Significance
SbcC-SbcD and ExoI belong to a class of highly conserved nucleases that are critical to genome stability, but whose cellular function remains poorly understood. Human homologs of these proteins are essential for viability and normal development, and lead to severe developmental abnormalities and cancer predisposition when mutated. Here we show that these enzymes process DNA intermediates at sites where replication forks converge and are required for chromosome replication to complete normally. Cells lacking these gene products are unable to complete replication normally, and rely on an aberrant recombinational mechanism to maintain viability that leads to genomic instability and amplifications at these sites, similar to that seen in human cancers where these genes have been found to be inactivated.
Keywords: SbcCD, ExoI, Mre11, Rad50, replication completion
Abstract
SbcC-SbcD are the bacterial orthologs of Mre11-Rad50, a nuclease complex essential for genome stability, normal development, and viability in mammals. In vitro, these enzymes degrade long DNA palindromic structures. When inactivated along with ExoI in Escherichia coli, or Sae2 in eukaryotes, palindromic amplifications arise and propagate in cells. However, long DNA palindromes are not normally found in bacterial or human genomes, leaving the cellular substrates and function of these enzymes unknown. Here, we show that during the completion of DNA replication, convergent replication forks form a palindrome-like structural intermediate that requires nucleolytic processing by SbcC-SbcD and ExoI before chromosome replication can be completed. Inactivation of these nucleases prevents completion from occurring, and under these conditions, cells maintain viability by shunting the reaction through an aberrant recombinational pathway that leads to amplifications and instability in this region. The results identify replication completion as an event critical to maintain genome integrity and cell viability, demonstrate SbcC-SbcD-ExoI acts before RecBCD and is required to initiate the completion reaction, and reveal how defects in completion result in genomic instability.
Accurate replication of the genome requires that cells tightly regulate the processes of initiation, elongation, and completion to ensure that each daughter cell inherits an identical copy of the genetic information. Although the mechanisms regulating initiation and elongation have been extensively characterized (1, 2), the process of how replication is completed has, until recently, remained unknown. Accurately completing replication necessitates that cells recognize replicated regions and join the strands of converging replication forks at the precise point where all sequences have doubled. This event occurs thousands of times per division along the chromosomes of human cells, and therefore must be remarkably efficient. Failure of even a single completion event would be expected to result in duplications, deletions, rearrangements, or cell death, depending on how the convergent strands are processed. Cells devote a large number of enzymes to maintain the fidelity of both replication initiation and elongation, and recent studies have shown that this final step, required to maintain genomic integrity, similarly requires enzymatic control and is tightly regulated (3, 4).
The completion step of DNA replication has been challenging to study in eukaryotic cells, in part because multiple origins are used with varying efficiencies and timing, making the genomic loci where forks meet highly variable between cells and cell cycles (5, 6). In comparison, Escherichia coli is uniquely suited to dissect this fundamental aspect of cellular metabolism because the event can be localized to a single ∼400-kb region of the chromosome, opposite to its bidirectional origin of replication (7). This region is flanked by ter sequences that bind the protein Tus, blocking replication forks in an orientation-specific manner (8). Although ter ensures that completion events occur within this region, they do not appear to be directly involved in the reaction, as chromosomes and plasmids lacking ter replicate normally and are stably maintained in the cell (9, 10).
In E. coli, the completion of DNA replication requires the RecBCD helicase–nuclease complex and occurs through a reaction that does not involve recombination or RecA (3, 4). In the absence of RecBCD, cultures either fail to replicate or fail to maintain the chromosome region where replication forks converge, leading to large segments of the chromosome remaining unreplicated or degraded in this region. The inability of recBCD mutants to replicate or maintain this region of the chromosome severely compromises growth and viability in these cultures (3, 4). In vitro, RecB and RecC interact with RecD to form a dual helicase–nuclease complex that unwinds and degrades double-strand DNA ends (11, 12). The enzymatic complex is capable of rapidly translocating and degrading DNA for tens to hundreds of kilobases at a rate of ∼1,000 bp/s, a processivity and rate that approaches that of the replisome’s ability to synthesize these sequences (13–15). Loss of RecB or RecC inactivates the complex, whereas loss of RecD inactivates the nuclease but retains helicase activity (16–18). How RecBCD promotes the completion of DNA replication, and what intermediate substrates are involved in this reaction, remain to be characterized.
Other enzymes that compromise the ability to complete replication have also been identified and include SbcC-SbcD and ExoI. In the absence of these enzymes, the region where replication forks converge is overreplicated or amplified (3, 19). Curiously, although chromosome replication is clearly abnormal in these mutants, the overrepresented amplification regions of the chromosome do not appear to compromise the growth or viability of cells in culture.
