCA powders (Sigma) were resuspended in alkaline TH, DMSO, and TH, respectively, at 200 mM.
Gene Expression Analysis in Mouse Tissues.
Mouse intestines were flushed with ice-cold PBS solution, and tissue samples were immediately disrupted in TRIzol reagent (Life Technologies) by using a Precellys homogenizer (Ozyme) for 2 × 15 s at a frequency of 6,500 rpm. Following RNA extraction, reverse transcription was performed by using SuperScriptII reverse transcriptase (Thermo Fisher Scientific) and random hexamers. Quantitative PCR experiments were performed by using Power SYBR Green mix (Applied) and 10 ng of cDNA. The primers used are listed in Table 1. Quantitative PCR experiments were performed with Power SYBR Green mix (Applied) and 10 ng of cDNA on a QuantiStudio Flex time PCR system (Thermo Fisher). Gene expression was calculated with the standard ΔCt method using Gapdh as the reference housekeeping gene.
Intestinal Bile Acid Profiling.
The quantification of excreted bile acids was performed by Metabolon. Lyophilized samples were shipped to Metabolon, samples were extracted with acidified methanol, and sample extracts were spiked with a solution of labeled internal standards and were evaporated to dryness in a gentle stream of nitrogen. The dried extracts were reconstituted and injected onto an Agilent 1290/Sciex QTrap 6500 LC-MS/MS system equipped with a C18 reverse-phase HPLC column. The mass spectrometer is operated in ESI negative ion mode. The peak area of each bile acid parent (pseudo-MRM mode) or product ion was measured against the peak area of the respective internal standard parent (pseudo-MRM mode) or product ion. Quantitation was performed by using a weighted linear least-squares regression analysis generated from fortified calibration standards prepared immediately before each run. Results were corrected for sample weight (i.e., dry lyophilized feces sample).
Supplementary Material
Acknowledgments
We thank Sylvia Fre (Institut Curie) for providing the Notch/APC mice and for insightful discussions; all the technicians working at the Pasteur Institute animal facility, and especially Eddie Maranghi and Thierry Angélique for their technical help in mice experiments; Camille Mayeur and Muriel Thomas (INRA, Jouy en Josas) for help in quantitative PCR experiments with 16S rRNA; Sylviane Hamon and Chantal Bizet (Centre de Ressources Biologiques de l’Institut Pasteur) for performing stool lyophilizion for the project; and Alexandra Gruss for critical reading of the abstract. This work was funded by La Ligue Nationale contre le Cancer, Association pour la Recherche sur le Cancer Foundation, the Research Applications and Industrial Partnerships Department of Pasteur Institute, the European Research Council (ERC), the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” Grant ANR-10-LABX-62-IBEID, ERC Grant DECRYPT 2013-ADG 339579 (to P.J.S.), and a gift from ONET Company. L.A. was supported by La Ligue Nationale contre le Cancer for 3 y and by ERC for 18 mo. P.J.S. is a Senior Foreign Scholar of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715112115/-/DCSupplemental.
References
- 1.Sobhani I, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogtmann E, et al. Colorectal cancer and the human gut microbiome: Reproducibility with whole-genome shotgun sequencing. PLoS One. 2016;11:e0155362. doi: 10.1371/journal.pone.0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: Inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010;9:249. doi: 10.1186/1476-4598-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic AD, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poyart C, Quesne G, Trieu-Cuot P. Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: Reclassification of ‘Streptococcus infantarius subsp. coli’ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype 11.2 as Streptococcus pasteurianus sp. nov. Int J Syst Evol Microbiol. 2002;52:1247–1255. doi: 10.1099/00207713-52-4-1247. [DOI] [PubMed] [Google Scholar]
- 9.Boleij A, van Gelder MMHJ, Swinkels DW, Tjalsma H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: Systematic review and meta-analysis. Clin Infect Dis. 2011;53:870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 10.Corredoira-Sánchez J, et al. Association between bacteremia due to Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis I) and colorectal neoplasia: A case-control study. Clin Infect Dis. 2012;55:491–496. doi: 10.1093/cid/cis434. [DOI] [PubMed] [Google Scholar]
- 11.zur Hausen H. Streptococcus bovis: Causal or incidental involvement in cancer of the colon? Int J Cancer. 2006;119:xi–xii. doi: 10.1002/ijc.22314. [DOI] [PubMed] [Google Scholar]
- 12.Abdulamir AS, Hafidh RR, Mahdi LK, Al-jeboori T, Abubaker F. Investigation into the controversial association of Streptococcus gallolyticus with colorectal cancer and adenoma. BMC Cancer. 2009;9:403. doi: 10.1186/1471-2407-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butt J, et al. Association of Streptococcus gallolyticus subspecies gallolyticus with colorectal cancer: Serological evidence. Int J Cancer. 2016;138:1670–1679. doi: 10.1002/ijc.29914. [DOI] [PubMed] [Google Scholar]
- 14.Boleij A, et al. Bacterial responses to a simulated colon tumor microenvironment. Mol Cell Proteomics. 2012;11:851–862. doi: 10.1074/mcp.M112.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta A, Madani R, Mukhtar H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis. 2010;12:164–171. doi: 10.1111/j.1463-1318.2009.01814.x. [DOI] [PubMed] [Google Scholar]
