Significance
NEET proteins belong to a unique family of iron-sulfur (Fe-S) proteins that regulate iron and reactive oxygen homeostasis and are involved in the progression of cancer, diabetes, neurodegeneration, and aging. Of the three human NEET proteins, the structure and function of the mitochondrial matrix-localized mitochondrial inner NEET protein (MiNT) are unknown. Here, we show that MiNT is a pseudosymmetrical monomeric protein that contains two distinct Fe-S cluster-binding motifs. MiNT transfers its clusters to the human mitochondrial ferredoxins FDX1/adrenodoxin and FDX2 and is required for regulating iron and reactive oxygen species levels in the mitochondria. Our study demonstrates that although MiNT differs in structure and localization from its homodimeric NEET counterparts, it nevertheless cooperates with them in the same important pathway.
Keywords: NEET proteins, mitochondria, iron homeostasis, iron-sulfur proteins, cancer
Abstract
The NEET family is a relatively new class of three related [2Fe-2S] proteins (CISD1–3), important in human health and disease. While there has been growing interest in the homodimeric gene products of CISD1 (mitoNEET) and CISD2 (NAF-1), the importance of the inner mitochondrial CISD3 protein has only recently been recognized in cancer. The CISD3 gene encodes for a monomeric protein that contains two [2Fe-2S] CDGSH motifs, which we term mitochondrial inner NEET protein (MiNT). It folds with a pseudosymmetrical fold that provides a hydrophobic motif on one side and a relatively hydrophilic surface on the diametrically opposed surface. Interestingly, as shown by molecular dynamics simulation, the protein displays distinct asymmetrical backbone motions, unlike its homodimeric counterparts that face the cytosolic side of the outer mitochondrial membrane/endoplasmic reticulum (ER). However, like its counterparts, our biological studies indicate that knockdown of MiNT leads to increased accumulation of mitochondrial labile iron, as well as increased mitochondrial reactive oxygen production. Taken together, our study suggests that the MiNT protein functions in the same pathway as its homodimeric counterparts (mitoNEET and NAF-1), and could be a key player in this pathway within the mitochondria. As such, it represents a target for anticancer or antidiabetic drug development.
The NEET family comprises a unique class of iron-sulfur (Fe-S) proteins that harbor a 3Cys-1His cluster-binding domain (1, 2). Also known as CDGSH proteins, due to their signature [2Fe-2S]-binding domain consensus sequence [C-X-C-X2-(S/T)-X3-P-X-C-D-G-(S/A/T)-H], these proteins are highly conserved from bacteria to humans (3). The 3Cys-1His cluster-binding site of NEET proteins houses a [2Fe-2S] redox-sensing cluster that can be transferred to apo-acceptor protein(s) when the cluster is oxidized (2, 4–7). The [2Fe-2S] clusters of NEET proteins can also participate in electron transfer reactions (8–11). Our initial analysis of the unique fold and structural features of mitoNEET (mNT) led us to propose potential functions of NEETs (12). Biological studies confirmed our initial proposals and led to the hypothesis that the NEETs participate in the regulation of different iron, Fe-S, and reactive oxygen/redox reactions in cells (2, 13). Indeed, NEET proteins play a key role in many cellular functions, as well as in different human diseases (2, 6, 13–18).
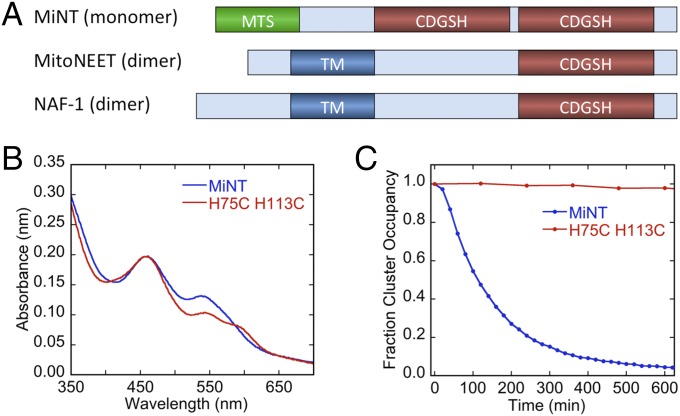
Three genes encode for NEET proteins in humans: CISD1, CISD2, and CISD3 (1, 3). The first human NEET protein discovered was mNT (CISD1), which was initially identified as a target for the antidiabetic drug pioglitazone (19) and, at that time, annotated as a zinc finger protein (1). This outer mitochondrial membrane (OMM)-tethered protein that faces the cytosol, defined a new class of Fe-S–binding proteins and introduced the unique NEET fold; a soluble, strand-swapped, homodimeric, two-domain (beta-cap and cluster-binding) structure (12, 20, 21). The strand-swapped structure has C2 symmetry with one [2Fe-2S] cluster bound per protomer and the swapped-strand beta-structure contributing to long-range allostery in the dimeric native protein (1, 12, 20, 22–25). The longevity factor protein NAF-1 (CISD2) is similar in structure and sequence to mNT (54% identical and 69% similar) (20), but is primarily localized to the cytosolic side of the endoplasmic reticulum (ER), OMM, and the membranes that connect the ER and the mitochondria (1, 20). In contrast to mNT and NAF-1, the protein encoded by CISD3 resides inside the mitochondrial matrix, and we term it MiNT for mitochondrial inner NEET protein (previously assigned as Miner2). MiNT differs from the other human NEETs as it contains two CDGSH cluster-binding motifs within a single polypeptide chain (1, 3, 26) (Fig. 1A). Both mNT and NAF-1 play key roles in regulating autophagy, apoptosis, and mitochondrial iron and reactive oxygen species (ROS) homeostasis (2, 13, 15, 16, 27). In addition, they are associated with the progression of diabetes, obesity, neurodegeneration, heart disease, and cancer (14, 16–18, 28). A missense mutation that abolishes NAF-1 function results in a genetic disease in humans called Wolfram syndrome 2, causing optical nerve atrophy, severe hearing impairment, early onset of diabetes, gastrointestinal ulcers, abnormal platelet aggregation that cumulatively leads to lower quality of life, and greatly diminished life expectancy (18, 29).