SbcC-SbcD and ExoI belong to a class of nucleases that are critical to genome stability but whose cellular function remains poorly understood. SbcC-SbcD are the bacterial orthologs of Mre11-Rad50 in humans and yeast (20, 21). They are both structurally and functionally conserved in all three domains of life (22, 23). All SbcC-SbcD orthologs form a heterotetrameric complex with a remarkably unique architecture, containing two long coiled–coil regions that extend up to 60 nm within the SbcC (Rad50) dimer where the ATPase and SbcD (Mre11) interaction site reside (22). When bound with the SbcD dimer, these complexes contain a prominent endonuclease activity that incises DNA hairpins, as well as single-strand endonuclease and double-strand 3′-5′ exonuclease activities (24). Human CtIP, yeast Sae2, and E. coli ExoI appear to be conserved at the functional level. Inactivation of these nucleases enhances the palindrome amplifications and genetic instabilities that arise in mutants lacking SbcC-D or Rad50-Mre11 (25–27). In humans, these genes are essential for viability and normal development, whereas hypomorphic mutations lead to severe developmental abnormalities and cancer predisposition (28–31). Mutations render cells in culture hypersensitive to double-strand breaks and perturb the normal progression through the cell cycle (29, 30, 32–34). These phenotypes are often attributed to defective repair of double-strand breaks, and the enzymes are most commonly proposed to function as initiators of recombinational repair, processing DNA ends before the Rad51 recombinase can load and facilitate recombination (35, 36). However, several enigmatic phenotypes associated with these enzymes are difficult to reconcile with this model. Rad51 foci form normally in the absence of Mre11 or Rad50 (37–39), implying that processing by these enzymes is not required to initiate recombinational Rad51-filaments. Curiously, the preferred substrate of the E. coli, yeast, and human enzymes are not double-stranded DNA ends, but long hairpin or palindromic DNA structures (40–42). Inactivation of Mre11 and CtIP in vivo leads to widespread genomic instabilities that include the appearance of palindromic duplications (25, 43). Similarly, in E. coli, palindromic sequences can only be maintained in mutants lacking SbcCD and ExoI (44, 45), indicating that, somehow, palindromic sequences are relevant to their in vivo function. However, long palindromic sequences are not encoded in eukaryotic or bacterial genomes, making it unclear why palindrome-specific nucleases would be essential in mammalian cells or how palindromic duplications rapidly appear in their absence. Thus, the substrates and essential function that these enzymes have in the cell has remained an elusive question.
In this study, we show that SbcC-SbcD and ExoI are required to initiate the completion of DNA replication on the chromosome and act at a step before RecBCD in the process. The enzymes process a proposed palindrome-like intermediate that forms at sites where replication forks converge. SbcCD and ExoI processing likely creates a substrate that allows RecBCD to bind and promote joining of convergent replication fork strands, either directly or indirectly. In the absence of SbcCD and ExoI, these structural intermediates persist and the normal completion of replication is prevented. Furthermore, we show that under these conditions, cells maintain viability by processing these intermediates through an aberrant recombinational mechanism that accounts for the widespread genomic instabilities and amplifications that are observed in mutants lacking these nucleases.
Results
SbcC-SbcD and ExoI Are Required for Replication to Complete Normally and Act Upstream of RecBCD in the Reaction.
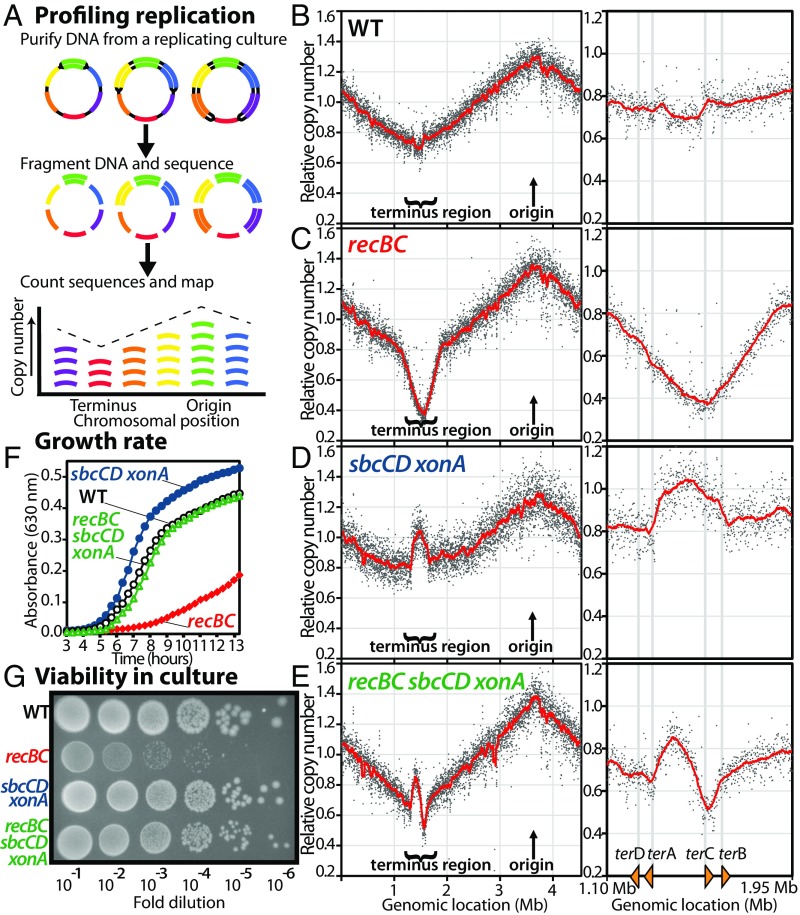
