Significance
Molecular mechanisms underlying distinct disabilities during neurological diseases may differ based on the neurological pathway involved. Multiple sclerosis (MS) is multifocal, characterized by distinct disabilities affecting walking, vision, cognition, and fatigue. Neuroprotective treatments tailored for each disability may be more effective than nonspecific treatments aiming to reduce a composite of disabilities in clinical trials. Here, we use the MS model to apply a cell-specific and region-specific gene expression approach to discover targets in distinct neuroanatomic regions. Altered cholesterol synthesis gene expression in astrocytes in spinal cord and optic nerve was identified as a potential target for walking and visual disabilities, respectively. This disability-specific discovery approach represents a strategy for finding neuroprotective treatments for multifocal neurodegenerative diseases.
Keywords: astrocytes, transcriptomics, experimental autoimmune encephalomyelitis, multiple sclerosis, cholesterol
Abstract
Changes in gene expression that occur across the central nervous system (CNS) during neurological diseases do not address the heterogeneity of cell types from one CNS region to another and are complicated by alterations in cellular composition during disease. Multiple sclerosis (MS) is multifocal by definition. Here, a cell-specific and region-specific transcriptomics approach was used to determine gene expression changes in astrocytes in the most widely used MS model, experimental autoimmune encephalomyelitis (EAE). Astrocyte-specific RNAs from various neuroanatomic regions were attained using RiboTag technology. Sequencing and bioinformatics analyses showed that EAE-induced gene expression changes differed between neuroanatomic regions when comparing astrocytes from spinal cord, cerebellum, cerebral cortex, and hippocampus. The top gene pathways that were changed in astrocytes from spinal cord during chronic EAE involved decreases in expression of cholesterol synthesis genes while immune pathway gene expression in astrocytes was increased. Optic nerve from EAE and optic chiasm from MS also showed decreased cholesterol synthesis gene expression. The potential role of cholesterol synthesized by astrocytes during EAE and MS is discussed. Together, this provides proof-of-concept that a cell-specific and region-specific gene expression approach can provide potential treatment targets in distinct neuroanatomic regions during multifocal neurological diseases.
The biology of cell types within the central nervous system (CNS) differs between neuroanatomic regions serving different functional neurological pathways (1–3). Channel distribution, neurotransmitters, and neurotrophic factors all differ by functional neuroanatomic pathway (2), and regional differences in astrocytes (1) and oligodendrocytes (3) have been shown. Thus, molecular mechanisms underlying distinct disabilities may differ based on the neurological pathway involved. Multiple sclerosis (MS) is by definition multifocal, characterized by a variety of disabilities affecting walking, vision, cognition, and fatigue, to name a few. The incidence and severity of each disability varies from patient to patient. Accordingly, we hypothesized that a “one size fits all” treatment may not be optimal to achieve neuroprotection in MS. Rather, treatments tailored for each specific disability may be more effective. This approach is challenging since elucidation of gene expression changes in neuroanatomic regions associated with each disability would be needed.
A neuroprotective treatment optimally designed for one disability may have maximal effects on one or two disabilities but have only modest effects on other disabilities, thereby diluting out effects on commonly used standard composites of all disabilities in MS clinical trials. Accordingly, the discovery of disability-specific targets should be accompanied by disability-specific biomarkers. To this end, our group has recently used MRI to create disability-specific atlases of gray matter loss in MS, which revealed distinct localized gray matter atrophy in clinically eloquent areas aligned with different disabilities (ref. 4; see comment in ref. 5).
Previous methods of identifying new molecular targets in neurological diseases have largely entailed examining gene expression in whole CNS tissues from disease versus normal controls in preclinical models of disease. Genes overexpressed or underexpressed in disease models were then investigated in humans with disease as potential targets for new treatments. A major limitation of this approach is that CNS tissues contain multiple different cell types and that the composition of these cell types is altered during disease. For example, in MS and its preclinical model EAE, there is infiltration of immune cells, proliferation and migration of progenitors, and damage or death of oligodendrocytes and neurons. Thus, a difference in gene expression during disease may reflect in part the difference in cellular composition, confounding mechanistic insights into the effect of disease on the biology of any given CNS cell. Examination of gene expression changes in whole CNS tissue may also miss important changes in a subset of cells. Laser microdissection of cells from the CNS and single cell RNA sequencing are valid alternative approaches but have their own set of challenges (6–8).
Here, to determine changes in gene expression in a cell-specific and region-specific manner during a multifocal neurological disease, we applied RiboTag technology (9) to the MS preclinical model, experimental autoimmune encephalomyelitis (EAE). Astrocytes were the cell of focus since the hallmark multiple sclerotic lesions are based on astrocytic scarring, and both beneficial and deleterious roles of astrocytes have been shown in neuroinflammatory diseases, including EAE and MS (1, 10–12). The RiboTag approach yields ribosomes with a tag [hemagglutinin (HA)] in a specific cell type using Cre-loxP recombination (9). RiboTag mice were crossed with mice of the mGFAP-Cre line 73.12 (13) to express HA-tagged ribosomal protein only in astrocytes, and then astrocyte-specific RNAs from different neuroanatomic regions were isolated and examined for changes in gene expression in astrocytes during EAE using RNA sequencing and bioinformatics analyses. Regional differences in the disease-related astrocyte transcriptome were observed, and treatment targets were discovered in EAE and confirmed in MS. Together, this represents an approach to neuroprotective treatment development in MS that is based on cell-specific and region-specific gene expression analyses.
Results
Specificity of Astrocyte mRNA Isolation.
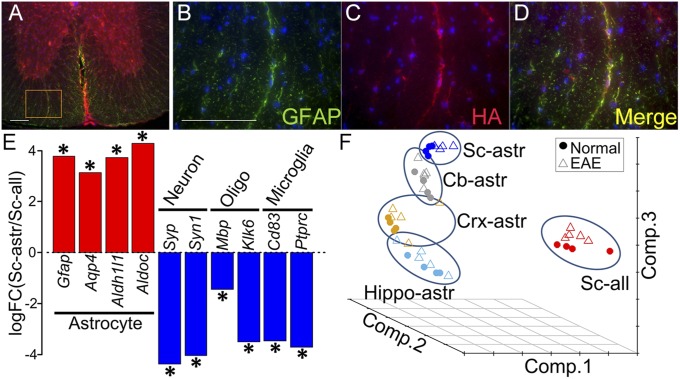
Colocalization of the HA tag specifically with astrocytes in astrocyte RiboTag mice was first shown at the protein level by immunohistochemistry (Fig. 1 A–D). Efficiency of HA colocalization to astrocytes in spinal cord at the protein level was 91.2%, as determined by the percent area of HA staining that colocalized with GFAP staining. RNA-seq analysis of astrocyte-enriched RNAs (immunoprecipitated by anti-HA antibody) confirmed enrichment of astrocyte-specific gene expression, and deenrichment of neuronal, oligodendroglial, and microglia-specific gene expression, shown as the log fold change calculated between spinal cord astrocyte RNA versus spinal cord total cell RNA (immunoprecipitated with control antibody, anti-RPL22) (13) (Fig. 1E).
