Significance
Cellular acidification occurs in response to many cellular events, including ischemia and apoptosis. The voltage-dependent anion channel (VDAC) is the most abundant protein in the outer mitochondrial membrane and has been implicated in mitochondrial regulation, cell death, and assembling protein complexes with Bcl2 family of apoptotic regulating proteins. During apoptosis, VDAC has demonstrated rapid oligomerization followed by cytochrome c release into the cytoplasm. We report a new dimeric model of VDAC induced by minor acidification with a pKa of 7.4, which is regulated by the protonation state of transmembrane residue E73. This study links a specific dimer formation with cellular acidification, which may be a salient feature of the molecular mechanism(s) involved in mitochondrial regulation.
Keywords: VDAC, dimerization, DEER, native mass spectrometry, cellular stress
Abstract
The voltage-dependent anion channel (VDAC) is the most abundant protein in the outer mitochondrial membrane and constitutes the primary pathway for the exchange of ions and metabolites between the cytosol and the mitochondria. There is accumulating evidence supporting VDAC’s role in mitochondrial metabolic regulation and apoptosis, where VDAC oligomerization has been implicated with these processes. Herein, we report a specific pH-dependent dimerization of murine VDAC1 (mVDAC1) identified by double electron–electron resonance and native mass spectrometry. Intermolecular distances on four singly spin-labeled mVDAC1 mutants were used to generate a model of the low-pH dimer, establishing the presence of residue E73 at the interface. This dimer arrangement is different from any oligomeric state previously described, and it forms as a steep function of pH with an apparent pKa of 7.4. Moreover, the monomer–dimer equilibrium affinity constant was determined using native MS, revealing a nearly eightfold enhancement in dimerization affinity at low pH. Mutation of E73 to either alanine or glutamine severely reduces oligomerization, demonstrating the role of protonated E73 in enhancing dimer formation. Based on these results, and the known importance of E73 in VDAC physiology, VDAC dimerization likely plays a significant role in mitochondrial metabolic regulation and apoptosis in response to cytosolic acidification during cellular stress.
Mitochondria are the power plants of eukaryotic cells, producing the vast majority of the universal cellular energy currency, ATP. The voltage-dependent anion channel (VDAC) is the most abundant protein in the outer mitochondrial membrane (OMM), forming a large pore composed of 19 β strands as revealed by high-resolution structures from X-ray crystallography and NMR (1–3). VDAC governs the flux of anions, cations, and metabolites between the mitochondria and the cytosol of the cell (4–6). In addition to its critical role in bioenergetics, VDAC is capable of modulating mitochondrial permeability (7–10) through interaction with the apoptosis regulating proteins in the Bcl-2 family, implicating VDAC in metabolic stress and mitochondria-mediated apoptotic cell death.
Although VDAC can function as a monomer (2), a common dimeric model—formed across beta strands β-1, β-17, β-18, and β-19—has been reported for zebra fish VDAC2 (zfVDAC2) (11), rat VDAC1 (12), and human VDAC1 (3). High-resolution atomic force microscopy revealed various oligomeric states of VDAC in native OMM (13, 14). Additional biophysical techniques including electron microscopy (15–17), fluorescence cross-correlation microscopy (18), X-ray crystallography (11), double electron–electron resonance (DEER) (11), and in vivo and in vitro cross-linking studies (3, 19) identified a diverse set of VDAC oligomers. The physiological role of VDAC oligomerization remains poorly understood; however, apoptosis and cellular stress, which coincide with cytosolic acidification (20–22), display a substantial increase in the population of VDAC oligomers (19, 23, 24).
The pH dependence of mVDAC1 oligomerization was characterized by DEER and native mass spectrometry (MS). DEER is a pulse EPR technique used to measure the magnetic dipolar interaction between coupled spins. In the DEER experiment, a spin echo amplitude is monitored as a function of time spacing between observe and pump pulses (Fig. S1A). The dipolar coupling modulates the echo amplitude as a function of the interspin distance and time between the two pulses. For a given total number of spins in a sample, the total amplitude of the echo modulation corrected for background effects (i.e., the depth of modulation, Δ) depends on the fraction of specifically interacting spin pairs that are excited by the microwave pulse. If singly labeled VDAC molecules are monomers with a random distribution of interspin distances, no specific pairwise spin interaction exist, and Δ = 0. If VDAC dimerizes and forms spin pairs, the modulation amplitude Δ will be proportional to the fraction of VDAC in dimer form (25, 26). Moreover, the DEER data provide the interspin distance distributions in the range of ∼15–80 Å (27, 28), and the distance constraints—from several singly labeled mutants—may be used to determine the relative orientation of subunits in the oligomer.
Native MS is an emerging biophysical approach whereby noncovalent interactions and native-like structure are preserved within the mass spectrometer. Recently, native MS has been used to determine equilibrium binding constants and binding thermodynamics for protein–ligand and membrane protein–lipid interactions (29). Together, DEER and native MS provide a powerful multidisciplinary approach for characterizing mVDAC1’s pH-induced oligomerization.
We report a specific pH-dependent dimer of mVDAC1 based on DEER and MS data of four singly spin-labeled mutants, revealing a dramatic increase in dimerization coupled to acidification. The mVDAC1 dimer model, generated from DEER distances and computational modeling, reveals membrane-exposed glutamate 73 (E73) buried in the dimer interface. Mutations of this residue, to alanine or glutamine, demonstrate the essential role of protonated E73 for pH-induced dimerization. It is noteworthy that E73 in VDAC1 plays an essential role in mitochondrial calcium handling (30) and is required for the binding of hexokinase to OMM (31), the latter of which is protective against mitochondrial-dependent apoptosis (32). Altogether, these data suggest that mVDAC1 pH-dependent oligomerization may be a regulatory mechanism that occurs in response to cellular acidification.