The ability to complete chromosome replication can be observed by profiling the copy number of DNA sequences across the genome of replicating cultures. In this approach, genomic DNA is purified from replicating cultures, fragmented, and sequenced. The replication profile is then determined by counting the number of sequences that align to each segment of the chromosome (Fig. 1A). In wild-type cultures, sequences surrounding the bidirectional origin replicate first, and are observed at higher frequencies relative to chromosome regions that replicate later. Overall, sequence frequencies decrease inversely with their distance from the origin until reaching the terminus region where replication forks converge and replication completes (ref. 3 and Fig. 1B). In recBCD mutants, a dramatic loss of sequences is observed in the region where replication forks converge (Fig. 1C). An identical result is observed in recBC mutants alone (3). The copy number of sequences in the terminus region is reduced approximately twofold relative to wild-type cultures. Assuming that more than half of sequence reads must correspond to parental DNA strands, one can infer that nearly all the cells in the recBCD population have difficulty replicating or maintaining this region of the chromosome. This severely impairs growth and viability in recBC cultures (3). The slope of the progressing replication forks away from the origin is similar in both recBCD and wild-type profiles (−30.0 vs. −28.8 normalized reads/Mb, respectively) before reaching the terminus region, where the recBCD slope rapidly deteriorates, relative to wild-type cultures (−133.8 vs. −13.9 normalized reads/Mb, respectively). The observation argues against the idea that overall fork progression is impaired or impeded in recBCD mutants and indicates that the replication defect localizes specifically to the region where replication forks converge in these mutants. In contrast, completion has previously been shown to occur normally in recA mutants (3, 4), demonstrating that the RecBCD-mediated reaction occurs independent of homologous recombination or double-strand repair and likely represents an intramolecular reaction.
Fig. 1.
Inactivation of SbcCD ExoI nucleases prevents processing of convergent replication forks and restores the terminus region in recBC mutants, indicating that SbcCD ExoI is required to process the overreplicated intermediate before RecBCD-mediated completion can occur. (A) Diagram of replication profile methodology. (B–E) Inactivation of the SbcCD ExoI nucleases restores the region where replication forks converge in recBC mutants. Genomic DNA from replicating cultures was purified, fragmented, and profiled using high-throughput sequencing. Normalized sequence read frequencies are plotted relative to genome position along with a 30-kb floating average in red. The terminus region of the chromosome, containing terD, terA, terC, and terB, is shown next to each plot. sbcCD xonA read frequencies were calculated from datasets appearing in ref. 3. (F and G) The ability to maintain the chromosome’s terminus region correlates with growth and viability in culture. Absorbance (630 nm) of cultures grown at 37 °C is plotted over time (F). Ten-microliter drops from 10-fold serial dilutions of overnight cultures were plated. Colonies were observed following overnight 37 °C incubation (G).
The inability of recBCD mutants to maintain the terminus region could arise either because RecBCD is required for replication forks to reach the terminus or because RecBCD is somehow required to allow DNA ends from convergent replication forks to be joined. In the latter possibility, persistent DNA ends from convergent replication forks would remain susceptible to nucleolytic attack, leading to extensive degradation and loss of DNA in the region where forks converge.
To differentiate between these two general mechanisms, we examined the replication profiles of recBC mutants that also lacked SbcCD and ExoI nucleases. ExoI, encoded by xonA, contains a processive 3′-5′ single-stranded exonuclease (46, 47), whereas SbcCD is a double-stranded exonuclease that also has endonucleolytic activity at DNA palindromes (24, 44). We reasoned that if RecBCD facilitates replication forks reaching the terminus, then the absence of exonucleases would have little effect on the depleted terminus region. Alternatively, if RecBCD action is required before DNA ends from convergent forks can be joined, then the absence of exonucleases may reduce or prevent the degradation occurring and partially restore the terminus region. We initially focused on SbcCD and ExoI as candidates because inactivation of these gene products has been shown to suppress recombination defects in recBC mutants during sexual processes, such as conjugation, transformation, or transduction (48, 49). In addition, SbcCD and Exo I appear to participate in the completion reaction, as mutants lacking these gene products exhibit an increased copy number of sequences in the terminus region (ref. 3 and Fig. 1D).