Fig. 1.
Enrichment of astrocyte-specific mRNA using RiboTag technology. (A–D) Double immunolabeling in mGFAP-Cre:RiboTag mice showed colocalization of ribosome associated HA-Tag (red) with the astrocyte-specific marker GFAP (green), with colocalization seen in Merge (yellow). Nuclei were counterstained with DAPI (blue) (40-μm-thick spinal cord sections). B–D are a magnification of Inset area in A. (Scale bars: 100 μm.) (E) Enrichment of astrocyte-specific gene expression and deenrichment of neuronal, oligodendroglial, and microglia-specific gene expression, shown as the log fold change calculated between spinal cord astrocyte RNAs immunoprecipitated by anti-HA antibody versus spinal cord total cell RNAs (including astrocytes) immunoprecipitated with control antibody, anti-RPL22. Asterisks show the significant enrichment or deenrichment (FDR < 0.1). (F) Principal component analysis of astrocyte-enriched RNAs from four different brain regions (Cb, cerebellum; Crx, cerebral cortex; Hippo, hippocampus; and Sc, spinal cord), as well as RNAs from heterogeneous cells from spinal cord (RNA from all cell types). The samples from all cells of spinal cord were markedly separated from astrocyte-enriched spinal cord samples. Within astrocyte-enriched samples, the samples from different brain regions were also separated.
Regional Differences in Astrocyte-Specific Gene Expression During EAE.
EAE was induced in astrocyte RiboTag mice. Clinical signs of EAE began at day 11 after disease induction, and mice were killed at day 45 (mean EAE score = 3.1), as were age- and sex-matched normal controls. Comparison was made between RNAs from spinal cord astrocytes versus RNAs from spinal cord all cell types using principal component analysis (PCA) graphed three-dimensionally. Separation between spinal cord astrocyte-specific RNAs versus spinal cord total cell RNAs was observed, underscoring the difference between a cell-specific versus a whole tissue gene expression approach (Fig. 1F). This was consistent with the difference in the number of genes with significantly [false discovery rate (FDR) < 0.1] different expression when comparing EAE versus healthy controls, with approximately twice as many significant genes from analysis of spinal cord astrocytes (n = 2,015) compared with spinal cord all cells (n = 1,168). Together, this demonstrated that many gene differences in astrocytes in spinal cord during EAE are missed using a whole tissue gene expression approach.
EAE-induced gene expression changes in astrocytes differed between neuroanatomic regions when comparing astrocytes from spinal cord, cerebellum, cerebral cortex, and hippocampus. Compared with spinal cord astrocytes during EAE, gene expression in cerebellar astrocytes was distinct, but more similar to spinal cord than cortical astrocytes and hippocampal astrocytes (Fig. 1F). This separation between astrocytes derived from various regions of the CNS was observed in both EAE and normal controls, consistent with the concept of regional heterogeneity of astrocytes in normals (1), with this being an observation of regional differences in gene expression in astrocytes during EAE. The number of genes with significantly (FDR < 0.1) different expression was highest in spinal cord astrocytes (n = 2,015) compared with the other brain regions (cerebellum, n = 197; hippocampus, n = 49; cerebral cortex, n = 20). See Dataset S1 for the list of significant gene differences with (FDR < 0.1) in astrocytes of each region. The FDR < 0.1 threshold for differentially expressed genes was based on previously published analyses of RNA-seq data from astrocyte RiboTag mice (13).
Up-Regulation of Immune Pathway Gene Expression in EAE.
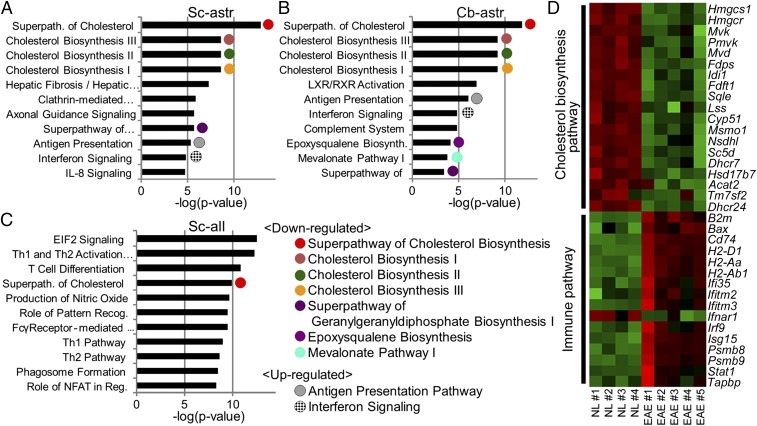
Canonical pathway analysis (Fig. 2 A–C and Table S1) of astrocyte-specific RNAs differentially expressed in EAE versus normal (FDR < 0.1) showed that genes involved in the antigen presentation pathway and the IFN signaling pathway were significantly different in astrocytes from spinal cord (Fig. 2A) and cerebellum (Fig. 2B). The change was in the direction of an increase in gene expression in EAE compared with normal (Fig. 2D). Since antigen presentation by astrocytes in spinal cords of EAE mice has been previously characterized (14, 15), this finding validated the RiboTag approach.
Fig. 2.
Expression of cholesterol synthesis pathway genes is decreased and of immune pathway genes is increased in spinal cord astrocytes during EAE. (A) Top canonical pathways are shown in spinal cord astrocytes (Sc-astr), (B) cerebellar astrocytes (Cb-astr), and (C) spinal cord all cells (Sc-all), each in descending order of significance. Astrocyte-specific RNAs differentially expressed in EAE versus normal (FDR < 0.1) showed that genes involved in several cholesterol synthesis pathways were significantly enriched in spinal cord and cerebellum (Table S1), with gene expression decreased in EAE (Tables S2 and S4). The antigen presentation pathway and IFN signaling pathway were also significantly enriched in both regions (Table S1), with gene expression increased in EAE (Table S4). Several pathways enriched in spinal cord astrocytes (Sc-astr) were not detectable in spinal cord all cells (Sc-all). n = 9 mice (5 EAE, 4 normal). Full names of pathways are listed in Table S1. (D) Heat map of direction of change of genes in pathways. A heat map of cholesterol synthesis and immune pathway genes that were significantly different between EAE and normal (NL) in astrocyte-specific RNAs (FDR < 0.1) revealed that cholesterol synthesis pathway genes were decreased in EAE (green) while immune pathway genes were increased (red). Green color represents lower expression, and red color represents higher expression. n = 9 mice (5 EAE, 4 normal). Significant differences in gene expression in spinal cord astrocytes were confirmed in another independent set of astrocyte RiboTag mice using RNA-seq analysis with n = 10 mice (5 EAE, 5 normal).