Fig. 1.
Domain organization of the NEET proteins and mutation of the cluster-coordinating His ligand of MiNT. (A) Domain organization. All members of the NEET family contain at least one CDGSH domain that binds a [2Fe-2S] cluster. MiNT has two CDGSH cluster-binding domains in a single polypeptide chain, while mNT and NAF-1 each have one domain per chain. MiNT lacks a transmembrane domain, but contains a classic cleavable mitochondrial targeting sequence. (B and C) Mutation of the cluster-coordinating His ligand to Cys stabilizes the [2Fe-2S] cluster. (B) UV-Vis absorption spectra of MiNT wild-type and H75C/H113C mutant proteins. (C) Relative stabilities of the [2Fe-2S] clusters of wild-type and H75C/H114C mutant MiNTs were measured by monitoring the decrease in absorbance of the 458-nm peak at pH 8.0 and 37 °C over time. Stability of the H75C/H113C mutant is greatly increased relative to wild-type MiNT under these conditions.
In contrast to mNT and NAF-1, very little is known about the structure and function of the human MiNT (CISD3). This lack of knowledge is in sharp contrast to the role MiNT plays in numerous cancers, as reflected by its high expression in human cancer cells (SI Appendix, Fig. S1). Indeed, MiNT is identified as an essential gene in high-resolution genetic vulnerability analyses in a recent study (30). Also, whereas most [2Fe-2S] clusters are destroyed by binding of nitric oxide (NO), MiNT is reported to stably bind NO (31). This feature of MiNT could suggest that it has a role in NO signaling. Proteomics studies have reported several protein partners of MiNT. Among those identified interaction partners, the proteins involved in import and processing of mitochondrial matrix proteins, including translocases of the inner and outer membrane and matrix peptidases, are consistent with its processing and localization. Potential functional partners include multiple components of respiratory complex I, ribosomal RNA-binding proteins, and GST (32).
In support of a possible role for MiNT in cancer, a recent study identified changes in CpG islands of methylation around the CISD3 gene, associated with its altered expression in aggressive pediatric brain tumors (33). To begin investigating the role of MiNT in cancer (SI Appendix, Fig. S1), as well as to provide new drug design targets associated with MiNT and other NEET proteins (34), we crystallized human MiNT and studied its interactions with apo-acceptor human proteins that reside within the mitochondrial matrix, as well as its function in cancer cells. We report the crystal structure of a stable form of the monomeric human MiNT [Protein Data Bank (PDB) ID code 6AVJ]. As opposed to mNT and NAF-1, the protein maintains a modified NEET fold but presents two distinctly different surfaces surrounding the clusters, and the two domains undergo distinctly asymmetrical dynamics in our computational model. Additionally, we show that MiNT transfers its [2Fe-2S] clusters to the human mitochondrial matrix ferredoxins, FDX1 (adrenodoxin) and FDX2, with high efficiency. Cellular studies of knockdown of MiNT in situ result in decreased mitochondrial membrane potential (MMP), as well as increased mitochondrial iron and ROS accumulation. Taken together, the similarity of the cellular effects of MiNT expression knockdown to those observed for the cytosolic-facing NAF-1 and mNT (15) and its ability to efficiently transfer [2Fe-2S] clusters indicate that MiNT coordinates a complementary role in mitochondrial iron and ROS regulation within the mitochondrial matrix.
Results
The NEET family of [2Fe-2S] proteins have been the focus of many structural/spectroscopic and biological studies since the unusual mNT was first implicated in diabetes (19); however, MiNT has remained largely uninvestigated until now. As MiNT resides inside the mitochondria, where iron accumulates under stress conditions, and its expression is linked to cancer progression, we sought to investigate the structure/function properties of this structurally distinct (on the domain motif level; Fig. 1) important protein. Bioinformatics studies indicate that MiNT carries two [2Fe-2S] cluster motifs on a single polypeptide chain that share the NEET hallmark CDGSH motif (Fig. 1A).
MiNT Is a Monomeric Protein Harboring Two [2Fe-2S] Clusters.