We found that inactivation of SbcCD and ExoI (sbcCD xonA) was sufficient to restore the DNA to the terminus region of recBC mutants. Surprisingly, loss of these nucleases resulted in overreplication of the terminus region, similar to that seen in sbcCD xonA mutants alone (Fig. 1E). The observation demonstrates that replication forks reach the terminus region normally in the absence of RecBCD and implies that convergent fork ends remain unjoined and susceptible to degradation in recBCD mutants. Notably, overreplication in sbcCD xonA is phenotypically dominant to degradation in recBC mutants, indicating that the structure created by converging replication forks must be processed by SbcCD-ExoI before RecBCD can facilitate resection and joining during the completion reaction.
The sbcCD and xonA mutations appear additive in their effect on overreplication in the terminus (Fig. S1). The observation could suggest that processing of the overreplicated region occurs by each of these enzymes, but that a double-stranded end required for RecBCD loading is only generated after both processing events have occurred.
Maintaining the Region Where Replication Forks Converge Correlates with the Growth and Viability of Cells in Culture.
The inability of recBC mutants to maintain the terminus region of the chromosome correlates with the impaired growth and reduced viability of these cultures relative to wild-type cells. Curiously, although the completion reaction is clearly abnormal in mutants lacking SbcCD and Exo I, the excess copy number or amplification of the terminus region does not impair the growth or viability of these mutants (Fig. 1 F and G). We noted that inactivation of SbcCD and ExoI, which prevents degradation of the terminus region in recBC mutants, also restores the growth and viability of these cultures (Fig. 1 F and G). The observation further supports the interpretation that the impaired growth and viability of recBC mutants is caused by an inability to maintain the terminus region on the chromosome and explains why inactivation of SbcCD and ExoI is able to suppress defects in recBC mutants. Importantly, however, although SbcCD and ExoI inactivation restores recBC viability, chromosomal abnormalities and amplifications persist in the region where forks converge, indicating that completion in the absence of these nucleases is occurring through an aberrant, alternative pathway.
Abnormalities in Completing Replication Can also Be Observed on Plasmids Replicating in These Mutants.
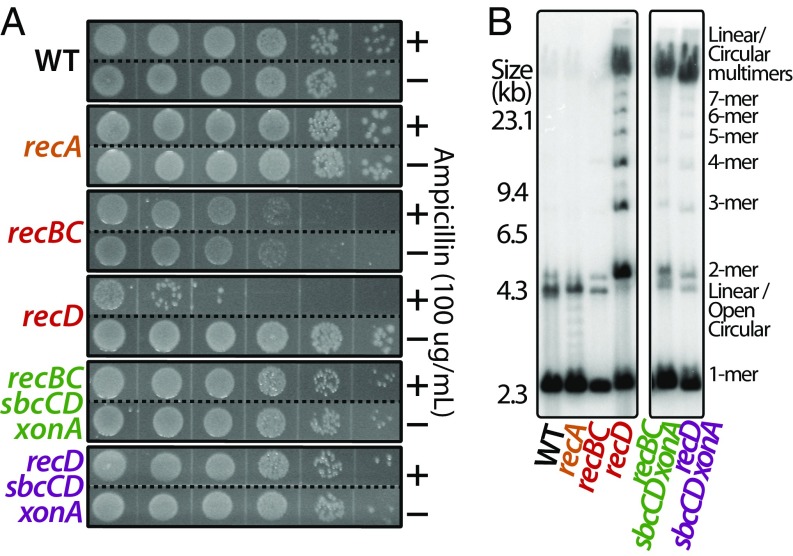
In recBC mutants, plasmids have a reduced copy number relative to wild-type cultures, suggesting that they replicate with a lower frequency of success. In recD mutants, which lack nuclease but retain helicase activity, plasmids continue to replicate past the doubling point, producing large quantities of both odd- and even-numbered multimeric circles as well as long linear multimers. These plasmids are not maintained and rapidly lost in culture (3, 50, 51). In contrast, in the absence of RecA, which is required for all recombination and double-strand break repair, plasmids remain stable and replicate normally. The observations argue that the plasmid instabilities in recBC and recD mutants are associated with an impaired ability to complete replication, rather than defects in recombination or double-strand break repair. We next examined how SbcCD and ExoI affected plasmid stability in recBC and recD mutants. We found that, similar to what is observed on the chromosome, inactivation of SbcCD and ExoI alleviates the low copy number phenotype in recBC mutants and improves plasmid stability in both recBC and recD mutants (Fig. 2A). Also, similar to what is seen on the chromosome, although stability is restored, large amounts of multimeric amplification products are produced and persist in these populations, indicating that under these conditions, the plasmids are maintained through an alternate, aberrant mechanism (Fig. 2B).
Fig. 2.