Down-Regulation of Cholesterol Synthesis Pathway Gene Expression in EAE.
In spinal cord astrocytes, several cholesterol synthesis pathways were significantly different in EAE compared with normal, with these pathways dominating the profile of top pathways in this cell type (Fig. 2A and Table S1). In contrast to an increase in immune pathway gene expression in spinal cord astrocytes, the direction of change for cholesterol synthesis pathway gene expression was a decrease in EAE compared with normal (Fig. 2D and Table S2). Cerebellar astrocytes also showed numerous cholesterol synthesis pathways that were significantly different during EAE compared with normal (Fig. 2B), again with gene expression decreased during EAE (Table S2). For cerebral cortex and hippocampus, the gene numbers with FDR < 0.1 significance level were too few to run for reliable enriched pathway analysis. The individual cholesterol synthesis genes that were down-regulated in astrocyte-specific RNAs during EAE in each region are listed in Table S2. When data were further analyzed using FDR < 0.01 and FDR < 0.05, the most significantly different pathways were the same as when FDR < 0.1 was used.
Regarding genes within the cholesterol synthesis pathway (Fig. S1), RNA-seq analyses showed robust down-regulation of 3-hydroxy-3-methylglutaryl-CoA synthase1 (Hmgcs1), farnesyl diphosphate synthase (Fdps), and farnesyl-diphosphate farnesyltransferase 1 (Fdft1) (Table S2). To validate these changes in gene expression in astrocytes of spinal cord, another independent set of RiboTag mice had EAE induced, and astrocyte gene expression in EAE versus control spinal cords was assessed by quantitative RT-PCR (qPCR). Expression of all three cholesterol synthesis genes was down-regulated in spinal cord astrocytes during EAE (Fig. 3A). Immunohistochemistry showed a decrease in expression in spinal cord astrocytes in WT mice during EAE at the protein level (Fig. 3B), and colocalization of HMGCS1, FDPS, and FDFT1 with astrocytes was demonstrated for each (Fig. 3C and Fig. S2). Finally, to further validate cell specificity of our findings in astrocytes, neuronal RiboTag mice (rNSEII-Cre:RiboTag) were created to assess gene expression in neurons during EAE, and the decreases in cholesterol gene expression observed in astrocytes did not occur in neurons (Fig. S3).
Fig. 3.
Cholesterol synthesis gene expression in spinal cord astrocytes during EAE. (A) Quantification of cholesterol synthesis gene expression by qPCR using astrocyte-specific RNAs from yet another set of astrocyte RiboTag mice with EAE at day 45 confirmed RNA-seq findings that Hmgcs1, Fdps, and Fdft1 expression were down-regulated in astrocytes during EAE (P = 0.0034 for Hmgcs1, P = 0.014 for Fdps, P = 0.0065 for Fdft1). (B) HMGCS1, FDPS, and FDFT1 were decreased in EAE spinal cord astrocytes of WT mice at the protein level using immunohistochemistry (P = 0.0007 for HMGCS1, P = 0.029 for FDPS, P = 0.0015 for FDFT1). (C) Colocalization of HMGCS1 (red) with GFAP (green) in astrocytes (yellow) in Merge. See Fig. S2 for colocalization of FDPS and FDFT1. Data are representative of two independent experiments. Shown are 10× images of ventral spinal cord white matter, yellow solid line Insets at 40×, and white dashed line Insets at 100×. (Scale bars: 100 μm.) ***P < 0.001, **P < 0.01, *P < 0.05. Four mice were examined for each group. Error bars indicate SEM between mice.
Focusing on White Matter Astrocytes: Optic Neuritis.
Regional differences in astrocyte gene expression during disease revealed that genes in cholesterol synthesis pathways were decreased more in spinal cord and cerebellum than in hippocampus and cerebral cortex (Table S2), despite the fact that hippocampus and cerebral cortex each have significant pathology during chronic EAE, including an increase in reactive astrocytes (16–21). The relative abundance of white matter in the first two regions compared with the latter two regions suggested that astrocytes from white matter might drive the observed changes in cholesterol synthesis gene expression during EAE. Since the anterior visual pathway is known to be affected by optic neuritis during EAE and contains only white matter, the astrocyte transcriptome of optic nerve during EAE was determined. First, colocalization of the HA label with GFAP in astrocytes of optic nerve of RiboTag mice was shown (Fig. S4 A–D). Then, to assess the enrichment of astrocyte-specific mRNA transcripts from optic nerve, qPCR analyses of cell-specific gene expression confirmed enrichment of astrocyte-specific gene expression and deenrichment of oligodendroglial, microglial, and axonal-specific gene expression (Fig. S4E).
Similar to astrocytes from spinal cord (Fig. 2A), astrocytes from optic nerve during EAE showed a difference compared with normal in several cholesterol synthesis pathways (Fig. 4A and Table S3). The direction of the change in cholesterol synthesis pathway genes was decreased in EAE compared with normal as shown in the heat map (Fig. 4B). This change in gene expression in astrocytes of optic nerve by RNA-seq was then confirmed in an independent set of RiboTag EAE mice where gene expression was assessed by qPCR. Hmgcs1 and Fdft1, two cholesterol synthesis genes in optic nerve that were found to be significantly down-regulated during EAE by RNA-seq, were confirmed to be decreased in EAE by qPCR (Fig. 5A). Quantification of expression in astrocytes in WT mice at the protein level also showed a decrease during EAE (Fig. 5 B and C). Immunohistochemistry showed colocalization of HMGCS1 and FDFT1 with astrocytes in optic nerve (Fig. 5D and Fig. S5). FDPS was not decreased in optic nerve during EAE by RNA-seq analysis and thus was not further assessed by qPCR and immunohistochemistry.
Fig. 4.
Cholesterol synthesis gene expression is decreased in optic nerve astrocytes during EAE. (A) Top canonical pathways in EAE optic nerve astrocytes are shown in descending order of significance. Astrocyte-specific RNAs differentially expressed in EAE versus normal (FDR < 0.1) showed that several cholesterol synthesis pathways were different in EAE (Table S3), with gene expression decreased in EAE (Table S4). Genes involved in the antigen presentation pathway and complement system pathway were also significantly different in EAE, but these were in the direction of an increase in EAE (Table S4). (B) A heat map of cholesterol synthesis pathway genes that were significantly different between EAE and normal (NL) in astrocyte-specific RNAs (FDR < 0.1) revealed that cholesterol synthesis pathway genes were decreased in EAE optic nerve (green). Green color represents lower expression and red color represents higher expression. n = 8 mice (4 EAE, 4 normal).