We expressed and purified wild-type MiNT. This protein was used for functional studies but was not amenable to our crystallization conditions. Based on our extensive knowledge of mNT and NAF-1 stability (4, 12, 20, 35), we mutated the coordinating His ligands to Cys (H75C, H113C), as our experience with mNT and NAF-1 indicates that these mutations significantly increase the [2Fe-2S] cluster stability with no significant effect on the global backbone fold (4, 35). A comparison of the UV-visible (Vis) spectra of wild-type and His-to-Cys mutant MiNT proteins shows the expected increase in absorption at 425 nm relative to the 458-nm peak, consistent with the change in the coordinating ligands of the [2Fe-2S] clusters (2, 35) (Fig. 1B). The cluster stability can be assessed by the time-dependent change of the absorption peak at 458 nm, as described previously (36). The change to a 4Cys coordination for each of the [2Fe-2S] clusters significantly enhances the stability of MiNT (Fig. 1C), making it suitable for crystallization under our conditions (discussed below). Interestingly, wild-type MiNT is significantly less stable than mNT and NAF-1 under the same conditions, with its clusters maintaining a half-life of ∼110 min at 37 °C and pH 8 for MiNT (Fig. 1C). For comparison, the cluster half-lives of mNT and NAF-1 are ≥10,000 min and ≥1,000 min, respectively (20). We analyzed the oligomeric state of both wild-type and mutant MiNT by analytical size exclusion chromatography (SEC; SI Appendix, Fig. S2); both are monomeric.
Crystal Structure of MiNT (H75C, H113C).
We performed parallel screens on wild-type and mutant MiNT (H75C, H113C) proteins. The stable, mutant protein was highly amenable to X-ray structure determination in our current studies. MiNT (H75C, H113C) crystallized in the monoclinic space group C121, and data were collected on a single crystal that diffracted to 1.9 Å (SI Appendix, Table S1). The protein structure was determined by molecular replacement. The asymmetric unit contained three copies of the protein, but it is monomeric in solution (SI Appendix, Fig. S2).
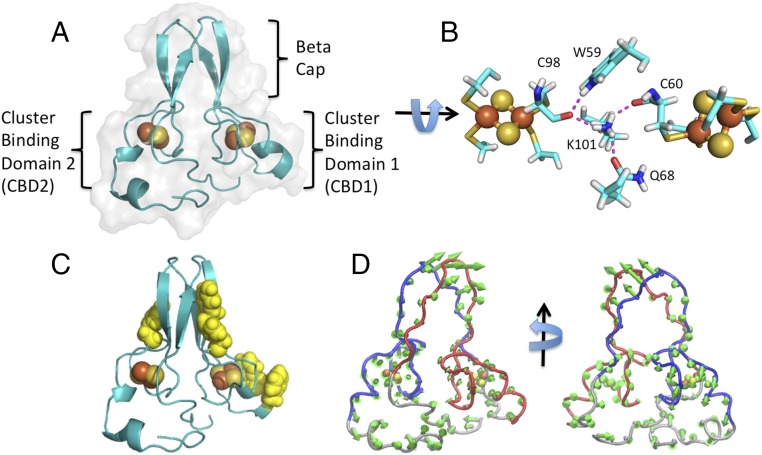
The human MiNT fold consists of a single polypeptide chain with a four-stranded beta-cap domain residing between the two CDGSH [2Fe-2S] cluster-binding domains (Fig. 2A). Two of the beta-strands are on the N-terminal side of the protein, while the other two are between the cluster-binding domains. Cluster-binding domain-1 (CBD1) consists of a loop region that starts from the end of the first two strands of the beta-cap and contains three of the ligating Cys residues (C59, C62, and C71), followed by a short five-residue alpha-helical segment that contains the mutant fourth ligand (H75C). A loop region connects this segment to the second pair of beta-strands. CBD2 has a loop that starts after the end of the beta-cap domain and also contains three of the ligating Cys residues (C98, C100, and C109) for the cluster. The other cluster ligand, Cys113 (H113C), resides within a short three-residue helical segment. This helix is disrupted by Ser115, which uses its side-chain hydroxyl to hydrogen-bond to the main chain of the preceding T111 and the following V118, inducing a sharp turn that is followed by a perpendicular five-residue helix. Each cluster-binding domain of MiNT contains a cis X-proline peptide bond that is critical for positioning the cluster-coordinating residues. These cis-proline peptide bonds are also conserved across human NEETs within the cluster-binding domain (12, 20). While most of the interactions in the core of the protein are hydrophobic, there are hydrogen bonds that connect the two cluster-binding domains. The Lys101 side chain from CBD2 forms hydrogen bonds with the carbonyls of cluster-coordinating Cys ligands (C60 and C98) from each cluster, as well as with Gln68 from CBD1. The indole ring of Trp59 from the bottom of the beta-cap on the CBD1 side of the protein also forms a hydrogen bond that connects to the carbonyl of Cys60 of CBD2. These connections may facilitate communication between the two [2Fe-2S] clusters (Fig. 2B).
Fig. 2.
Structural organization of monomeric MiNT highlights the C2 pseudosymmetry and the asymmetrical surface. (A) MiNT contains a four-stranded beta-cap domain composed of two antiparallel hydrogen-bonded stranded sheets that dock to form the four-stranded cap domain. Each contains a structured loop and a turn of helix that contains the [2Fe-2S] cluster-coordinating ligands. However, CBD2 has additional helix and long N- and C-terminal loop extensions. (B) Close-up of the cluster-coordinating ligands as well as the hydrogen-bond network that connects the two clusters across the pseudosymmetrical axis. Residue numbers are indicated. (C) Ribbon diagram of MiNT with surface aromatics highlighted. The clear asymmetry in the surface of MiNT is evident in this representation. (D) Displacements around the average structure for the first principal component of simulated native-state dynamics, scaled 10-fold.