Results
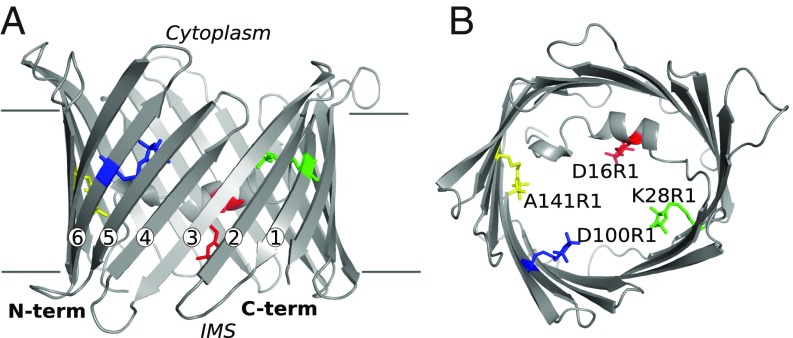
DEER was used to characterize the oligomerization state of mVDAC1 induced by acidification. Four single-cysteine mutants (D16C, K28C, D100C, and A141C) were engineered in the mVDAC1 cysless-construct (33) at sites facing inside the pore and reacted with a methanethiosulfonate spin label to generate the R1 side chain (Fig. 1). D16R1 is part of the pore-lining N-terminal helix, whereas K28R1, D100R1, and A141R1 are located in the center of β strands 1, 6, and 9, respectively. mVDAC1 was purified as previously reported using the detergent LDAO (2), and subsequent DEER measurements were performed both on detergent solubilized protein and in a lipidic bicelle environment (34) (where specified). Continuous-wave electron paramagnetic resonance (CW EPR) spectra for the four mutants exhibited only a small pH dependence (Fig. S2), indicating that the local protein environment of the spin-labeled residues is relatively insensitive to changes in pH. The oligomerization level of the protein at various pH values was assessed using the modulation depth of the DEER data as described previously (25, 26), as well as native MS. A model of the mVDAC1 dimer was constructed using the intramolecular distances at pH 5 for each of the four single-spin labeled mutants.
Fig. 1.
Structure of mVDAC1 with modeled nitroxide spin labels. Shown are cartoon representations of mVDAC1 3EMN showing the position of the four single mutations—D16R1 (red), K28R1 (green), D100R1 (blue), and A141R1 (yellow)—presented on the same VDAC molecule. (A) View from the membrane plane. (B) View from the cytoplasm down to the intermembrane space (IMS). R1 labels are presented in stick representation with the β strands numbered in A.
Acidification Triggers mVDAC1’s Dimerization.
DEER measurements were performed on all four mutants in detergent micelles (LDAO) at pH 8 and pH 5 (Fig. 2 and Fig. S3). For all mutants, the data clearly show an increase in the modulation depth (triangles, Fig. 2) when the pH decreases, indicating oligomerization of mVDAC1 is driven by acidification. The modulation depth was used to calculate the average number of spins per nanoobject (Materials and Methods), allowing the oligomeric state to be determined. There was some variation in modulation depth between the D16R1, K28R1, D100R1, and A141R1 samples at pH 8, which can be attributable to differences in labeling efficiency and protein concentration.
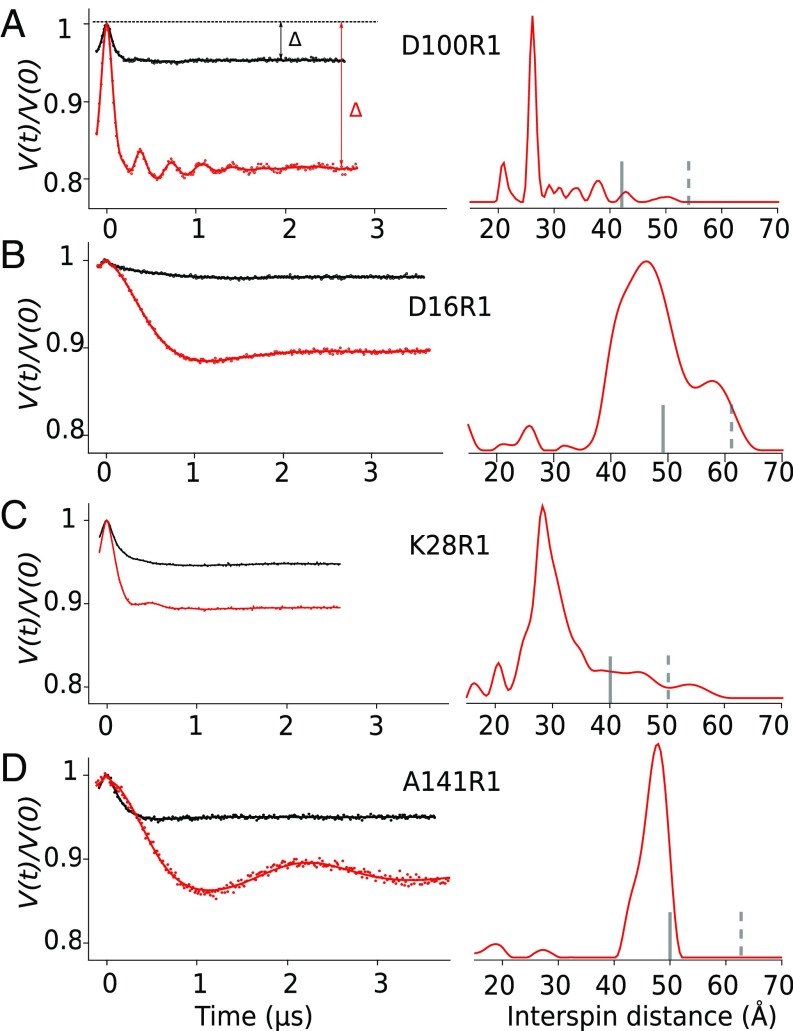
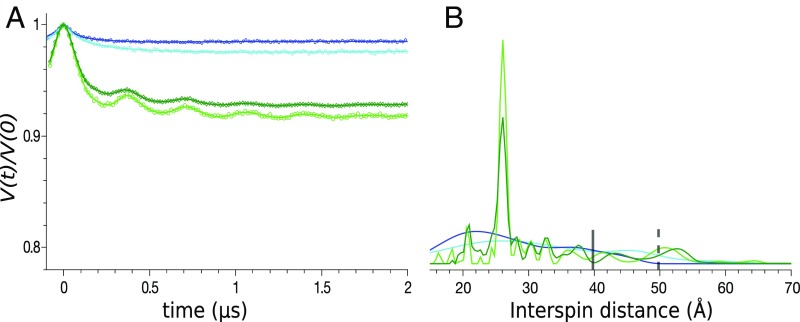
Fig. 2.
pH reduction induces an increase of the modulation depth and well-defined prominent peaks in the distance distribution for each spin-labeled VDAC. (Left) Normalized, background-corrected dipolar evolutions (A) D100R1, (B) D16R1, (C) K28R1, and (D) A141R1 in LDAO at pH 8 (black) and pH 5 (red). The triangles in A represents the modulation depth of the sample at pH 8 (black) and pH 5 (red). (Right) Normalized DEER distance distribution at pH 5. The vertical bars indicate the upper limit of reliable width (solid line) and position (dotted line) of the distances in the distribution (Materials and Methods).