Fig. 5.
Validation of the decrease in cholesterol synthesis gene expression in optic nerve astrocytes during EAE. (A) Quantification of cholesterol synthesis gene expression by qPCR using astrocyte-specific RNAs from an additional set of astrocyte RiboTag mice with EAE at day 45 confirmed RNA-seq findings that Hmgcs1 and Fdft1 expression was down-regulated in astrocytes during EAE (P = 0.034 for Hmgcs1; P = 0.0017 for Fdft1). (B and C) HMGCS1 and FDFT1 were decreased in EAE optic nerve astrocytes of WT mice at the protein level using immunohistochemistry (P = 0.0378 for HMGCS1; P = 0.0046 for FDFT1). n = 3 to 4 mice in each group. (D) Colocalization of HMGCS1 (red) with GFAP (green) in astrocytes (yellow) in Merge. Shown are 10× images of coronal optic nerve sections. See Fig. S5 for colocalization of FDFT1. Data are representative of two independent experiments. (Scale bars: 10× images, 100 μm; Insets, 50 μm.) **P < 0.01, *P < 0.05. Error bars indicate SEM between mice.
Regarding inflammatory pathways in astrocytes in optic nerve during EAE, astrocytes from optic nerve showed an increase in expression of genes in the antigen presentation pathway as the top pathway (Fig. 4A and Table S3), consistent with the antigen presentation pathway being one of the top pathways in spinal cord and cerebellar astrocytes (Fig. 2 A and B and Table S1). However, unlike spinal cord and cerebellar astrocytes, optic nerve astrocytes showed a highly significant difference in expression of genes in the complement system pathway (Fig. 4A and Table S3), revealing region-specific differences in inflammation in astrocytes during EAE.
Changes in Astrocyte Gene Expression in Early Versus Late EAE.
While our focus was on astrocytes during late EAE, we next asked whether gene expression changes in astrocytes that were observed late were also observed earlier during EAE. Using qPCR for mRNAs from an additional set of astrocyte RiboTag mice, we found that cholesterol synthesis gene expression was not significantly decreased early during EAE (day 17) but was decreased late during EAE (day 50), in both spinal cord and optic nerve, with each confirmed at the protein level by immunohistochemistry in WT mice (Fig. S6 A, B, D, and E). Expression of MHC class II of the antigen presentation pathway in astrocytes was markedly increased in early EAE and then decreased late, albeit still higher than normal (Fig. S6C). Early immune activation, with some persistence late, is consistent with previous literature showing that immune responses in the CNS during EAE are not quiescent in chronic EAE in the C57BL/6 model (22–24). Interestingly, optic nerve astrocytes showed an increase in MHC class II early, and this did not decrease late (Fig. S6F).
Targeting Cholesterol Homeostasis as a Treatment for EAE.
The astrocyte-specific molecular signature of decreased cholesterol synthesis pathways in EAE spinal cord, cerebellum, and optic nerve suggested a novel target for treatment in EAE, namely cholesterol homeostasis. During adulthood, astrocytes are the main CNS cells producing cholesterols, with transport via apolipoprotein E (ApoE) to neurons to make membranes and synapses and to oligodendrocytes to make myelin (25). We hypothesized that decreased cholesterol synthesis in astrocytes during EAE could lead to decreased cholesterol transport. Thus, we investigated CS-6253, an agonist for ATP-binding cassette transporter A1 (ABCA1) that is known to increase efflux of cholesterol to extracellular ApoE (26, 27). Since it was not known whether treatment with CS-6253 could affect cholesterol synthesis gene expression, astrocyte-RiboTag mice that either had EAE or were normal were treated with either CS-6253 or vehicle, and qPCR determined if CS-6253 treatment affected cholesterol synthesis gene expression in astrocytes in spinal cords. Significant increases in Hmgcs1, Fdps, and Fdft1 were observed in spinal cords of CS-6253–treated EAE mice and also in normal controls (Fig. 6A). Immunohistochemistry showed increased expression of HMGCS1, FDPS, and FDFT1 at the protein level in CS-6253-treated versus vehicle-treated WT EAE mice, with CS-6253–treated EAE mice demonstrating levels as high as those in normal mice (Fig. 6B). Expression was principally in astrocyte cell bodies, but also in astrocyte processes (Fig. 6C). Finally, CS-6253–treated EAE mice had less severe EAE disability compared with vehicle-treated, as shown by motor function using standard EAE walking scores (Fig. 6D) and rotarod testing performance (Fig. 6E).
Fig. 6.
Treatment targeting cholesterol homeostasis during EAE. EAE (blue) and normal (NL, black) mice were treated with either Vehicle (Veh, solid) or CS-6253 (CS, striped). (A) Quantification of cholesterol synthesis gene expression by qPCR using astrocyte-specific RNAs from CS or vehicle-treated astrocyte RiboTag mice with EAE at day 43 showed that Hmgcs1, Fdps, and Fdft1 expression were up-regulated in astrocytes in CS-treated normal mice (P = 0.0117 for Hmgcs1, P = 0.0324 for Fdps, P = 0.0475 for Fdft1) and in CS-treated EAE mice (P = 0.0059 for Hmgcs1, P = 0.0093 for Fdps, P = 0.005 for Fdft1). n = 5 in each group. (B) HMGCS1, FDPS, and FDFT1 were also increased in CS-treated EAE spinal cord astrocytes in WT mice at the protein level using immunohistochemistry (P = 0.0008 for HMGCS1, P = 0.0002 for FDPS, P = 0.00195 for FDFT1). n = 4 in each group. (C) Colocalization of HMGCS1 (red) with GFAP (green) in astrocytes (yellow) in Merge, showing the CS-mediated increase in HMGCS expression in astrocyte cell bodies and processes. Shown are 10× images of ventral spinal cord white matter, Insets at 40×. (Scale bars: 100 μm.) (D and E) CS treatment resulted in a reduction in clinical EAE severity scores, which assess walking disability (P = 0.005 cumulative disease index after EAE day 30, P = 0.013 all days; CS group n = 6, vehicle group n = 7), and an increase in the number of seconds that mice could stay on the rotarod (P = 0.0077), respectively, each compared with vehicle treatment in EAE. For confirmation, an additional set of EAE mice underwent treatment with CS-6253 or vehicle, again demonstrating improvement in EAE scores (P < 0.0001 cumulative disease index after EAE day 30, P = 0.0006 all days; CS group n = 7, vehicle group n = 9).
Translation to Multiple Sclerosis.