The overall domain organization of MiNT is similar to the dimeric NEET proteins, with two cluster-binding domains and a beta-cap domain. However, mNT and NAF-1 have six total strands that make up their beta-cap domains, three strands in each sheet that contains an interprotomer swapped strand, while MiNT has only four beta-strands with no swapping across the symmetry axis. Additionally, the MiNT fold has C2 pseudosymmetry (because of the differences in amino acid composition of each side), while the dimeric NEETs have true C2 symmetry (12, 20). In contrast to the backbone fold, the side chains on the surface of each side of MiNT are distinctly different. The CBD1 side of MiNT has four aromatic side chains on its surface, with a Phe residue just before the CDGSH sequence and two consecutive Phe residues immediately after the last [2Fe-2S] ligand, as well as a Tyr on the beta-cap surface. In contrast, CBD2 only has a single Tyr on the beta-cap surface and no surface aromatics near the [2Fe-2S] cluster-binding domain (Fig. 2C). The homodimeric mNT and NAF-1 proteins each have a symmetrical arrangement of surface aromatics. mNT has four on each side, similar to the one surface of MiNT CBD1, while NAF-1 has only two aromatics on each surface (12, 20).
Simulations of Native-State Dynamics.
A structure-based model (SBM) (37–39) of MiNT was generated from our crystal structure of MiNT (H75C, H113C). SBM interactions are assigned based on the insight from energy-landscape theory that native-state protein interactions are uniquely favorable, and must possess the property of minimal frustration to make the protein foldable (37). The resulting simple model enables efficient simulations of folding and functional native-state dynamics (39–43). Here, we used an all-atom representation (39) that explicitly included the [2Fe-2S] clusters. We simulated the dynamics of folded MiNT at constant temperature. For the set of CA atoms, [2Fe-2S] cluster atoms, and coordinating Cys SG atoms, principal component analysis (PCA) (44, 45) was used to extract the robust, slow component of the dynamics from the simulated trajectories. SI Appendix, Fig. S3A shows the components of the first eigenvector of the covariance matrix. The corresponding displacements from the average structure are shown as green arrows in Fig. 2D, scaled by a factor of 10 for visibility. The symmetry of motions by the two domains can be assessed easily, because the y axis of the chosen coordinate system for the PCA corresponds to the pseudo-C2 rotation axis of the protein, and the z axis is parallel to the vector connecting the centers of mass of the two domains. For motions that preserve the C2 symmetry, displacements of corresponding residues in the two domains should have equal magnitudes but opposite signs in both the X and Z directions. Instead, the most prominent feature is a peak in the Z component that is negative for both domains, corresponding to a joint motion of the two cluster-binding domains. Furthermore, the overall motions of the two cluster-binding regions are also different in magnitude, as can be seen in SI Appendix, Fig. S3B, where the amplitude of fluctuations due to the first principal component is plotted.
MiNT Transfers Its [2Fe-2S] Clusters to Human Mitochondrial Matrix Ferredoxins.
We discovered that both mNT and NAF-1 can transfer their [2Fe-2S] clusters to apo-acceptor proteins, including the human cytosolic Anamorsin (5) as well as the universal acceptor protein ferredoxin from Mastigocladus laminosus (mFd) (7, 46). Humans have two [2Fe-2S] cluster-binding ferredoxins (FDX1/adrenodoxin and FDX2) that both localize to the mitochondrial matrix (47). Because MiNT is also localized to the matrix, we wanted to determine if it could donate its [2Fe-2S] clusters to these important proteins (47–50). These studies were carried out with wild-type MiNT at 25 °C to allow for easy monitoring of the transfer reaction with UV-Vis absorption spectroscopy. Wild-type MiNT has the characteristic peak at 458 nm, while both ferredoxins have an additional peak around 420 nm. We use the ratio between these two absorbance peaks (420 nm and 458 nm) to track the progress of the transfer from holo-MiNT to each apo-ferredoxin (7, 46). The transfer reaction is very fast; the first spectra collected after the ∼5–10 s of dead time between MiNT addition and collection of the first spectrum was more than 40% complete for transfer to FDX1 and more than 30% complete for transfer to FDX2 (Fig. 3). MiNT can transfer to FDX1 and FDX2 with a half-time of transfer of ca. 10 min and 15 min, respectively. MiNT can also transfer to mFd with a transfer half-time of <10 min (SI Appendix, Fig. S4). In contrast, both mNT and NAF-1 transfer much more slowly to ferredoxin, with a half-time of between 100 and 200 min at 37 °C (7, 46).
Fig. 3.
MiNT donates its [2Fe-2S] clusters to human mitochondrial ferredoxins. The [2Fe-2S] cluster transfer from holo-MiNT to human apo-FDX1 and to apo-FDX2 was monitored by UV-Vis absorption spectroscopy. Spectra from select time points of the transfer to apo-FDX1 (A) and to apo-FDX2 (C) are shown. Transfer progress as determined by the ratio of absorbance at 420 nm to 458 nm vs. time is shown for the transfer to apo-FDX1 (B) and to apo-FDX2 (D). The traces shown were obtained with 15 μM MiNT (30 μM [2Fe-2S] clusters) and 60 μM apo-FDX1 or apo-FDX2 at 25 °C. AU, absorbance units.
Suppression of MiNT Expression Impairs Mitochondrial Function and Enhances Mitochondrial Iron and ROS Accumulation.