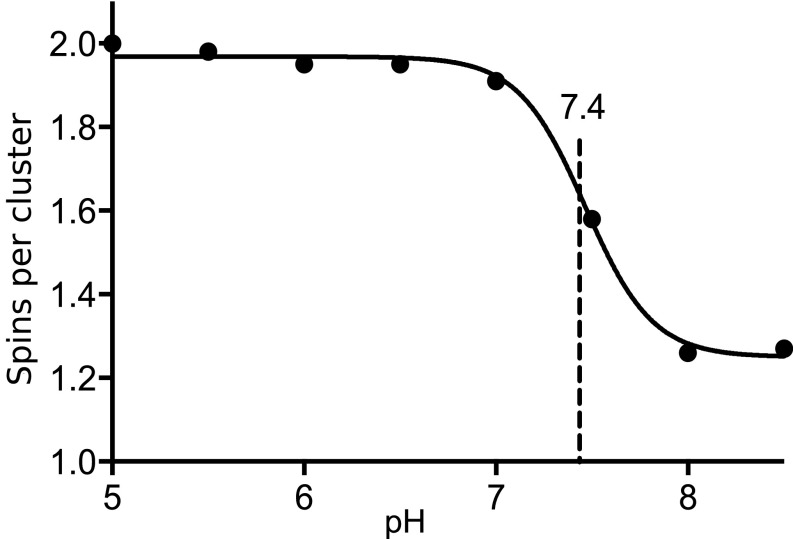
A titration curve of fully labeled mVDAC1 D100R1 (Fig. S4) from eight different pH values (Fig. 3 and Fig. S5) was generated using modulation depth. At pH 8, the D100R1 mutant has a modulation depth of 0.05, corresponding to an average of 1.22 interacting spins per objects as calculated using Eq. 1 (Materials and Methods). This value indicates that the protein is primarily monomeric with a small fraction in various oligomeric forms. The corresponding distance distribution is extremely broad and multimodal (Figs. S3 and S5), implying that there is more than one structurally distinct oligomeric state in the small population, which all contribute to the average number of interacting spins. When the pH is lowered, the modulation depth increases, plateauing at 0.22, corresponding to two interacting spins. This indicates that acidification triggers a rearrangement of mVDAC1 from a mainly monomeric population to a mainly dimeric population with a sharp transition (within 1 pH unit) at an apparent pKa of 7.4.
Fig. 3.
mVDAC1 forms a dimer at low pH. Titration curve of mVDAC1 D100R1 at eight different pH values. All sample were measured at 100 μM, and the modulation depth was used to calculate the number of spins per cluster using Eq. 3. Data were fitted using PRISM software, revealing a sharp transition with a midpoint value of 7.4. The dipolar evolution functions and distance distributions are displayed in Fig. S5.
In addition to the increased modulation depth at lower pH, a common trend observed for the four engineered R1-labeled samples is the appearance of a single dominant population in the distance distribution (Fig. 2, Right). All four individually spin-labeled mVDAC1 samples independently suggest dimerization, due to the appearance of a single predominant peak in the distance distribution at pH 5, and taken together strongly indicate that the dimer adopts a single, stable conformation. The narrowest peaks occur for labels at positions K28R1, D100R1, and A141R1, which adopt separation distances of 26, 48, and 26 Å, respectively. For D16R1 a wider distribution was observed with the most probable distance at 46 Å. The broader distribution for D16R1 may be attributed to increased flexibility of the protein and/or spin label side chain at this site as reflected in the CW spectra (Fig. S2).
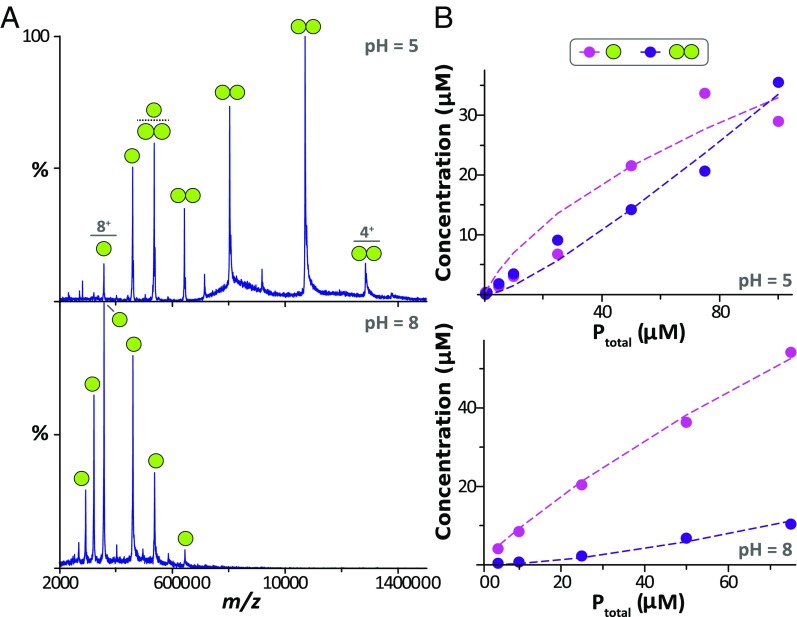
The pH-dependent oligomerization of mVDAC1 was further characterized using native MS to analyze mVDAC1 D100R1 in LDAO detergent micelles (Fig. 4). Native mass spectra were initially recorded at a protein concentration of 10 μM at pH 5 or 8 (Fig. 4A). The experimental masses for the monomer and dimer were 32,173 ± 18 and 64,230 ± 40.09 Da, which coincide nicely with the theoretical monomer value of the sample labeled with the R1 spin label (32,175.17 Da). The mass spectrum for the sample at a pH of 8 is predominately composed of signals for the monomer population (∼85%) with a small component for the dimer. In contrast, the mass spectrum at pH 5 primarily consists of signals for dimeric species (∼70%). These observations are consistent with DEER measurements that demonstrate an enhancement in dimerization of mVDAC1 at low pH.
Fig. 4.
Native MS reveals monomer–dimer equilibrium binding constants (KD) for mVDAC1 D100R1 at pH 5 and 8. (A) Representative mass spectra for mVDAC1 D100R1 acquired at 10 μM in either (Top) pH 5 or (Bottom) pH 8 buffer. Labeled are the ions corresponding to either monomer (one sphere) or dimer (dual spheres). (B) Plot of monomer (pink dots) and dimer (purple dots) concentration of VDAC1 D100R1 as a function of total protein concentration (Ptotal) at (Top) pH 5 and (Bottom) pH 8. Shown are the experimental concentrations (dots) and resulting fit from a monomer–dimer binding model (dashed lines), revealing a KD of 32 and 247 μM at pH 5 and 8, respectively. Fitting statistics are reported in Table S1.