Translating our findings in optic neuritis in EAE to humans, optic neuritis is common in MS. Therefore, gene expression analyses of white matter tracts of the anterior visual pathway in MS optic chiasm tissues were examined using MS brain autopsy tissues. We compared gene expression in five MS and five healthy age-matched controls (MS mean age = 57.6 y, healthy controls mean age = 56.2 y; MS disease duration mean = 20.4 y; MS and healthy control all female; see SI Materials and Methods for additional subject characteristics). Cholesterol synthesis pathways were significantly different in MS optic chiasm compared with healthy control optic chiasm (Fig. 7A and Table S3), with cholesterol synthesis gene expression decreased in MS compared with healthy controls (Fig. 7B).
Fig. 7.
Cholesterol synthesis gene expression is decreased in optic chiasm in MS. (A) Top canonical pathways in MS optic chiasm tissues. Pathway analysis of optic chiasm from MS and healthy control patients showed that cholesterol synthesis pathways were significantly different. n = 10 human (5 MS, 5 normal). Full names of pathways are listed in Table S3. (B) Heat map of cholesterol synthesis pathway genes that were significantly different between MS and normal (NL) RNAs (FDR < 0.1) revealed that cholesterol synthesis pathway genes were decreased in MS optic chiasm (green). Green color represents lower expression and red color represents higher expression. n = 10 human (5 MS, 5 normal).
Discussion
RiboTag technology was applied to EAE to determine astrocyte-specific gene expression networks in multiple regions of the CNS. Our results provide proof of concept that unbiased bioinformatics analyses of the molecular signature of gene expression during disease in a cell-specific and region-specific manner can provide unique insights. Here, the study of astrocytes in EAE unexpectedly pointed to cholesterol homeostasis. To explore how a decrease in cholesterol synthesis in astrocytes might affect demyelinating autoimmune disease, we reviewed known cholesterol actions in the CNS. Peripheral cholesterol does not pass through the blood–brain barrier; instead, cholesterols in the brain must be synthesized de novo. The predominant CNS cell that synthesizes cholesterol changes during development, with synthesis by neurons at the embryonic stage, oligodendrocytes at the postnatal stage, and astrocytes at the adult stage (25). In adult brain, cholesterols synthesized in astrocytes are transported via ATP-binding cassette transporter (ABCA1) to apolipoprotein E (ApoE) to neurons and oligodendrocytes. Neurons use cholesterol to make cell membranes and synapses (28, 29) while oligodendrocytes use cholesterol to make myelin (25, 30) (Fig. 8A). Thus, we hypothesize that reduced cholesterol synthesis in astrocytes during EAE may lead to reduced cholesterol available for transport to neurons and/or oligodendrocytes. While demyelination and synaptic loss are due to the autoimmune attack in EAE, limited reparative synaptic plasticity and remyelination may be due in part to reduced synthesis of cholesterol by astrocytes (Fig. 8B). Further investigation of this hypothesis is now warranted.
Fig. 8.
Hypothetical effect of reduced cholesterol synthesis in astrocytes during EAE. (A) Peripheral cholesterols cannot enter into the CNS due to the blood–brain barrier; thus, cholesterols in the CNS are synthesized de novo. In adults, astrocytes are the main cells producing cholesterols (25), with transport via ATP-binding cassette transporter (ABCA1) to apolipoprotein E (ApoE) to neurons to make membranes and synapses (28, 29), and to oligodendrocytes to make myelin (25, 30). (B) In EAE, there is synaptic loss, axonal damage, and demyelination (71). Here, we hypothesize that less cholesterol synthesis in astrocytes during EAE may lead to reduced cholesterol for transport to neurons and oligodendrocytes, thereby reducing reparative synaptic plasticity and remyelination.
Other effects of decreased cholesterol synthesis in astrocytes during EAE are possible and not mutually exclusive. Another purpose of the de novo synthesis of cholesterol in astrocytes is neurosteroid production. Cholesterol is a precursor for estradiol, progesterone, and testosterone. Neurosteroid production in the CNS is known to be a neuroprotective response to brain injury (31–34), and expression of neurosteroid receptors is increased in MS brain (35). Sex steroids delivered systemically are lipophilic, cross the blood–brain barrier, and are neuroprotective in MS models (36–38). Thus, exogenous sex hormone treatments may compensate for decreased neurosteroid production from cholesterol in astrocytes during disease in MS models and perhaps in MS clinical trials (39–43).
Consistent with our region-specific transcriptomics data, clinical studies in MS have suggested that altered levels of cholesterol precursors and oxysterols in blood and cerebrospinal fluid may be biomarkers for neurodegeneration (44, 45). Treatment with statins in MS clinical trials has focused on antiinflammatory effects, but whether statins can enter the CNS during disease to affect cholesterol homeostasis in astrocytes in a deleterious or beneficial manner warrants investigation (46–48). Together, our data in spinal cord and optic nerve suggest that targeting cholesterol synthesis in astrocytes in MS may affect walking and vision, respectively.
Regarding other MS disabilities, there are implications for cholesterol metabolism in the hippocampus, given the association of apolipoprotein E (apoE) allele inheritance in cognitive disability in MS (49–51). Cognition is frequently affected in MS, including hippocampal-dependent verbal and spatial memory (52). Hippocampal atrophy and neuropathology have been shown in EAE (18, 21, 53–55) and MS (56–58). However, we found relatively few cholesterol gene expression changes in astrocytes in hippocampus in EAE. That said, an effect of disease on cholesterol pathways in another cell type (neurons, oligodendrocytes) in hippocampus during EAE remains possible.
Findings herein also have implications for the role of astrocytes in the immunopathogenesis of MS and EAE. It is known that there are regional differences in local inflammatory infiltrates in MS and EAE. There is diffuse and localized microglial and astrocyte activation with high levels of T lymphocyte and macrophage infiltration in white matter while there is diffuse microglial and astrocyte activation with a relative paucity of localized T lymphocyte and macrophage infiltration in gray matter, each accompanied by axonal and/or synaptic loss (11, 21, 59, 60). Inflammatory infiltrates in EAE white matter can also vary qualitatively from one CNS region to another (61–63). A central question is whether differences in local inflammatory infiltrates are responsible for regional differences in astrocyte gene expression, or whether regional differences in astrocyte gene expression are responsible for differences in local inflammatory infiltrates. Either is possible as a causal driver of the other. Since differences in gene expression in astrocytes exist in healthy states in the absence of inflammation (1), regional differences in astrocytes could drive regional differences in immune infiltration during EAE. For example, lipocalin2 (LCN2), an autocrine mediator of astrocyte activation (64), drives expression of CCL2 and CXCL10 to promote microglia activation and peripheral immune infiltration in optic nerve and spinal cord during EAE (65–67). Here, the astrocyte transcriptome during EAE showed that Lcn2 gene expression was increased in astrocytes from white, but not gray, matter (Dataset S1). Conversely, differential production of IFNγ and interleukin-17 (IL-17) in peripheral immune cells has led to differential inflammation in white matter regions during autoimmune inflammation (62, 63), and LCN2 expression in astrocytes could be induced by IFNγ, IL-17, and tumor necrosis factor α (TNFα) (68, 69). Thus, these mechanisms are not mutually exclusive and may coexist in a self-perpetuating feedback loop. Regardless of causality, regional differences in astrocyte gene expression and immune infiltration may provide a rationale for a therapeutic strategy optimally targeting each region and its associated disability.