Inactivation of SbcCD-ExoI improves plasmid stability in recBC and recD mutants, although aberrant plasmid species accumulate and persist. (A) Inactivation of SbcCD-ExoI improves the stability of plasmids in recBC and recD mutants. Cultures containing plasmid pBR322 were grown for 30 generations without selection before plating 10-µL drops of 10× serial dilutions, with and without ampicillin selection, to determine the fraction of cells that maintained the plasmid in each strain. (B) Plasmids in recBC and recD mutants lacking SbcCD-Exo I are maintained through an aberrant alternative pathway. Total DNA was prepared from replicating cultures containing pBR322 and examined by Southern analysis after agarose gel electrophoresis, using 32P-labeled pBR322 as a probe.
When Normal Completion Is Impaired, the Ability to Maintain Cell Growth and the Region Where Forks Converge Becomes Dependent on an Aberrant Recombinational Mechanism.
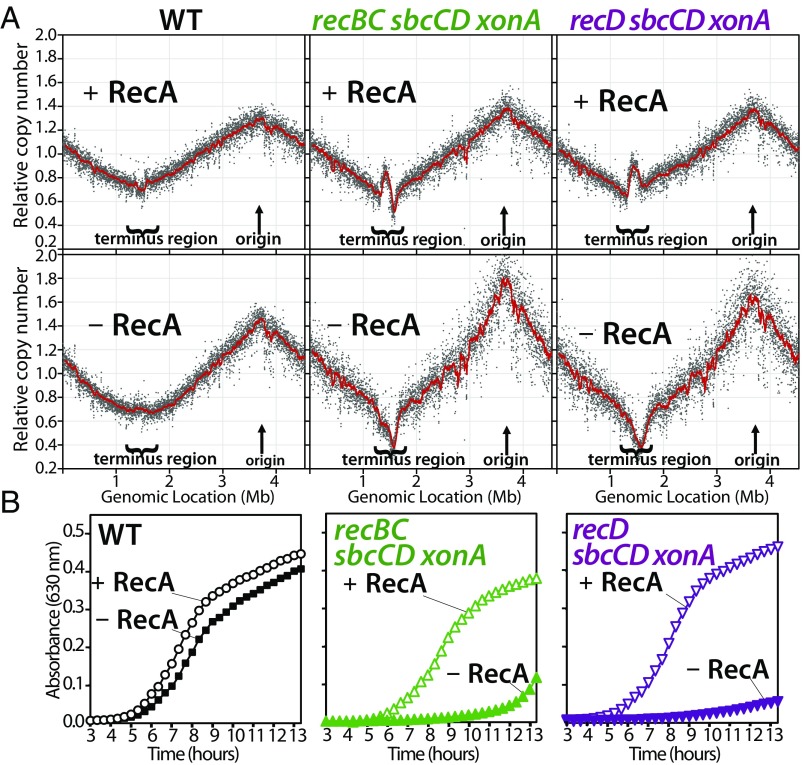
To further investigate how cell viability and the terminus region are maintained under these aberrant conditions, we examined whether this alternative process depended on recombination. The completion of replication on the chromosome normally occurs through a mechanism that is independent of recombination and RecA (3). In mutants lacking RecA, the region where forks converge is maintained and cultures grow at rates comparable to wild-type cultures (Fig. 3). However, we observed that in mutants where the normal mechanism of completion is prevented, the cell’s ability to maintain growth and the chromosome region where forks converge becomes dependent on recombination and RecA (Fig. 3 and Fig. S2). In the absence of RecA, sbcCD xonA mutations no longer suppress the defects in recBC cultures, as the terminus region is not maintained, and growth is severely compromised. Similarly, in sbcCD xonA recD mutants, the ability to maintain the terminus region and grow depends entirely on RecA. Thus, the normal RecBCD-mediated completion reaction occurs in the absence of recombination. However, when the normal completion reaction is prevented, the reaction is shunted through an aberrant, recombination-dependent mechanism that prevents loss of this chromosome region but is associated with amplifications and genomic instability at these locations. Strains that lose the ability to maintain the region where replication forks converge are no longer able to grow, and viability is severely reduced.
Fig. 3.
In the absence of SbcCD-ExoI processing, normal completion is prevented, and maintaining the region where forks converge becomes dependent on recombination. (A) Replication completes normally in the absence of RecA. However, after SbcCD-ExoI inactivation, recBC and recD mutants become dependent on RecA to complete replication. (B) Maintaining the chromosome region where forks converge correlates with the ability of cells to grow in culture. Replication profiles and growth were performed as in Fig. 1.