To obtain the monomer–dimer equilibrium binding constants (KD), titrations at different pH values were performed, varying the total protein concentration from 5 to 100 μM, and their native mass spectra were recorded (Fig. 4B and Fig. S6). The concentration of monomer and dimer, at each protein concentration, was determined by deconvolution of the mass spectra (Materials and Methods) (35) and fitted employing a monomer–dimer binding model to determine the KD at different pH values (Fig. 4B, Fig. S7, and Table S1). The KD values at pH 5 and 8 are 32 and 247 μM, respectively. The enhancement in dimerization at low pH corresponds to nearly an eightfold increase in the equilibrium affinity constant. Additional sampling between pH values of 6 and 7 supports the notion that mVDAC1 D100R1 adopts a primarily dimeric conformation below the experimentally determined pKa of 7.4. Further, both native MS and DEER results have independently revealed the presence of an ensemble of dimer structures present at high pH and enhancement of dimerization at low pH.
mVDAC1 Forms a Parallel Dimer at Low pH.
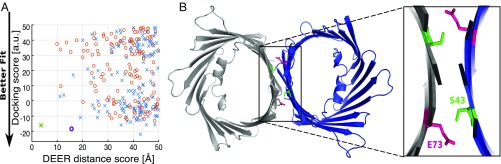
Using the most probable distances from the DEER measurements—recorded at pH 5—for the four R1 label sites, we constructed a structural model of the pH-induced dimer (Materials and Methods). An important aspect to investigate was the relative orientation of the two monomers forming a dimer within the membrane plane because both parallel (in which both proteins are inserted in the same orientation) and antiparallel (in which the proteins are inserted in the opposite orientation) dimer organizations have been described for VDAC (3, 11, 36). Starting with two copies of the mVDAC1 structure (Protein Data Bank ID: 3EMN), random rigid body displacements in the plane of the membrane were performed followed by arbitrary rotations about the pore axis to generate thousands of putative dimers. Both parallel and antiparallel orientation models were ranked based on surface complementarity (docking score; Materials and Methods) and the agreement with the most probable interspin distance from DEER measurements (DEER distance score) (Table S2). A scatterplot of the docking score and DEER distance score, for all parallel (cross) and antiparallel (circle) models, reveals that only a parallel model satisfies both constraints simultaneously (Fig. 5A, green cross). Additionally, the antiparallel models failed to satisfy the DEER constraints, even under conditions where surface complementarity was ignored and physically unreasonable overlaps between the two channels were permitted (Fig. S8A).
Fig. 5.
Model of mVDAC1 pH-induced dimer. (A) Dimer models were generated and assessed based on the DEER distances using mVDAC1 3EMN structure as a rigid body (Materials and Methods). Parallel dimers are displayed as crosses, and antiparallel are displayed as circles. The green and purple symbols represent the best possible model for parallel (cross) and antiparallel (circle) dimer, respectively. (B) mVDAC1 dimer model (green cross) displayed in cartoon representation with E73 represented in pink sticks. Expanded view shows putative coordination of E73 (pink) and S43 (green) at the dimer interface.
The parallel model achieves a nearly perfect fit for both the DEER distance constraints and surface complementarity as shown in Fig. 5B (green cross). A parallel model generated solely based on the DEER distance constraints without taking into account steric overlap was nearly identical (Fig. S8B). The model presented in Fig. 5B reveals a dimerization interface composed of β strands 2 through 5, where the side chain of E73 (β strand 4) is buried in the dimerization interface interacting with S43 (β strand 2) from neighboring molecule. The best parallel model produced from this search strategy simultaneously satisfies all four distance constraints (Table S2) and generates very little steric clash at the dimer interface.
Glutamate 73 Is Essential for Dimerization.
To investigate the role of E73—a highly polar or charged residue buried within the membrane—in the dimerization process, two mutants were engineered into the cysless-mVDAC1 D100R1 background: E73A and E73Q, where the D100C mutation was used to attach a nitroxide spin label for DEER. The alanine mutation was selected to replace the polar and potentially charged glutamic acid residue with a small hydrophobic residue more compatible with the membrane environment of residue 73. The glutamine mutation was selected as a polar but electrically neutral mimic of glutamic acid that is roughly isosteric and maintains hydrogen bonding capabilities. Notably, VDAC3 shares an 80% sequence similarity with VDAC1 and VDAC 2 but carries a glutamine at position 73, making the E73Q mutant a reasonable mimic for VDAC3.
DEER measurements of D100R1 in the presence of the E73A and E73Q mutations reveal a striking variation in the oligomerization pattern compared with native E73 (Fig. 6). The modulation depth of native E73 increases from 0.05 to 0.22 when pH is decreased from 8 to 5 (Fig. 2A) corresponding to an ∼100% dimer population under acidic conditions. For the E73A mutant, low modulation depths (0.015–0.024) at both pH 8 and pH 5 were observed with a broad and featureless distance distribution, indicating no detectable dimer described above. For the E73Q mutant, the modulation depth was larger than the native E73 sample at pH 8 (0.07), and the dimerization peak at 26 Å was present in the distance distribution, attesting to the formation of the dimer population even at higher pH. Based on the modulation depth, the dimeric population was estimated to be ∼35% using Eq. 2. It is noteworthy that under acidic conditions, the modulation depth of the E73Q mutant hardly changes compared with pH 8, revealing a negligible pH dependency in the dimerization of the protein. Hence, the E73A mutation reveals the crucial role of E73 in the dimerization process, and the E73Q mutant indicates that the pH dependence of the observed dimerization is tied to the protonation state of E73.
Fig. 6.
E73 has a striking effect on the dimerization of mVDAC1. (A) Normalized, background-corrected dipolar evolutions and (B) normalized DEER distance distribution of mVDAC1 D100R1/E73A at pH 8 (dark blue) and pH 5 (light blue) and D100R1/E73Q at pH 8 (dark green) and pH 5 (light green). The vertical bars indicate the upper limit of reliable width (solid line) and position (dotted line) of the distances in the distribution (Materials and Methods).
mVDAC1 Forms a Parallel Dimer Under Acidic Conditions in a Lipidic Environment.