In conclusion, discovery of cell-specific and region-specific molecular signatures of disease in preclinical models can provide novel targets tailored for each neurological pathway in MS as a strategy to optimize neuroprotective treatment development. Accordingly, a clinical trial designed to reverse the molecular signature within a specific neurological pathway should have a primary outcome measure sensitive to change in a specific disability, rather than a composite that pools different disabilities. This disability-specific approach that begins with discovery and ends with clinical trial design represents a strategy for finding neuroprotective treatments, not only for MS but also for other multifocal neurodegenerative diseases.
Materials and Methods
Mice.
RiboTag mice (9) were purchased from The Jackson Laboratory. Mice expressing HA-tagged ribosomal protein RPL22 in astrocytes were generated by crossing RiboTag mice with GFAP-Cre mice (13). All animal experiments were approved by the University of California, Los Angeles Animal Research Committee. Informed consent was attained for research involving human autopsy tissues.
Active EAE Induction, Clinical Scoring, and Treatment.
GFAP-Cre RiboTag mice and WT C57BL/6 mice were induced with active EAE and scored, as described (16). CS-6253 or vehicle treatment was started 4 d before EAE induction, as described (70).
RNA Coimmunoprecipitation.
Immunoprecipitation using mouse monoclonal anti-HA antibody or rabbit anti-RPL22 control were as described (13), with minor modification.
Quantitative RT-PCR and Immunohistochemistry.
Standard procedures were used to quantify gene expression (see SI Materials and Methods for primer sequences) and to colocalize cholesterol synthesis gene expression with GFAP, as described (10).
High Throughput Sequencing and Statistical Analysis.
Astrocyte-specific transcriptome (mouse) and whole tissue transcriptome (human) analyses were performed using a high throughput sequencing approach.
Data Availability.
Datasets are available in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (SuperSeries accession no. GSE100330).
Further details on methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Giovanni Coppola and Dr. Riki Kawaguchi for discussions regarding RNA sequencing and bioinformatics analyses, as well as Kimberly Kwan and Krystyna Maruszko for laboratory assistance. Tissue specimens were obtained from the Human Brain and Spinal Fluid Resource Center, Veterans Administration Healthcare System (Los Angeles), which is sponsored by the National Institutes of Health, the National Multiple Sclerosis Society, and the US Department of Veterans Affairs. This work was supported by Conrad N. Hilton Foundation Grants 20130231 and 20150232, NIH Grant R01NS096748, and California Community Foundation Grant BAPP-15-118094 (each to R.R.V.), and funding from the Tom Sherak MS Hope Foundation, the Rhoda Goetz Foundation for Multiple Sclerosis, and other partners of the University of California, Los Angeles MS Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Datasets generated during this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ [SuperSeries accession no. GSE100330) and accession nos. GSE100329 (astrocyte-specific and region-specific transcriptomes in control and EAE mice; 45 samples; paired end), GSE100294 (optic nerve astrocyte-specific transcriptomes in control and EAE mice; 8 samples; single end), and GSE100297 (human optic chiasm from healthy controls and multiple sclerosis patients; 10 samples; single end)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716032115/-/DCSupplemental.
References
- 1.Khakh BS, Sofroniew MV. Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci. 2015;18:942–952. doi: 10.1038/nn.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko Y, et al. Cell type-specific genes show striking and distinct patterns of spatial expression in the mouse brain. Proc Natl Acad Sci USA. 2013;110:3095–3100. doi: 10.1073/pnas.1222897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viganò F, Möbius W, Götz M, Dimou L. Transplantation reveals regional differences in oligodendrocyte differentiation in the adult brain. Nat Neurosci. 2013;16:1370–1372. doi: 10.1038/nn.3503. [DOI] [PubMed] [Google Scholar]
- 4.MacKenzie-Graham A, et al. Disability-specific atlases of gray matter loss in relapsing-remitting multiple sclerosis. JAMA Neurol. 2016;73:944–953. doi: 10.1001/jamaneurol.2016.0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Racke MK, Imitola J. Cortical volume loss and neurologic dysfunction in multiple sclerosis. JAMA Neurol. 2016;73:910–912. doi: 10.1001/jamaneurol.2016.1535. [DOI] [PubMed] [Google Scholar]
- 6.Burbach GJ, Dehn D, Del Turco D, Staufenbiel M, Deller T. Laser microdissection reveals regional and cellular differences in GFAP mRNA upregulation following brain injury, axonal denervation, and amyloid plaque deposition. Glia. 2004;48:76–84. doi: 10.1002/glia.20057. [DOI] [PubMed] [Google Scholar]
- 7.Burbach GJ, Dehn D, Nagel B, Del Turco D, Deller T. Laser microdissection of immunolabeled astrocytes allows quantification of astrocytic gene expression. J Neurosci Methods. 2004;138:141–148. doi: 10.1016/j.jneumeth.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Matcovitch-Natan O, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 9.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spence RD, et al. Neuroprotection mediated through estrogen receptor-alpha in astrocytes. Proc Natl Acad Sci USA. 2011;108:8867–8872. doi: 10.1073/pnas.1103833108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voskuhl RR, et al. Reactive astrocytes form scar-like perivascular barriers to leukocytes during adaptive immune inflammation of the CNS. J Neurosci. 2009;29:11511–11522. doi: 10.1523/JNEUROSCI.1514-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosnan CF, Raine CS. The astrocyte in multiple sclerosis revisited. Glia. 2013;61:453–465. doi: 10.1002/glia.22443. [DOI] [PubMed] [Google Scholar]
- 13.Anderson MA, et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature. 2016;532:195–200. doi: 10.1038/nature17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becher B, Prat A, Antel JP. Brain-immune connection: Immuno-regulatory properties of CNS-resident cells. Glia. 2000;29:293–304. [PubMed] [Google Scholar]