To determine the role that MiNT plays in mitochondria, we suppressed its expression in human breast cancer (MDA-MB-231) cells using shRNA of CISD3. Suppression of CISD3 expression resulted in a decrease in MMP [measured with tetramethylrhodamine ethyl ester (TMRE)], indicating that MiNT is important for mitochondrial function (Fig. 4). Suppression of MiNT expression further resulted in enhanced accumulation of mitochondrial labile iron (mLI; measured with B-[(1,10-phenanthrolin-5-yl) aminocarbonyl] benzyl ester [RPA]), as well as in a build-up of mitochondrial ROS (mROS) levels [measured with the fluorescent superoxide (SOX) indicator mitoSOX], demonstrating that MiNT is essential for maintaining mitochondrial iron and ROS homeostasis (Fig. 4). Interestingly, all of the above measured alterations in mitochondrial metabolism, due to suppression of MiNT expression, could be corrected by the addition of the iron chelator deferiprone (DFP), demonstrating that MiNT suppression affected mitochondrial function primarily through increasing the levels of labile iron (Fig. 4). The outcome of suppressing MiNT expression in cells (Fig. 4) was similar to the effects of suppressing mNT and/or NAF-1 in breast cancer cells (15, 51), suggesting that all three NEET proteins are involved in maintaining mitochondrial iron and ROS homeostasis in mammalian cells.
Fig. 4.
Impaired mitochondrial function and enhanced mitochondrial iron and ROS accumulation in cancer cells with suppressed expression of MiNT. (A) Decreased MMP in MDA-MB-231 cancer cells with suppressed MiNT expression. (Top) Confocal microscopy images of control cells (transformed with scrambled vector), and two different cell lines with suppressed expression of MiNT [CISD3(−)A and CISD3(−)D], stained with the MMP indicator TMRE (0.1 μM). (Bottom) Bar graph showing quantification of TMRE fluorescence in the different cell lines calculated for n = 3 independent experiments, each with 90 cells per group. ***P < 0.001. Pretreatment with DFP (50 μM) for 16 h prevents the effect of CISD3 suppression on MMP. r.u., relative units. (B) Enhanced accumulation of mLI in cancer cells with suppressed CISD3 expression. (Left) Bar graph showing a decrease (quenching) in RPA fluorescence (indicative of high mLI levels) in two different cell lines with suppressed expression of MiNT [CISD3(−)A and CISD3(−)D], compared with control. Pretreatment with DFP (100 μM) for 60 min prevents the effect of CISD3 suppression on mLI. (Right) Bar graph showing the relative RPA fluorescence change upon addition of DFP, indicative of the relative change in mLI induced by the iron chelator. Results are calculated for n = 3 independent experiments each with 90 cells per group. ***P < 0.001. Δr.u., change in relative units. (C) Enhanced accumulation of mROS in cancer cells with suppressed MiNT expression compared with control. mROS was measured following 60 min of incubation with mitoSOX (5 μM). Pretreatment with DFP (100 μM) for 60 min prevents the effect of CISD3 suppression.
Discussion
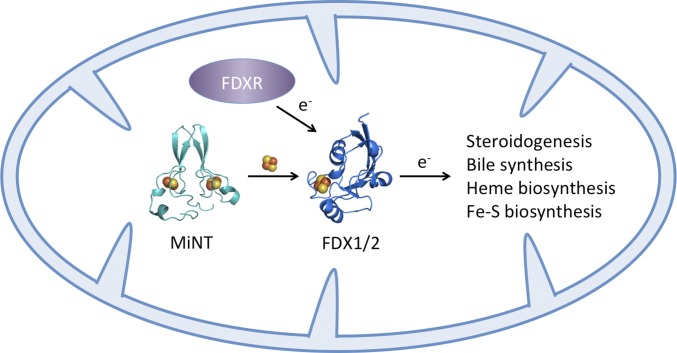
Cellular iron levels are tightly controlled to prevent toxic iron and subsequent unregulated ROS accumulation. We discovered that the NEET family proteins are key regulators of iron and ROS homeostasis (2, 13, 15, 21). An open question to be addressed was whether MiNT was also involved in Fe-S biology and disease mechanisms (3). We now show that MiNT transfers [2Fe-2S] clusters to the mitochondrial matrix acceptor proteins FDX1 and FDX2. Both FDX1 and FDX2 function in assembly of Fe-S clusters. Each receives electrons from ferredoxin reductase (FDXR) and supplies those electrons for Fe-S cluster assembly on the iron-sulfur assembly scaffold (ISCU) (49, 50) (Fig. 5). In addition to Fe-S cluster biosynthesis, FDX1 and FDX2 are essential for heme biosynthesis (47, 49). FDX1 also supplies electrons used for steroidogenesis and bile synthesis (47). Knockdown of FDX1, FDX2, or FDXR results in mitochondrial iron accumulation (47, 49), a similar phenotype to what we observed for CISD3 knockdown (Fig. 4). This indicates an important role for MiNT in regulation of mitochondrial iron levels. Reduction of MiNT expression would decrease the level of active ferredoxins inhibiting production of Fe-S clusters and heme in the mitochondria, causing increased levels of free iron that would otherwise be incorporated into these prosthetic groups.
Fig. 5.