In addition to detergent solubilized samples, DEER measurements were collected on channels embedded in a lipidic bicelle environment composed of DMPC and CHAPSO (Fig. S9) (34). DEER measurements were performed at pH 5, 6, 7, and 8 for D100R1 and at pH 5 for the D16R1, K28R1, and A141R1 spin-labeled mutants. The most probable distance values at pH 5—for all samples—were the same in lipidic bicelles as they were in detergent micelles (Fig. 2 and Fig. S9), indicating that the same dimeric state is formed in both environments. In contrast, the DEER data at pH 8 (D100R1; Fig. S9) exhibit a large modulation depth in lipidic bicelles compared with detergent micelles. This indicates an increased protein–protein association in lipidic bicelle environment, and the multimodal distance distribution suggests that these are low-specificity interactions from various oligomeric forms.
Although the modulation depth for D100R1 spin-labeled mutant has minimal pH dependency, there is a strong pH dependency observed in the distance distribution. At elevated pH in lipidic bicelles, the D100R1 spin-labeled mutant has a significant peak at ∼55 Å (black arrow, Fig. S9), which is consistent with a different dimer identified in zfVDAC2 (11). Under acidic conditions, the peak at ∼55 Å disappears, and a narrow peak at 26 Å, corresponding to the dimer described in this study, is present. In total, the average amount of protein–protein interaction stays constant upon acidification, but the distance distribution indicates a reorganization of mVDAC1 from various oligomeric forms to predominately the highly ordered dimer shown in Fig. 5.
Discussion
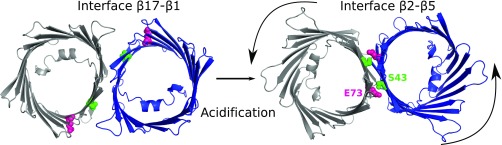
The current study identifies a unique dimeric model for mVDAC1 with a protein–protein interface occurring between β strands 2 and 5. The association of two monomeric channels is driven by acidification, wherein E73’s protonation state regulates the dimerization process. VDAC channels are known to organize into several oligomeric states (3, 11, 13, 19) as reported previously by atomic force microscopy on native OMM surfaces and consistent with the high modulation depth observed for mVDAC1 in lipidic bicelles at neutral pH (Fig. S9). In lipidic bicelles at pH 8, two prominent peaks are observed at 55 and 26 Å. The peak at 55 Å is compatible with the previously described dimer of zfVDAC2 where beta strands β-1, β-17, β-18, and β-19 form the dimeric interface (11). Upon cellular acidification, E73 becomes protonated and may H-bond with S43 resulting in a VDAC1 reorganization, which primarily forms a high-affinity dimer between β strands 2 and 5 corresponding to the peak at 26 Å in the DEER distance distributions (Fig. 7 and Fig. S9). This newly formed dimer is now the primary population and replaces all other oligomeric states. Together, these data suggest that VDAC1 undergoes a specific molecular reorganization in response to acidification and that this organization is likely formed during incidents like ischemia and apoptosis where cytosolic acidification is known to occur.
Fig. 7.
Acidification triggers a reorganization of mVDAC1. (Left) Dimer described for zfVDAC2 (11), corresponding to the peak at 55 Å in the lipidic bicelle samples (Fig. S9). (Right) Parallel dimer modeled in this study, corresponding to the peak at 26 Å in the lipidic bicelle samples (Fig. S9). E73 and S43 are represented in pink and green spheres, respectively. Upon acidification, mVDAC1 rearranges into a stable dimer with a dimer interface between β strands 2 and 5.
The acidic amino acid E73 is embedded in the membrane’s hydrophobic core and is critical for pH-induced dimerization. Although it is energetically unfavorable for charged or polar residues to face the membrane core, there have now been several reports of proteins having charged residues residing within the membrane, which may have mechanistic implications for their function (37–41). For mVDAC1, mutations of E73 reveal its crucial role in the dimerization process and strongly indicate that the association is coupled to the protonation state of E73 with an apparent pKa of 7.4 (Fig. 3). This experimental result nicely coincides with theoretical calculations of glutamate residues residing in different dielectric environments, where solvent exposed glutamates are negatively charged at neutral pH having a pKa ∼ 4.25. However, when placed in a low-dielectric environment, such as the core of a membrane, theoretical calculations suggest that the pKa dramatically increases by ∼3 units (41, 42), in excellent agreement with our measured pKa value. Our work suggests that upon acidification, E73 accepts a proton at an elevated pKa of 7.4 to become neutral, facilitating dimerization of the channel.
Glutamine substitution is frequently used to mimic a protonated glutamate. As expected, the E73Q mutant displayed no pH dependency; however, there were some dramatic changes to the dimerization profile. Where wild-type mVDAC1 completely dimerizes under acidic conditions, the E73Q mutant has a dimeric fraction of only 35% at all pH values. It suggests that polarity of the side chain, as the protonated carboxylate group has a higher polarity than the acetamide group, is an important factor for dimerization. However, in both subunits of the dimer S43 is in close proximity to E73, and it is a likely interaction partner (Fig. 5B). It is worthy to note that VDAC3 features a glutamine at position 73; thus, the results of the E73Q mutant suggest an isoform specific behavior toward dimerization and cellular acidification.
The pKa of E73 is closely tuned to the physiological pH of the cell’s cytosol (pH ∼ 7.4). Cellular stress, from events like ischemia and apoptosis, often results in acidification corresponding to reduction in cytosolic pH by 0.3–0.4 units (22), which according to our titration results (Figs. 3 and 4) would result in a significant shift in VDAC1’s dimeric population. VDAC’s involvement in the apoptotic cascade by generating a large protein complex with members of Bcl-2 family of apoptotic proteins remains a controversial subject (10, 43, 44). This view is propagated by a number of studies implicating VDAC’s oligomeric state as a component of the mitochondrial apoptosis pathway (18, 19, 45) and furthered through its identification as a critical component for cytochrome c release (46). Although our study does not resolve this controversy, our results allow us to apprehend previous results from a different angle. Indeed, the cytochrome c release assays were only successful under acidic condition (pH 5.2) where our results suggest that a dimeric form of VDAC1 would be present. Another prevailing hypothesis links hexokinase binding to the OMM through interactions of VDAC1’s E73 (31, 47). It plausible that a VDAC1–hexokinase complex would shield the oligomerization interface preventing VDAC’s dimerization resulting in an antiapoptotic response. Taken altogether, the VDAC monomer–dimer equilibrium is conveniently tuned to fall precisely in the range that could respond to physiological changes in cytoplasmic pH and a responsive mechanism for cellular stress and apoptosis.