- 15.Sofroniew MV. Astrocyte barriers to neurotoxic inflammation. Nat Rev Neurosci. 2015;16:249–263. doi: 10.1038/nrn3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du S, et al. XY sex chromosome complement, compared with XX, in the CNS confers greater neurodegeneration during experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2014;111:2806–2811. doi: 10.1073/pnas.1307091111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKenzie-Graham A, et al. Purkinje cell loss in experimental autoimmune encephalomyelitis. Neuroimage. 2009;48:637–651. doi: 10.1016/j.neuroimage.2009.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nisticò R, et al. Inflammation subverts hippocampal synaptic plasticity in experimental multiple sclerosis. PLoS One. 2013;8:e54666. doi: 10.1371/journal.pone.0054666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen S, et al. Persistent activation of microglia is associated with neuronal dysfunction of callosal projecting pathways and multiple sclerosis-like lesions in relapsing–Remitting experimental autoimmune encephalomyelitis. Brain. 2007;130:2816–2829. doi: 10.1093/brain/awm219. [DOI] [PubMed] [Google Scholar]
- 20.Spence RD, et al. Bringing CLARITY to gray matter atrophy. Neuroimage. 2014;101:625–632. doi: 10.1016/j.neuroimage.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziehn MO, Avedisian AA, Tiwari-Woodruff S, Voskuhl RR. Hippocampal CA1 atrophy and synaptic loss during experimental autoimmune encephalomyelitis, EAE. Lab Invest. 2010;90:774–786. doi: 10.1038/labinvest.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarkson BD, et al. CCR2-dependent dendritic cell accumulation in the central nervous system during early effector experimental autoimmune encephalomyelitis is essential for effector T cell restimulation in situ and disease progression. J Immunol. 2015;194:531–541. doi: 10.4049/jimmunol.1401320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soulika AM, et al. Initiation and progression of axonopathy in experimental autoimmune encephalomyelitis. J Neurosci. 2009;29:14965–14979. doi: 10.1523/JNEUROSCI.3794-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiwari-Woodruff S, Morales LB, Lee R, Voskuhl RR. Differential neuroprotective and antiinflammatory effects of estrogen receptor (ER)alpha and ERbeta ligand treatment. Proc Natl Acad Sci USA. 2007;104:14813–14818. doi: 10.1073/pnas.0703783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saher G, Stumpf SK. Cholesterol in myelin biogenesis and hypomyelinating disorders. Biochim Biophys Acta. 2015;1851:1083–1094. doi: 10.1016/j.bbalip.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Boehm-Cagan A, et al. Differential effects of apoE4 and activation of ABCA1 on brain and plasma lipoproteins. PLoS One. 2016;11:e0166195. doi: 10.1371/journal.pone.0166195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boehm-Cagan A, et al. ABCA1 agonist reverses the ApoE4-driven cognitive and brain pathologies. J Alzheimers Dis. 2016;54:1219–1233. doi: 10.3233/JAD-160467. [DOI] [PubMed] [Google Scholar]
- 28.Fester L, et al. Cholesterol-promoted synaptogenesis requires the conversion of cholesterol to estradiol in the hippocampus. Hippocampus. 2009;19:692–705. doi: 10.1002/hipo.20548. [DOI] [PubMed] [Google Scholar]
- 29.Mauch DH, et al. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- 30.Saher G, et al. High cholesterol level is essential for myelin membrane growth. Nat Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- 31.Giatti S, et al. Acute experimental autoimmune encephalomyelitis induces sex dimorphic changes in neuroactive steroid levels. Neurochem Int. 2010;56:118–127. doi: 10.1016/j.neuint.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Melcangi RC, Garcia-Segura LM, Mensah-Nyagan AG. Neuroactive steroids: State of the art and new perspectives. Cell Mol Life Sci. 2008;65:777–797. doi: 10.1007/s00018-007-7403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson RS, Saldanha CJ, Schlinger BA. Rapid upregulation of aromatase mRNA and protein following neural injury in the zebra finch (Taeniopygia guttata) J Neuroendocrinol. 2001;13:317–323. doi: 10.1046/j.1365-2826.2001.00647.x. [DOI] [PubMed] [Google Scholar]
- 34.Luchetti S, Huitinga I, Swaab DF. Neurosteroid and GABA-A receptor alterations in Alzheimer’s disease, Parkinson’s disease and multiple sclerosis. Neuroscience. 2011;191:6–21. doi: 10.1016/j.neuroscience.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Luchetti S, et al. Gender differences in multiple sclerosis: Induction of estrogen signaling in male and progesterone signaling in female lesions. J Neuropathol Exp Neurol. 2014;73:123–135. doi: 10.1097/NEN.0000000000000037. [DOI] [PubMed] [Google Scholar]
- 36.Hussain R, et al. The neural androgen receptor: A therapeutic target for myelin repair in chronic demyelination. Brain. 2013;136:132–146. doi: 10.1093/brain/aws284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spence RD, Voskuhl RR. Neuroprotective effects of estrogens and androgens in CNS inflammation and neurodegeneration. Front Neuroendocrinol. 2012;33:105–115. doi: 10.1016/j.yfrne.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voskuhl RR, Gold SM. Sex-related factors in multiple sclerosis susceptibility and progression. Nat Rev Neurol. 2012;8:255–263. doi: 10.1038/nrneurol.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurth F, et al. Neuroprotective effects of testosterone treatment in men with multiple sclerosis. Neuroimage Clin. 2014;4:454–460. doi: 10.1016/j.nicl.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sicotte NL, et al. Testosterone treatment in multiple sclerosis: A pilot study. Arch Neurol. 2007;64:683–688. doi: 10.1001/archneur.64.5.683. [DOI] [PubMed] [Google Scholar]
- 41.Sicotte NL, et al. Treatment of multiple sclerosis with the pregnancy hormone estriol. Ann Neurol. 2002;52:421–428. doi: 10.1002/ana.10301. [DOI] [PubMed] [Google Scholar]
- 42.Voskuhl R, Wang H, Elashoff RM. Why use sex hormones in relapsing-remitting multiple sclerosis?–Authors’ reply. Lancet Neurol. 2016;15:790–791. doi: 10.1016/S1474-4422(16)00129-0. [DOI] [PubMed] [Google Scholar]
- 43.Voskuhl RR, et al. Estriol combined with glatiramer acetate for women with relapsing-remitting multiple sclerosis: A randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 2016;15:35–46. doi: 10.1016/S1474-4422(15)00322-1. [DOI] [PubMed] [Google Scholar]
- 44.Teunissen CE, Dijkstra C, Polman C. Biological markers in CSF and blood for axonal degeneration in multiple sclerosis. Lancet Neurol. 2005;4:32–41. doi: 10.1016/S1474-4422(04)00964-0. [DOI] [PubMed] [Google Scholar]