MiNT provides a link between FDXR and ferredoxins (FDX1 and FDX2) in the mitochondrial matrix. MiNT provides [2Fe-2S] clusters produced inside the mitochondria by the iron-sulfur cluster assembly system (ISC) to FDX1 and FDX2. MiNT can provide parallel routes linking the ISC to mitochondrial matrix processes. Matrix-produced clusters are important for cell metabolism, maintenance, and proliferation.
Both mNT and NAF-1 are up-regulated in breast cancer cells, and knocking down either of their protein levels reduces tumor growth and causes accumulation of iron and ROS in the mitochondria (15). We have shown that these proteins can be targeted with small-molecule anticancer compounds that cause release of their [2Fe-2S] clusters (34). Like the other NEETs, MiNT levels are up-regulated in breast cancer (SI Appendix, Fig. S1) as well as other cancers, including ependymoma (33), lymphoma, and liver cancer (SI Appendix, Fig. S1; The Human Protein Atlas) (52). In this study, we knocked down MiNT and observed a similar accumulation of iron and ROS in the mitochondria. Together, this shows that MiNT is another possible target for anticancer drugs that affect its ability to regulate iron levels.
Our structural analysis of MiNT reveals that its overall architecture resembles that of NAF-1 and mNT in their homodimeric forms. While MiNT is a monomer, it maintains the overall “NEET-fold” containing two main domains: a beta-cap and two cluster-binding domains (Fig. 2). However, in contrast to mNT and NAF-1, both the surfaces and the motions of the two cluster-binding domains of MiNT are asymmetrical. This suggests that the two MiNT clusters likely have different functions, stabilities, and binding partners. Additionally, the intramolecular coordination of the two clusters may play an important role in MiNT’s function. Although MiNT and the bacterial homolog (mmCISD) show a high degree of conservation (26), they have important differences. MiNT contains longer beta-strands in the beta-cap region as well as additional helical content. There is an alpha-helical region within the C terminus of MiNT that is absent in mmCISD, as well as an additional short helical region in CBD1. Furthermore, the surfaces are very different as MiNT has an asymmetrical arrangement of aromatic residues on its surface, with two surface Phe residues immediately following the [2Fe-2S] cluster in CBD1 that are absent in mmCISD.
Taken together, our study indicates that MiNT functions within the mitochondria to regulate levels of toxic iron and ROS similar to its cytosolic-facing counterparts, mNT and NAF-1. Despite its distinct localization and structural differences, MiNT still maintains a similar function. As such, it may be an attractive target for anticancer or antidiabetic drug designs.
Materials and Methods
Protein Expression, Purification, and SEC Analysis.
Procedures for expression and purification of MiNT, FDX1, FDX2, and mFd, as well as SEC analysis of MiNT, can be found in SI Appendix, SI Materials and Methods.
Crystallization, X-Ray Data Collection, and Structure Determination.
Crystals of MiNT H75C/H113C were prepared by sitting-drop vapor diffusion. Three microliters of reservoir solution [100 mM Bis-Tris propane (pH 8.5), 2.5 M ammonium sulfate, and 3% vol/vol polyethylene glycol 400] were added to 3 μL of 750 μM protein. Single crystals appeared after 2 d at 20 °C. Crystals were cryoprotected in reservoir solution with 10% glycerol and frozen at 77 K. Diffraction data were collected using a Micromax-007HF copper anode X-ray source with Varimax HF optics (Rigaku) and a Platinum 135 CCD detector (Bruker). Data were processed with APEX3 software (Bruker). The structure was determined by molecular replacement using a homolog from the bacterium Magnetospirillum magneticum (PDB ID code 3TBN) as a search model. A partial model was built using Phenix Autobuild and completed using Coot. The model was refined using Phenix and verified with MOLProbity.
Measurement of [2Fe-2S] Cluster Stability and Transfer Kinetics.
All UV-Vis absorption spectra were recorded from 350 to 700 nm on a Cary 50 spectrophotometer (Varian). Stability of the [2Fe-2S] cluster of wild-type MiNT and H75C/H113C mutant was determined by monitoring the decrease in absorbance of the [2Fe-2S] cluster at 458 nm as a function of time at 37 °C in 50 mM Bis-Tris propane and 100 mM NaCl at pH 8.0. Data were normalized and plotted with Kaleidagraph (Synergy Software).
For cluster transfer measurement, 60 μM apo-FDX1, FDX2, or mFd was incubated in 50 mM Bis-Tris propane, 100 mM NaCl, 1 mM EDTA, and 2 mM DTT at pH 8.0 at 37 °C for 1 h. The temperature was adjusted to 25 °C, and 15 μM MiNT was added to start the reaction. The ratio of the absorbance at 420 nm to 458 nm was used to monitor transfer progress. Data were normalized and fit to a single exponential rise.
Structure-Based Molecular Dynamics Simulations.
An SBM (37–39, 53) of MiNT H75C H113C was prepared, using the SMOG-2 software tool (54). All heavy atoms were represented, including the [2Fe-2S] clusters, which were defined as a new ligand type in a custom model template created as an extension of the standard all-atom SBM parameters (39, 54). Iron was assigned a reduced mass of 4, and all Fe-S connections were treated as covalent bonds, represented by harmonic potentials. The model was simulated using a customized version of gromacs 4.5 (53, 55). Four independent runs were started from the crystal structure and propagated in the folded state. A constant reduced temperature of 0.91 T* (where T* is defined by the condition kBT = 1 at T = T*, where kB is the Boltzmann constant and T is temperature) was maintained using a Langevin thermostat with a time constant of 2 ps. Each run was propagated for 200 million steps with a step size of 2 fs, giving an overall simulation time of 1.6 μs. PCA of the trajectories was carried out using the gromacs tools g_govar and g_anaeig, and results were visualized using MATLAB and Visual molecular dynamics (VMD) (56).