Materials and Methods
Cloning of pQE9-mVDAC1-C127A/C232A and Single-Cysteine Mutants.
Mutants were prepared using the QuikChange site-directed Mutagenesis Kit from Stratagene (Agilent Technologies). A cysless-mVDAC1 was engineered by mutating Cys-127 and 232 into Alanines (33). Cysless-mVDAC1 was used as a template to introduce point mutations D16C, K28C, D100C, and A141C. The cysless-mVDAC1 D100C mutant was then used as a template to make the D100C/E73A and D100C/E73Q mutants. Each mutant was verified by DNA sequencing.
Expression, Purification, and Spin Labeling of mVDAC1.
Expression and purification of recombinant proteins was described previously (2). Briefly, M15 Escherichia coli cells were used to express mVDAC1 in inclusion bodies. The protein was isolated by a two-step purification where solubilized IB were purified using Talon Metal affinity column, and eluted protein was refolded by a three-step dialysis. Refolded protein was then centrifuged (200,000 g for 45 min), and DTT was removed using a PD10 desalting column and the buffer exchanged to 20 mM Hepes, pH 7.0, 150 mM NaCl, 0.1% LDAO to facilitate labeling. The protein was incubated with a 1.5-fold molar excess of the spin label reagent 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate for 2 h at 4 °C to generate the R1 side chain (48–50). The protein was then concentrated using an Amicon Ultra-50k (Millipore) and injected on a size exclusion chromatography column (superdex 200; GE Healthcare) in 150 mM NaCl, 20 mM Tris⋅HCl, pH 8.0, 0.1% LDAO to remove excess spin label and misfolded proteins. The pH was lowered by washing and concentrating using the following buffers made with D2O: 150 mM NaCl, 0.1% LDAO, and 50 mM Hepes, pH 8.5, pH 8.0, pH 7.5, pH 7.0; 50 mM Pipes, pH 6.5, pH 6.0; and 50 mM Mes, pH 5.5, pH 5.0. After an ultracentrifugation step to remove protein aggregates (200,000 g for 45 min), 20% glycerol-D8 was added to the sample. Some of the spin-labeled mVDAC1 was reconstituted in bicelles by mixing the protein at a 4:1 protein:bicelle ratio with a (2.8:1) DMPC/CHAPSO bicellar solution (34). All of the samples for the same mutants were concentration-normalized (80–100 μM depending on mutants) to compare the modulation depths and the distance distributions.
CW EPR and DEER Experiments.
CW EPR spectra in LDAO and bicelles were recorded at room temperature on a Varian E-109 spectrometer fitted with a two-loop one-gap resonator (Medical Advances) (51); samples of 5 μL were loaded into a borosilicate capillary tube (0.6-mm inner diameter, 0.84-mm outer diameter; VitroCom Inc.). Spectra were recorded at X band (∼9.4 GHz) over a 100 G field range using 2 mW incident microwave power and 100 kHz field modulation with an amplitude of 1 G.
DEER data were acquired at Q band (∼33.5 GHz) on a Bruker Elexsys E580 spectrometer equipped with a SuperQFT bridge and AmpQ 10-W amplifier and fitted with an ER 5107D2 resonator (Bruker BioSpin). A dead-time free four-pulse sequence was used with an eight-step averaging cycle for suppression of deuterium nuclear modulations and eight-step phase cycling (50, 52). A rectangular 32-ns pump pulse was applied at the maximum of the absorption spectrum, and rectangular 18-ns π/2 and 36-ns π observe pulses were applied at a −50-MHz offset. For DEER experiments, 14 μL of spin-labeled protein reconstituted in LDAO or bicelles containing 20% (vol/vol) glycerol were loaded into a borosilicate capillary tube (1.4-mm inner diameter × 1.7-mm outer diameter; VitroCom) and subsequently flash-frozen in liquid nitrogen before loading into the resonator for data acquisition at 80 K.
DEER data were analyzed using the program LongDistances, written in LabVIEW and available for download at www.biochemistry.ucla.edu/Faculty/Hubbell/. For protein in LDAO micelles, a background contribution from remote spins was fitted assuming a 3D distribution. For protein in DMPC/CHAPSO bicelles a 2D distribution of spins was assumed for the background correction. Distance distributions were determined from the background-corrected signal using a regularization procedure similar to the commonly used Tikhonov regularization (27). The regularization parameter for distance distribution determination was selected using the L-curve criterion (53). The upper limit of reliable mean distance (r) and width determination (σ) in nanometers was calculated for each mutant according to ref. 27.
DEER was used to determine the average number of spins per nanoobject () as previously described (25, 28) using the equation
| [1] |
where Δ is the modulation depth and C is a constant dependent on the resonator, the pump pulse profile, and the spectral line shape of the nitroxide spin label. The fractional dimerization may then be calculated according to the equation
| [2] |
where fD is the fractional dimerization and a is the spin-labeling efficiency. The spin-labeling efficiency was assessed for D100R1 using Tetramethylrhodamine-5-maleimide (TMRM), a fluorescent probe that binds to free cysteines, on SDS-Polyacrylamide gel (Fig. S9), indicating complete labeling (). Thus, was taken to be equal to 1 to solve for fD.
Native Mass Spectrometry.
Samples of mVDAC D100R1 were buffer exchanged into MS buffer (200 mM ammonium acetate supplemented with 2× CMC LDAO) following established methods (54). The pH of MS buffer was adjusted with either acetic acid or ammonium hydroxide. Native mass spectrometry experiments were carried out on a Synapt G1 (Waters Corporation) instrument tuned to preserve noncovalent interactions and native-like structure following established methods (29, 54). Briefly, the instrument was set a capillary voltage of 1.8 kV, sampling cone voltage of 50 V, extractor cone voltage of 5 V, and argon flow rate at 10 mL/min (5.2 × 10–2 mbar). The T-wave settings for trap (300 ms–1/2.0 V), IMS (300 ms–1/20 V) and transfer (100 ms–1/10 V), source temperature (90 °C), and trap bias (35.1 V) were also optimized. Native MS data were deconvoluted using UniDec to obtain intensities and converted to mole fraction to determine the concentration of monomer (M) and dimer (D) at equilibrium:
| [3] |
and
| [4] |
where FM,exp and FD,exp are the experimental mole fraction for monomer and dimer, respectively, and Ptotal is total protein concentration. To obtain the monomer–dimer equilibrium constant (KA), we applied the following monomer–dimer model:
| [5] |
and
| [6] |
Ptotal in the system can be expressed as
| [7] |
Eq. 6 can be rearranged and substituted into Eq. 7:
| [8] |
To calculate the concentration of P, Eq. 8 can be solved, and taking the positive solution,
| [9] |
Eq. 7 can be rearranged to obtain D:
| [10] |
The monomer–dimer binding model was coded into Python2.7 (https://www.python.org), making use of libraries scipy (55), numpy (56), and matplotlib (57). To obtain KA, the data collected at different Ptotal concentrations were globally fit through least square minimization of the pseudoχ2 function:
| [11] |
where n is the total number of Ptotal concentrations, Mi,exp and Di,exp and Mi,calc and Di,calc are the experimental and calculated concentrations at a given Ptotal concentration (i).