- 45.van de Kraats C, et al. Oxysterols and cholesterol precursors correlate to magnetic resonance imaging measures of neurodegeneration in multiple sclerosis. Mult Scler. 2014;20:412–417. doi: 10.1177/1352458513499421. [DOI] [PubMed] [Google Scholar]
- 46.Chataway J, et al. Effect of high-dose simvastatin on brain atrophy and disability in secondary progressive multiple sclerosis (MS-STAT): A randomised, placebo-controlled, phase 2 trial. Lancet. 2014;383:2213–2221. doi: 10.1016/S0140-6736(13)62242-4. [DOI] [PubMed] [Google Scholar]
- 47.Giovannoni G, Baker D, Schmierer K. Simvastatin in patients with progressive multiple sclerosis. Lancet. 2014;384:952. doi: 10.1016/S0140-6736(14)61621-4. [DOI] [PubMed] [Google Scholar]
- 48.Youssef S, et al. The HMG-CoA reductase inhibitor, atorvastatin, promotes a Th2 bias and reverses paralysis in central nervous system autoimmune disease. Nature. 2002;420:78–84. doi: 10.1038/nature01158. [DOI] [PubMed] [Google Scholar]
- 49.Benedict RH, Zivadinov R. Risk factors for and management of cognitive dysfunction in multiple sclerosis. Nat Rev Neurol. 2011;7:332–342. doi: 10.1038/nrneurol.2011.61. [DOI] [PubMed] [Google Scholar]
- 50.Shi J, Zhao CB, Vollmer TL, Tyry TM, Kuniyoshi SM. APOE epsilon 4 allele is associated with cognitive impairment in patients with multiple sclerosis. Neurology. 2008;70:185–190. doi: 10.1212/01.wnl.0000264004.62612.44. [DOI] [PubMed] [Google Scholar]
- 51.Shi J, Han P, Kuniyoshi SM. Cognitive impairment in neurological diseases: Lessons from apolipoprotein E. J Alzheimers Dis. 2014;38:1–9. doi: 10.3233/JAD-130623. [DOI] [PubMed] [Google Scholar]
- 52.Chiaravalloti ND, DeLuca J. Cognitive impairment in multiple sclerosis. Lancet Neurol. 2008;7:1139–1151. doi: 10.1016/S1474-4422(08)70259-X. [DOI] [PubMed] [Google Scholar]
- 53.Di Filippo M, et al. Effects of central and peripheral inflammation on hippocampal synaptic plasticity. Neurobiol Dis. 2013;52:229–236. doi: 10.1016/j.nbd.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Ziehn MO, Avedisian AA, Dervin SM, O’Dell TJ, Voskuhl RR. Estriol preserves synaptic transmission in the hippocampus during autoimmune demyelinating disease. Lab Invest. 2012;92:1234–1245. doi: 10.1038/labinvest.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziehn MO, et al. Therapeutic testosterone administration preserves excitatory synaptic transmission in the hippocampus during autoimmune demyelinating disease. J Neurosci. 2012;32:12312–12324. doi: 10.1523/JNEUROSCI.2796-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLuca GC, Yates RL, Beale H, Morrow SA. Cognitive impairment in multiple sclerosis: Clinical, radiologic and pathologic insights. Brain Pathol. 2015;25:79–98. doi: 10.1111/bpa.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koenig KA, et al. Hippocampal volume is related to cognitive decline and fornicial diffusion measures in multiple sclerosis. Magn Reson Imaging. 2014;32:354–358. doi: 10.1016/j.mri.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sicotte NL, et al. Regional hippocampal atrophy in multiple sclerosis. Brain. 2008;131:1134–1141. doi: 10.1093/brain/awn030. [DOI] [PubMed] [Google Scholar]
- 59.Bø L, Vedeler CA, Nyland H, Trapp BD, Mørk SJ. Intracortical multiple sclerosis lesions are not associated with increased lymphocyte infiltration. Mult Scler. 2003;9:323–331. doi: 10.1191/1352458503ms917oa. [DOI] [PubMed] [Google Scholar]
- 60.Prins M, et al. Pathological differences between white and grey matter multiple sclerosis lesions. Ann N Y Acad Sci. 2015;1351:99–113. doi: 10.1111/nyas.12841. [DOI] [PubMed] [Google Scholar]
- 61.Pierson E, Simmons SB, Castelli L, Goverman JM. Mechanisms regulating regional localization of inflammation during CNS autoimmunity. Immunol Rev. 2012;248:205–215. doi: 10.1111/j.1600-065X.2012.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoolman JS, Duncker PC, Huber AK, Segal BM. Site-specific chemokine expression regulates central nervous system inflammation and determines clinical phenotype in autoimmune encephalomyelitis. J Immunol. 2014;193:564–570. doi: 10.4049/jimmunol.1400825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stromnes IM, Cerretti LM, Liggitt D, Harris RA, Goverman JM. Differential regulation of central nervous system autoimmunity by T(H)1 and T(H)17 cells. Nat Med. 2008;14:337–342. doi: 10.1038/nm1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee S, et al. Lipocalin-2 is an autocrine mediator of reactive astrocytosis. J Neurosci. 2009;29:234–249. doi: 10.1523/JNEUROSCI.5273-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chun BY, et al. Pathological Involvement of astrocyte-derived lipocalin-2 in the demyelinating optic neuritis. Invest Ophthalmol Vis Sci. 2015;56:3691–3698. doi: 10.1167/iovs.15-16851. [DOI] [PubMed] [Google Scholar]
- 66.Lee S, et al. Lipocalin-2 is a chemokine inducer in the central nervous system: Role of chemokine ligand 10 (CXCL10) in lipocalin-2-induced cell migration. J Biol Chem. 2011;286:43855–43870. doi: 10.1074/jbc.M111.299248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nam Y, et al. Lipocalin-2 protein deficiency ameliorates experimental autoimmune encephalomyelitis: The pathogenic role of lipocalin-2 in the central nervous system and peripheral lymphoid tissues. J Biol Chem. 2014;289:16773–16789. doi: 10.1074/jbc.M113.542282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen F, Hu Z, Goswami J, Gaffen SL. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J Biol Chem. 2006;281:24138–24148. doi: 10.1074/jbc.M604597200. [DOI] [PubMed] [Google Scholar]
- 69.Zhao P, Stephens JM. STAT1, NF-κB and ERKs play a role in the induction of lipocalin-2 expression in adipocytes. Mol Metab. 2013;2:161–170. doi: 10.1016/j.molmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bielicki JK, et al. A new HDL mimetic peptide that stimulates cellular cholesterol efflux with high efficiency greatly reduces atherosclerosis in mice. J Lipid Res. 2010;51:1496–1503. doi: 10.1194/jlr.M003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Itoh N, et al. Bedside to bench to bedside research: Estrogen receptor beta ligand as a candidate neuroprotective treatment for multiple sclerosis. J Neuroimmunol. 2017;304:63–71. doi: 10.1016/j.jneuroim.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets are available in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (SuperSeries accession no. GSE100330).
Further details on methods are provided in SI Materials and Methods.