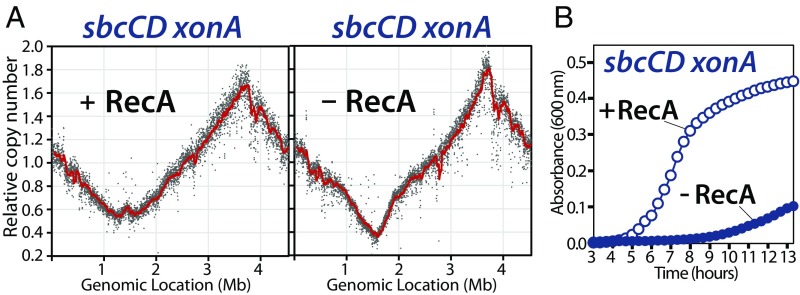
Inactivation of SbcCD and ExoI, in an otherwise wild-type background, leads to amplification and genomic instability in the terminus region (Fig. 1). To determine whether the absence of these nucleases, by themselves, prevent replication from completing normally, we asked whether maintaining the terminus region and cell growth in sbcCD xonA mutants depended on recombination. We found that the absence of SbcCD ExoI alone prevented replication from completing normally, and that in the absence of recombination, cells were unable to maintain the region where replication forks converge, and growth was severely impaired (Fig. 4). Thus, in the absence of the nucleases that process the intermediate created by convergent replication forks, normal completion of replication is prevented, and the reaction is shunted through an aberrant recombinational process that prevents loss of these chromosome regions, but leads to genetic instability and amplifications at these sites.
Fig. 4.
In the absence of SbcCD-ExoI processing, normal completion is prevented, and growth and viability become dependent on recombination. (A) In an SbcCD-ExoI deficient strain, the aberrant recombinational pathway facilitates completion. (B) Growth and viability of the sbcCD-xonA mutant correlate with the ability to maintain the region where forks converge. Replication profiles and growth were performed as in Fig. 1.
Discussion
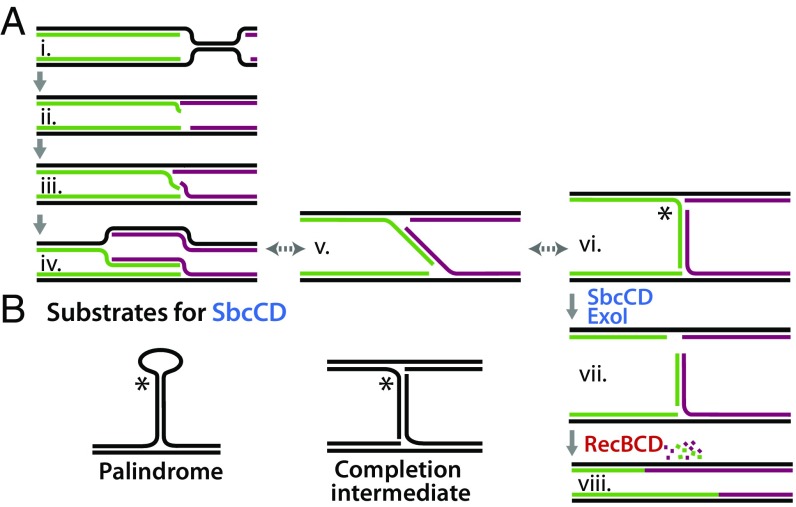
Taken together, the data presented here demonstrate that the completion of DNA replication occurs through a transient overreplication of the sequences where replication forks converge. This overreplication can be observed both in vivo and in vitro (3, 52). SbcCD and ExoI are required to process the intermediate produced at sites where replication forks converge before RecBCD can resect these regions and facilitate joining of convergent DNA ends (Fig. 5A). In the absence of SbcCD-ExoI, completion cannot occur and cell growth becomes dependent on an aberrant recombination mechanism to resolve these chromosomes that leads to genomic instability and amplification in this region. We also show that replication forks reach the terminus normally in the absence of RecBCD, but that joining of the nascent convergent forks fails to occur, leading to extensive degradation in these regions. SbcC-SbcD, and its human homolog, Mre11-Rad50, specifically incise long DNA palindromes both in vitro and in vivo (24, 25, 44, 53). However, long palindromic sequences are not found in these genomes, and identifying the cellular substrate these enzymes act on has remained elusive. When considering the substrate created by the transient overreplication at sites where replication forks converge, it is apparent that the intermediate bears a striking similarity to the palindrome-like structure that is known to be cleaved by SbcC-SbcD and Mre11-Rad50 in vitro (Fig. 5B). Here, we show that these sequences are substrates for SbcCD and ExoI in vivo. Furthermore, SbcCD-ExoI processing of this intermediate is essential for the normal completion of replication to occur, and results in genomic instabilities and amplifications when processing is impaired or prevented.
Fig. 5.
Model for completing replication. (A, i–vi) Replication forks continue past their meeting point, creating a palindromic substrate. (A, vi) SbcCD-ExoI cleave and process the overreplicated intermediate. (A, vii) RecBCD-mediated resection and joining of the DNA ends completes replication. (B) In vitro and proposed in vivo substrates for SbcCD, respectively.