Dimer Modeling.
VDAC dimers were created and ranked based on satisfaction of DEER distance restraints (S1) and surface complementarity (S2) (58). Starting with the high-resolution structure of the mouse VDAC1 structure (2), we replaced the four labeled residues employed in the DEER studies with the R1 side chain. Starting with two copies of the channel centered at the origin with both pores aligned to the z axis (parallel model) or one pore oriented along the −z axis (antiparallel model), we rigidly displaced the second channel in the x–y plane (x0,y0) followed by rigid rotation about the pore axis (θ0). The distance scoring function was
| [12] |
where Di is the experimentally measured residue–residue distance between labeled residues i and Do is distance between the nitroxide oxygen atoms in the model.
Surface complementarity was calculated by determining for all heavy atoms in model 2 the closest atom in model 1. For each atom, call this distance di, then the score is given by
and the total score for the model is given by the sum over all N heavy atoms:
| [13] |
The complementarity weights were chosen to give equal weight to the DEER and complementarity score.
Models were generated with two methods. First, the parameter set {x0, y0, θ0} was randomly generated by picking uniformly from −50 to 50 Å for both x0 and y0 values and then from 0 to 360° for θ0. We generated and scored 1,000 random models for the parallel model and 1,000 for the antiparallel model. Second, we used a Nelder–Mead simplex method (59) to identify the optimal solution for the parallel and antiparallel configurations using the scoring function of (F = S1 + S2).
Supplementary Material
Acknowledgments
We thank Drs. Aviv Paz, Thorsten Althoff, and Matthias Elgeti for fruitful discussions. This work was supported by NIH Grant DP2GM123486 and startup funds from the Department of Chemistry, Texas A&M University (to A.L.); Grant R01GM089740 (to M.G.); Grants R01EY005216 and P30EY00331 and the Jules Stein Professorship Endowment (to W.L.H.); and Grant R01GM078844 and University of California, Los Angeles, Cardiovascular Theme Discovery Award (to J.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715464115/-/DCSupplemental.
References
- 1.Hiller S, et al. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ujwal R, et al. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayrhuber M, et al. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schein SJ, Colombini M, Finkelstein A. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from paramecium mitochondria. J Membr Biol. 1976;30:99–120. doi: 10.1007/BF01869662. [DOI] [PubMed] [Google Scholar]
- 5.Rostovtseva T, Colombini M. VDAC channels mediate and gate the flow of ATP: Implications for the regulation of mitochondrial function. Biophys J. 1997;72:1954–1962. doi: 10.1016/S0006-3495(97)78841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komarov AG, Graham BH, Craigen WJ, Colombini M. The physiological properties of a novel family of VDAC-like proteins from Drosophila melanogaster. Biophys J. 2004;86:152–162. doi: 10.1016/S0006-3495(04)74093-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemasters JJ, Holmuhamedov E. Voltage-dependent anion channel (VDAC) as mitochondrial governator—Thinking outside the box. Biochim Biophys Acta. 2006;1762:181–190. doi: 10.1016/j.bbadis.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Tsujimoto Y. Bcl-2 family of proteins: Life-or-death switch in mitochondria. Biosci Rep. 2002;22:47–58. doi: 10.1023/a:1016061006256. [DOI] [PubMed] [Google Scholar]
- 9.Zamzami N, Kroemer G. The mitochondrion in apoptosis: How Pandora’s box opens. Nat Rev Mol Cell Biol. 2001;2:67–71. doi: 10.1038/35048073. [DOI] [PubMed] [Google Scholar]
- 10.Naghdi S, Várnai P, Hajnóczky G. Motifs of VDAC2 required for mitochondrial Bak import and tBid-induced apoptosis. Proc Natl Acad Sci USA. 2015;112:E5590–E5599. doi: 10.1073/pnas.1510574112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schredelseker J, et al. High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J Biol Chem. 2014;289:12566–12577. doi: 10.1074/jbc.M113.497438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geula S, Naveed H, Liang J, Shoshan-Barmatz V. Structure-based analysis of VDAC1 protein: Defining oligomer contact sites. J Biol Chem. 2012;287:2179–2190. doi: 10.1074/jbc.M111.268920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonçalves RP, Buzhynskyy N, Prima V, Sturgis JN, Scheuring S. Supramolecular assembly of VDAC in native mitochondrial outer membranes. J Mol Biol. 2007;369:413–418. doi: 10.1016/j.jmb.2007.03.063. [DOI] [PubMed] [Google Scholar]
- 14.Hoogenboom BW, Suda K, Engel A, Fotiadis D. The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J Mol Biol. 2007;370:246–255. doi: 10.1016/j.jmb.2007.04.073. [DOI] [PubMed] [Google Scholar]
- 15.Mannella CA. Conformational changes in the mitochondrial channel protein, VDAC, and their functional implications. J Struct Biol. 1998;121:207–218. doi: 10.1006/jsbi.1997.3954. [DOI] [PubMed] [Google Scholar]
- 16.Mannella CA. Structure of the outer mitochondrial membrane: Ordered arrays of porelike subunits in outer-membrane fractions from Neurospora crassa mitochondria. J Cell Biol. 1982;94:680–687. doi: 10.1083/jcb.94.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolder M, et al. Crystallization of the human, mitochondrial voltage-dependent anion-selective channel in the presence of phospholipids. J Struct Biol. 1999;127:64–71. doi: 10.1006/jsbi.1999.4141. [DOI] [PubMed] [Google Scholar]
- 18.Betaneli V, Petrov EP, Schwille P. The role of lipids in VDAC oligomerization. Biophys J. 2012;102:523–531. doi: 10.1016/j.bpj.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keinan N, Tyomkin D, Shoshan-Barmatz V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol Cell Biol. 2010;30:5698–5709. doi: 10.1128/MCB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemasters JJ, et al. The pH paradox in ischemia-reperfusion injury to cardiac myocytes. EXS. 1996;76:99–114. doi: 10.1007/978-3-0348-8988-9_7. [DOI] [PubMed] [Google Scholar]