Cell Culture and Fluorescence Microscopy.
MDA-MB-231 cells were grown as described by Sohn et al. (15). Plasmid for suppressing MiNT expression [shRNA-MiNT/CISD3] was pGFP-RS vector. Genjuice (EMD Millipore) was used for transfection (15). Stable cell lines were obtained by FACS sorting. MDA-MB-231 cells were cultured in glass-bottom microscope dishes for assessing MMP, mitochondrial iron, and ROS using confocal microscopy (Nikon A1R) with gallium arsenide phosphide detectors and four lasers (15, 27). MMP was measured with TMRE (15). Accumulation of mLI was determined by RPA (15). Accumulation of mROS was determined with mitoSOX Red (M36008; Invitrogen) (15). The iron chelator DFP (ferriprox, 1,2-dimethyl-3-hydroxypyridin-4-one; Apo Pharma) was used to determine the dependency of MMP, Mli, and mROS on iron levels in cells (6, 15).
Supplementary Material
Acknowledgments
Work at the laboratory of P.A.J. is supported by NIH Grant GM101467. Work at the Center for Theoretical Biological Physics is sponsored by the National Science Foundation (NSF; Grants PHY-1427654 and CHE-1614101). R.N. and R.M. acknowledge the support of NSF-Binational Science Foundation (BSF) Grant NSF-MCB-1613462 (to R.M.) and BSF Grant 2015831 (to R.N.). Work at the laboratory of R.N. is also supported by funding from the Israel Science Foundation (Grant ISF-865/13). The funders had no role in the design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6AVJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715842115/-/DCSupplemental.
References
- 1.Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc Natl Acad Sci USA. 2007;104:5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamir S, et al. Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim Biophys Acta. 2015;1853:1294–1315. doi: 10.1016/j.bbamcr.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Inupakutika MA, et al. Phylogenetic analysis of eukaryotic NEET proteins uncovers a link between a key gene duplication event and the evolution of vertebrates. Sci Rep. 2017;7:42571. doi: 10.1038/srep42571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamir S, et al. A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties. Acta Crystallogr D Biol Crystallogr. 2014;70:1572–1578. doi: 10.1107/S1399004714005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipper CH, et al. Cancer-related NEET proteins transfer 2Fe-2S clusters to anamorsin, a protein required for cytosolic iron-sulfur cluster biogenesis. PLoS One. 2015;10:e0139699. doi: 10.1371/journal.pone.0139699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darash-Yahana M, et al. Breast cancer tumorigenicity is dependent on high expression levels of NAF-1 and the lability of its Fe-S clusters. Proc Natl Acad Sci USA. 2016;113:10890–10895. doi: 10.1073/pnas.1612736113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuris JA, et al. Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc Natl Acad Sci USA. 2011;108:13047–13052. doi: 10.1073/pnas.1109986108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landry AP, Cheng Z, Ding H. Reduction of mitochondrial protein mitoNEET [2Fe-2S] clusters by human glutathione reductase. Free Radic Biol Med. 2015;81:119–127. doi: 10.1016/j.freeradbiomed.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry AP, et al. Flavin nucleotides act as electron shuttles mediating reduction of the [2Fe-2S] clusters in mitochondrial outer membrane protein mitoNEET. Free Radic Biol Med. 2017;102:240–247. doi: 10.1016/j.freeradbiomed.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergner M, et al. Model of the mitoNEET [2Fe-2S] cluster shows proton coupled electron transfer. J Am Chem Soc. 2017;139:701–707. doi: 10.1021/jacs.6b09180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Landry AP, Ding H. The mitochondrial outer membrane protein mitoNEET is a redox enzyme catalyzing electron transfer from FMNH2 to oxygen or ubiquinone. J Biol Chem. 2017;292:10061–10067. doi: 10.1074/jbc.M117.789800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paddock ML, et al. MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc Natl Acad Sci USA. 2007;104:14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mittler RD-YM, et al. NEET proteins: A new link between iron metabolism, ROS and cancer. Antioxid Redox Signal, in press. [DOI] [PMC free article] [PubMed]
- 14.Kusminski CM, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18:1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sohn YS, et al. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci USA. 2013;110:14676–14681. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29:606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YF, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23:1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danielpur L, et al. GLP-1-RA corrects mitochondrial labile iron accumulation and improves β-cell function in type 2 Wolfram syndrome. J Clin Endocrinol Metab. 2016;101:3592–3599. doi: 10.1210/jc.2016-2240. [DOI] [PubMed] [Google Scholar]
- 19.Colca JR, et al. Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol Endocrinol Metab. 2004;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- 20.Conlan AR, et al. Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram syndrome 2. J Mol Biol. 2009;392:143–153. doi: 10.1016/j.jmb.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nechushtai R, et al. Characterization of Arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell. 2012;24:2139–2154. doi: 10.1105/tpc.112.097634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Zhou T, Ye K, Wang J. Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins. Proc Natl Acad Sci USA. 2007;104:14640–14645. doi: 10.1073/pnas.0702426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou X, et al. Crystallographic studies of human mitoNEET. J Biol Chem. 2007;282:33242–33246. doi: 10.1074/jbc.C700172200. [DOI] [PubMed] [Google Scholar]