Several observations suggest the conserved Mre11-Rad50 and CtIP function similarly in humans and other eukaryotes (20, 21). When processing of convergent replication fork intermediates is prevented in E. coli, abnormalities are observed on the chromosome. Although only one completion event occurs per cell cycle in E. coli, human cells require that that this event occur thousands of times along the chromosome each time the cell divides, and may explain why these genes are essential and lead to severe developmental defects and cancer when their function is impaired (25, 29, 30). Consistent with this interpretation, lethality in mutants with compromised Mre11-Rad50-Nbs1 function arises not during, but after, replication through S-phase, as a result of a failure to resolve DNA bridges that persist between chromosomes attempting to separate during mitosis (33). On the basis of the results presented here, we propose that these bridges represent unresolved completion events, such as depicted in Fig. 5 A, vi.
We show that when processing of the convergent replication forks is prevented, the overreplicated regions create homologous substrates that can alternatively be resolved through recombinational mechanisms (Fig. S3). However, when completion occurs under these conditions, it produces chromosome instabilities and amplifications that are readily apparent in the replication profiles of mutants. In human cells, where completion events occur throughout the genome, this compromised fidelity would be expected to provide cancerous cells with a broad selective mechanism by which they may achieve a growth advantage through amplification. Direct and palindromic amplifications are correlated with tumor initiation, metastasis, and the development of drug resistance, sometimes in regions of known oncogenes (54–60). These mutational signatures match those appearing on the sbcCD xonA chromosome, and are similar to the widespread palindromic amplifications and gross chromosome rearrangements arising in yeast after inactivation of Mre11 and Sae2 (25, 43).
Finally, the aberrant, recombination-mediated mechanism that operates to maintain the chromosome in E. coli when the normal mechanism of completion is prevented has a number of similarities to the mechanisms that maintain chromosomes in immortalized eukaryotic cells. Similar to bacteria, telomerase-negative immortalized cells maintain the telomeric regions of their chromosomes through a recombinational process that is required for the growth and viability of these populations (25, 33, 43, 61–64). However, activation of this pathway leads to high rates of chromosome rearrangements and amplifications across the genome. SbcC-SbcD, ExoI and Mre11-Rad50, CtIP are conserved at the structural, biochemical, and phenotypic levels, making it highly likely that the role of these bacterial enzymes in completing replication, reported here, is likely to be similar for the mammalian enzymes and may account for their essential role in the cell.
Materials and Methods
All strains are derived from SR108 (Table S1). Bacterial culturing, genomic profile analysis, plasmid stability, genomic and plasmid DNA extractions, and Southern analysis have been described previously (3, 65, 66). Further details are provided in SI Material and Methods.
Supplementary Material
Acknowledgments
Supported by the National Science Foundation (Grant MCB0130486) and National Institute of Environmental Health Sciences (Grant R15ES025953).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data reported in this paper have been deposited in the Sequence Read Archive (SRA), www.ncbi.nlm.nih.gov/Traces/sra (accession nos. SRP107355 and SRP047195).
See Commentary on page 237.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715960114/-/DCSupplemental.
References
- 1.Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wendel BM, Courcelle CT, Courcelle J. Completion of DNA replication in Escherichia coli. Proc Natl Acad Sci USA. 2014;111:16454–16459. doi: 10.1073/pnas.1415025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courcelle J, Wendel BM, Livingstone DD, Courcelle CT. RecBCD is required to complete chromosomal replication: Implications for double-strand break frequencies and repair mechanisms. DNA Repair (Amst) 2015;32:86–95. doi: 10.1016/j.dnarep.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heichinger C, Penkett CJ, Bähler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006;25:5171–5179. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission yeast. Cell. 2009;136:852–864. doi: 10.1016/j.cell.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill TM. Arrest of bacterial DNA replication. Annu Rev Microbiol. 1992;46:603–633. doi: 10.1146/annurev.mi.46.100192.003131. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T, Hidaka M, Horiuchi T. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J. 1989;8:2435–2441. doi: 10.1002/j.1460-2075.1989.tb08374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggin IG, Wake RG, Bell SD, Hill TM. The replication fork trap and termination of chromosome replication. Mol Microbiol. 2008;70:1323–1333. doi: 10.1111/j.1365-2958.2008.06500.x. [DOI] [PubMed] [Google Scholar]
- 10.Roecklein B, Pelletier A, Kuempel P. The tus gene of Escherichia coli: Autoregulation, analysis of flanking sequences and identification of a complementary system in Salmonella typhimurium. Res Microbiol. 1991;142:169–175. doi: 10.1016/0923-2508(91)90026-7. [DOI] [PubMed] [Google Scholar]