- 21.Das S, Steenbergen C, Murphy E. Does the voltage dependent anion channel modulate cardiac ischemia-reperfusion injury? Biochim Biophys Acta. 2012;1818:1451–1456. doi: 10.1016/j.bbamem.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagadic-Gossmann D, Huc L, Lecureur V. Alterations of intracellular pH homeostasis in apoptosis: Origins and roles. Cell Death Differ. 2004;11:953–961. doi: 10.1038/sj.cdd.4401466. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Hail D, Shoshan-Barmatz V. VDAC1-interacting anion transport inhibitors inhibit VDAC1 oligomerization and apoptosis. Biochim Biophys Acta. 2016;1863:1612–1623. doi: 10.1016/j.bbamcr.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Hail D, et al. Novel compounds targeting the mitochondrial protein VDAC1 inhibit apoptosis and protect against mitochondria dysfunction. J Biol Chem. 2016;291:24986–25003. doi: 10.1074/jbc.M116.744284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bode BE, et al. Counting the monomers in nanometer-sized oligomers by pulsed electron-electron double resonance. J Am Chem Soc. 2007;129:6736–6745. doi: 10.1021/ja065787t. [DOI] [PubMed] [Google Scholar]
- 26.Hilger D, et al. Assessing oligomerization of membrane proteins by four-pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E. coli. Biophys J. 2005;89:1328–1338. doi: 10.1529/biophysj.105.062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 28.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: A case study of myoglobin. Biochemistry. 2012;51:6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong X, et al. Determining membrane protein-lipid binding thermodynamics using native mass spectrometry. J Am Chem Soc. 2016;138:4346–4349. doi: 10.1021/jacs.6b01771. [DOI] [PubMed] [Google Scholar]
- 30.Israelson A, Abu-Hamad S, Zaid H, Nahon E, Shoshan-Barmatz V. Localization of the voltage-dependent anion channel-1 Ca2+-binding sites. Cell Calcium. 2007;41:235–244. doi: 10.1016/j.ceca.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 31.De Pinto V, al Jamal JA, Palmieri F. Location of the dicyclohexylcarbodiimide-reactive glutamate residue in the bovine heart mitochondrial porin. J Biol Chem. 1993;268:12977–12982. [PubMed] [Google Scholar]
- 32.Pastorino JG, Hoek JB. Regulation of hexokinase binding to VDAC. J Bioenerg Biomembr. 2008;40:171–182. doi: 10.1007/s10863-008-9148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaptal V, et al. Fluorescence detection of heavy atom labeling (FD-HAL): A rapid method for identifying covalently modified cysteine residues by phasing atoms. J Struct Biol. 2010;171:82–87. doi: 10.1016/j.jsb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marty MT, et al. Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ujwal R, Cascio D, Chaptal V, Ping P, Abramson J. Crystal packing analysis of murine VDAC1 crystals in a lipidic environment reveals novel insights on oligomerization and orientation. Channels (Austin) 2009;3:167–170. doi: 10.4161/chan.3.3.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perozo E, Kloda A, Cortes DM, Martinac B. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat Struct Biol. 2002;9:696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Elferich J, Gouaux E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat Struct Mol Biol. 2012;19:212–219. doi: 10.1038/nsmb.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fillingame RH, Jiang W, Dmitriev OY. Coupling H(+) transport to rotary catalysis in F-type ATP synthases: Structure and organization of the transmembrane rotary motor. J Exp Biol. 2000;203:9–17. doi: 10.1242/jeb.203.1.9. [DOI] [PubMed] [Google Scholar]
- 40.Valiyaveetil F, Hermolin J, Fillingame RH. pH dependent inactivation of solubilized F1F0 ATP synthase by dicyclohexylcarbodiimide: pK(a) of detergent unmasked aspartyl-61 in Escherichia coli subunit c. Biochim Biophys Acta. 2002;1553:296–301. doi: 10.1016/s0005-2728(01)00251-1. [DOI] [PubMed] [Google Scholar]
- 41.Marcoline FV, Bethel N, Guerriero CJ, Brodsky JL, Grabe M. Membrane protein properties revealed through data-rich electrostatics calculations. Structure. 2015;23:1526–1537. doi: 10.1016/j.str.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teixeira VH, Vila-Viçosa D, Reis PB, Machuqueiro M. pK(a) values of titrable amino acids at the water/membrane interface. J Chem Theory Comput. 2016;12:930–934. doi: 10.1021/acs.jctc.5b01114. [DOI] [PubMed] [Google Scholar]
- 43.Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol. 2007;9:550–555. doi: 10.1038/ncb1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shoshan-Barmatz V, Keinan N, Zaid H. Uncovering the role of VDAC in the regulation of cell life and death. J Bioenerg Biomembr. 2008;40:183–191. doi: 10.1007/s10863-008-9147-9. [DOI] [PubMed] [Google Scholar]
- 45.Zalk R, Israelson A, Garty ES, Azoulay-Zohar H, Shoshan-Barmatz V. Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem J. 2005;386:73–83. doi: 10.1042/BJ20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu S, Narita M, Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 47.Shoshan-Barmatz V, Zakar M, Rosenthal K, Abu-Hamad S. Key regions of VDAC1 functioning in apoptosis induction and regulation by hexokinase. Biochim Biophys Acta. 2009;1787:421–430. doi: 10.1016/j.bbabio.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 49.Fleissner MR, et al. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc Natl Acad Sci USA. 2011;108:16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 51.Hubbell WL, Froncisz W, Hyde JS. Continuous and stopped flow EPR spectrometer based on a loop gap resonator. Rev Sci Instrum. 1987;58:1879–1886. [Google Scholar]
- 52.Polyhach Y, et al. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys Chem Chem Phys. 2012;14:10762–10773. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- 53.Jeschke G, et al. DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 54.Laganowsky A, Reading E, Hopper JT, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones EOE, et al. 2001 SciPy: Open Source Scientific Tools for Python. Available at https://www.scipy.org/. Accessed December 12, 2017.
- 56.van der Walt S, Colbert SC, Varoquaux G. The NumPy array: A structure for efficient numerical computation. Comput Sci Eng. 2011;13:22–30. [Google Scholar]
- 57.Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- 58.Katchalski-Katzir E, et al. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Press WH. 2007. Numerical Recipes: The Art of Scientific Computing (Cambridge Univ Press, New York), 1235 p.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.