- 24.Baxter EL, Jennings PA, Onuchic JN. Strand swapping regulates the iron-sulfur cluster in the diabetes drug target mitoNEET. Proc Natl Acad Sci USA. 2012;109:1955–1960. doi: 10.1073/pnas.1116369109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxter EL, et al. Allosteric control in a metalloprotein dramatically alters function. Proc Natl Acad Sci USA. 2013;110:948–953. doi: 10.1073/pnas.1208286110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Zhang L, Lai S, Ye K. Structure and molecular evolution of CDGSH iron-sulfur domains. PLoS One. 2011;6:e24790. doi: 10.1371/journal.pone.0024790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holt SH, et al. Activation of apoptosis in NAF-1-deficient human epithelial breast cancer cells. J Cell Sci. 2016;129:155–165. doi: 10.1242/jcs.178293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du X, et al. NAF-1 antagonizes starvation-induced autophagy through AMPK signaling pathway in cardiomyocytes. Cell Biol Int. 2015;39:816–823. doi: 10.1002/cbin.10453. [DOI] [PubMed] [Google Scholar]
- 29.Amr S, et al. A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am J Hum Genet. 2007;81:673–683. doi: 10.1086/520961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart T, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Z, Landry AP, Wang Y, Ding H. Binding of nitric oxide in CDGSH-type [2Fe-2S] clusters of the human mitochondrial protein Miner2. J Biol Chem. 2017;292:3146–3153. doi: 10.1074/jbc.M116.766774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floyd BJ, et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Ramírez M, et al. Genomics and epigenetics: A study of ependymomas in pediatric patients. Clin Neurol Neurosurg. 2016;144:53–58. doi: 10.1016/j.clineuro.2016.02.041. [DOI] [PubMed] [Google Scholar]
- 34.Bai F, et al. The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc Natl Acad Sci USA. 2015;112:3698–3703. doi: 10.1073/pnas.1502960112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conlan AR, et al. Mutation of the His ligand in mitoNEET stabilizes the 2Fe-2S cluster despite conformational heterogeneity in the ligand environment. Acta Crystallogr D Biol Crystallogr. 2011;67:516–523. doi: 10.1107/S0907444911011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiley SE, et al. The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J Biol Chem. 2007;282:23745–23749. doi: 10.1074/jbc.C700107200. [DOI] [PubMed] [Google Scholar]
- 37.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 38.Noel J, Onuchic J. 2012. The many faces of structure-based potentials: From protein folding landscapes to structural characterization of complex biomolecules. Computational Modeling of Biological Systems, Biological and Medical Physics, Biomedical Engineering, ed Dokholyan NV (Springer, New York), pp 31–54.
- 39.Whitford PC, et al. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields. Proteins. 2009;75:430–441. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavez LL, Onuchic JN, Clementi C. Quantifying the roughness on the free energy landscape: Entropic bottlenecks and protein folding rates. J Am Chem Soc. 2004;126:8426–8432. doi: 10.1021/ja049510+. [DOI] [PubMed] [Google Scholar]
- 41.Clementi C, García AE, Onuchic JN. Interplay among tertiary contacts, secondary structure formation and side-chain packing in the protein folding mechanism: All-atom representation study of protein L. J Mol Biol. 2003;326:933–954. doi: 10.1016/s0022-2836(02)01379-7. [DOI] [PubMed] [Google Scholar]
- 42.Lammert H, Noel JK, Haglund E, Schug A, Onuchic JN. Constructing a folding model for protein S6 guided by native fluctuations deduced from NMR structures. J Chem Phys. 2015;143:243141. doi: 10.1063/1.4936881. [DOI] [PubMed] [Google Scholar]
- 43.Lammert H, Noel JK, Onuchic JN. The dominant folding route minimizes backbone distortion in SH3. PLoS Comput Biol. 2012;8:e1002776. doi: 10.1371/journal.pcbi.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amadei A, Linssen ABM, Berendsen HJC. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 45.García AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 46.Tamir S, et al. Nutrient-deprivation autophagy factor-1 (NAF-1): Biochemical properties of a novel cellular target for anti-diabetic drugs. PLoS One. 2013;8:e61202. doi: 10.1371/journal.pone.0061202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheftel AD, et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci USA. 2010;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouault TA. Mammalian iron-sulphur proteins: Novel insights into biogenesis and function. Nat Rev Mol Cell Biol. 2015;16:45–55. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- 49.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai K, Tonelli M, Frederick RO, Markley JL. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry. 2017;56:487–499. doi: 10.1021/acs.biochem.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karmi O, et al. Interactions between mitoNEET and NAF-1 in cells. PLoS One. 2017;12:e0175796. doi: 10.1371/journal.pone.0175796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 53.Lammert H, Schug A, Onuchic JN. Robustness and generalization of structure-based models for protein folding and function. Proteins. 2009;77:881–891. doi: 10.1002/prot.22511. [DOI] [PubMed] [Google Scholar]
- 54.Noel JK, et al. SMOG 2: A versatile software package for generating structure-based models. PLoS Comput Biol. 2016;12:e1004794. doi: 10.1371/journal.pcbi.1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.