- 11.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 12.Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- 13.Taylor A, Smith GR. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980;22:447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- 14.Bianco PR, et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 15.Dillingham MS, Webb MR, Kowalczykowski SC. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J Biol Chem. 2005;280:37069–37077. doi: 10.1074/jbc.M505520200. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhury AM, Smith GR. A new class of Escherichia coli recBC mutants: Implications for the role of RecBC enzyme in homologous recombination. Proc Natl Acad Sci USA. 1984;81:7850–7854. doi: 10.1073/pnas.81.24.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: The gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbour SD, Clark AJ. Biochemical and genetic studies of recombination proficiency in Escherichia coli. I. Enzymatic activity associated with recB+ and recC+ genes. Proc Natl Acad Sci USA. 1970;65:955–961. doi: 10.1073/pnas.65.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG. Avoiding chromosome pathology when replication forks collide. Nature. 2013;500:608–611. doi: 10.1038/nature12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 21.Connelly JC, Leach DR. Tethering on the brink: The evolutionarily conserved Mre11-Rad50 complex. Trends Biochem Sci. 2002;27:410–418. doi: 10.1016/s0968-0004(02)02144-8. [DOI] [PubMed] [Google Scholar]
- 22.Hopfner KP, et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 23.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 24.Connelly JC, de Leau ES, Leach DR. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 1999;27:1039–1046. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng SK, Yin Y, Petes TD, Symington LS. Mre11-Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol Cell. 2015;60:500–508. doi: 10.1016/j.molcel.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darmon E, et al. E. coli SbcCD and RecA control chromosomal rearrangement induced by an interrupted palindrome. Mol Cell. 2010;39:59–70. doi: 10.1016/j.molcel.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo G, et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang J, Bronson RT, Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J. 2002;21:1447–1455. doi: 10.1093/emboj/21.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 32.D’Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruhn C, Zhou ZW, Ai H, Wang ZQ. The essential function of the MRN complex in the resolution of endogenous replication intermediates. Cell Rep. 2014;6:182–195. doi: 10.1016/j.celrep.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Ajimura M, Leem SH, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeeles JT, Dillingham MS. The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair (Amst) 2010;9:276–285. doi: 10.1016/j.dnarep.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 36.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51:195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoa NN, et al. Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines. Genes Cells. 2015;20:1059–1076. doi: 10.1111/gtc.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto K, et al. Kinase-dead ATM protein is highly oncogenic and can be preferentially targeted by Topo-isomerase I inhibitors. Elife. 2016;5:e14709. doi: 10.7554/eLife.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeda S, Hoa NN, Sasanuma H. The role of the Mre11-Rad50-Nbs1 complex in double-strand break repair-facts and myths. J Radiat Res (Tokyo) 2016;57:i25–i32. doi: 10.1093/jrr/rrw034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 43.Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chalker AF, Leach DR, Lloyd RG. Escherichia coli sbcC mutants permit stable propagation of DNA replicons containing a long palindrome. Gene. 1988;71:201–205. doi: 10.1016/0378-1119(88)90092-3. [DOI] [PubMed] [Google Scholar]
- 45.Gibson FP, Leach DR, Lloyd RG. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J Bacteriol. 1992;174:1222–1228. doi: 10.1128/jb.174.4.1222-1228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips GJ, Prasher DC, Kushner SR. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J Bacteriol. 1988;170:2089–2094. doi: 10.1128/jb.170.5.2089-2094.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehman IR, Nussbaum AL. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase) J Biol Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 48.Lloyd RG, Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985;164:836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbour SD, Nagaishi H, Templin A, Clark AJ. Biochemical and genetic studies of recombination proficiency in Escherichia coli. II. Rec+ revertants caused by indirect suppression of rec- mutations. Proc Natl Acad Sci USA. 1970;67:128–135. doi: 10.1073/pnas.67.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biek DP, Cohen SN. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bassett CL, Kushner SR. Exonucleases I, III, and V are required for stability of ColE1-related plasmids in Escherichia coli. J Bacteriol. 1984;157:661–664. doi: 10.1128/jb.157.2.661-664.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hill TM, Marians KJ. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci USA. 1990;87:2481–2485. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitada K, Yamasaki T. The MDR1/ABCB1 regional amplification in large inverted repeats with asymmetric sequences and microhomologies at the junction sites. Cancer Genet Cytogenet. 2007;178:120–127. doi: 10.1016/j.cancergencyto.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 55.Kitada K, Yamasaki T, Aikawa S. Amplification of the ABCB1 region accompanied by a short sequence of 200 bp from chromosome 2 in lung cancer cells. Cancer Genet Cytogenet. 2009;194:4–11. doi: 10.1016/j.cancergencyto.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka H, Yao MC. Palindromic gene amplification: An evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer. 2009;9:216–224. doi: 10.1038/nrc2591. [DOI] [PubMed] [Google Scholar]
- 58.Marotta M, et al. Palindromic amplification of the ERBB2 oncogene in primary HER2-positive breast tumors. Sci Rep. 2017;7:41921. doi: 10.1038/srep41921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slamon DJ, et al. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 60.Waddell N, et al. Australian Pancreatic Cancer Genome Initiative Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 62.Zhong ZH, et al. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 63.Lombard DB, Guarente L. Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 2000;60:2331–2334. [PubMed] [Google Scholar]
- 64.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- 66.Courcelle J, Carswell-Crumpton C, Hanawalt PC. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.