- 16.Zarkin BA, et al. The triad of Streptococcus bovis bacteremia, colonic pathology, and liver disease. Ann Surg. 1990;211:786–791. doi: 10.1097/00000658-199006000-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grinberg M, Mansur AJ, Ferreira DO, Bellotti G, Pileggi F. [Endocarditis caused by Streptococcus bovis and colorectal neoplasms] Arq Bras Cardiol. 1990;54:265–269. [PubMed] [Google Scholar]
- 18.Hill MJ, et al. Faecal bile-acids and clostridia in patients with cancer of the large bowel. Lancet. 1975;1:535–539. doi: 10.1016/s0140-6736(75)91556-1. [DOI] [PubMed] [Google Scholar]
- 19.Owen RW, Henly PJ, Thompson MH, Hill MJ. Steroids and cancer: Faecal bile acid screening for early detection of cancer risk. J Steroid Biochem. 1986;24:391–394. doi: 10.1016/0022-4731(86)90088-9. [DOI] [PubMed] [Google Scholar]
- 20.Luk WK, et al. Biliary tract infection due to bile-soluble bacteria: An intriguing paradox. Clin Infect Dis. 1998;26:1010–1012. doi: 10.1086/517638. [DOI] [PubMed] [Google Scholar]
- 21.Fre S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martins M, et al. Streptococcus gallolyticus Pil3 pilus is required for adhesion to colonic mucus and for colonization of mouse distal colon. J Infect Dis. 2015;212:1646–1655. doi: 10.1093/infdis/jiv307. [DOI] [PubMed] [Google Scholar]
- 23.Nissen-Meyer J, Oppegård C, Rogne P, Haugen HS, Kristiansen PE. Structure and mode-of-action of the two-peptide (Class-IIb) bacteriocins. Probiotics Antimicrob Proteins. 2010;2:52–60. doi: 10.1007/s12602-009-9021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani HC, Hu H, Worobo RW, Russell JB. Bovicin HC5, a bacteriocin from Streptococcus bovis HC5. Microbiology. 2002;148:3347–3352. doi: 10.1099/00221287-148-11-3347. [DOI] [PubMed] [Google Scholar]
- 25.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 26.Middendorp S, et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. 2014;32:1083–1091. doi: 10.1002/stem.1655. [DOI] [PubMed] [Google Scholar]
- 27.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 28.Dumke J, Vollmer T, Akkermann O, Knabbe C, Dreier J. Case-control study: Determination of potential risk factors for the colonization of healthy volunteers with Streptococcus gallolyticus subsp. gallolyticus. PLoS One. 2017;12:e0176515. doi: 10.1371/journal.pone.0176515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansom OJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son JS, et al. Altered interactions between the gut microbiome and colonic mucosa precede polyposis in APCMin/+ mice. PLoS One. 2015;10:e0127985. doi: 10.1371/journal.pone.0127985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawid S, Roche AM, Weiser JN. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun. 2007;75:443–451. doi: 10.1128/IAI.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao H, et al. The secondary bile acid, deoxycholate accelerates intestinal adenoma-adenocarcinoma sequence in Apc (min/+) mice through enhancing Wnt signaling. Fam Cancer. 2014;13:563–571. doi: 10.1007/s10689-014-9742-3. [DOI] [PubMed] [Google Scholar]
- 33.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 34.Nissen-Meyer J, Holo H, Håvarstein LS, Sletten K, Nes IF. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauge HH, Nissen-Meyer J, Nes IF, Eijsink VG. Amphiphilic alpha-helices are important structural motifs in the alpha and beta peptides that constitute the bacteriocin lactococcin G–Enhancement of helix formation upon alpha-beta interaction. Eur J Biochem. 1998;251:565–572. doi: 10.1046/j.1432-1327.1998.2510565.x. [DOI] [PubMed] [Google Scholar]
- 36.Hauge HH, Mantzilas D, Eijsink VG, Nissen-Meyer J. Membrane-mimicking entities induce structuring of the two-peptide bacteriocins plantaricin E/F and plantaricin J/K. J Bacteriol. 1999;181:740–747. doi: 10.1128/jb.181.3.740-747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami Y, Tanabe S, Suzuki T. High-fat diet-induced intestinal hyperpermeability is associated with increased bile acids in the large intestine of mice. J Food Sci. 2016;81:H216–H222. doi: 10.1111/1750-3841.13166. [DOI] [PubMed] [Google Scholar]
- 38.Barker GM, et al. Biliary bile acid profiles in patients with familial adenomatous polyposis before and after colectomy. Br J Surg. 1994;81:441–444. doi: 10.1002/bjs.1800810340. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, et al. An association between genetic polymorphisms in the ileal sodium-dependent bile acid transporter gene and the risk of colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2001;10:931–936. [PubMed] [Google Scholar]
- 40.Raufman J-P, et al. Slc10a2-null mice uncover colon cancer-promoting actions of endogenous fecal bile acids. Carcinogenesis. 2015;36:1193–1200. doi: 10.1093/carcin/bgv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar R, et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 2017;13:e1006440. doi: 10.1371/journal.ppat.1006440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reikvam DH, et al. Depletion of murine intestinal microbiota: Effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danne C, et al. Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J Infect Dis. 2011;204:1960–1970. doi: 10.1093/infdis/jir666. [DOI] [PubMed] [Google Scholar]
- 44.Rusniok C, et al. Genome sequence of Streptococcus gallolyticus: Insights into its adaptation to the bovine rumen and its ability to cause endocarditis. J Bacteriol. 2010;192:2266–2276. doi: 10.1128/JB.01659-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tettelin H, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 46.Lux T, et al. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J Bacteriol. 2007;189:7741–7751. doi: 10.1128/JB.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.