Significance
Autophagy mediates the routing of cytoplasmic components to degradative membrane-bound compartments. During infection by intracellular pathogens, autophagy proteins orchestrate several antimicrobial responses by marking pathogen-containing vacuoles and protecting the host cytosol from invaders. However, intracellular pathogens such as Listeria monocytogenes circumvent the autophagy machinery to promote pathogenesis. By combining bacterial and host mutants, we have dissected the role of two distinct autophagy-related processes in controlling L. monocytogenes growth in macrophages. Our results showed that L. monocytogenes is oblivious to the initial marking of its vacuole by autophagy proteins, but that subsequent autophagic targeting restricts bacterial growth in the host cytosol. We suggest that processes coordinated by the autophagy machinery constitute a multilayered network of cell-autonomous defenses.
Keywords: LC3-associated phagocytosis, phospholipases, ActA, bacteria, macrophage
Abstract
Xenophagy is a selective macroautophagic process that protects the host cytosol by entrapping and delivering microbes to a degradative compartment. Both noncanonical autophagic pathways and xenophagy are activated by microbes during infection, but the relative importance and function of these distinct processes are not clear. In this study, we used bacterial and host mutants to dissect the contribution of autophagic processes responsible for bacterial growth restriction of Listeria monocytogenes. L. monocytogenes is a facultative intracellular pathogen that escapes from phagosomes, grows in the host cytosol, and avoids autophagy by expressing three determinants of pathogenesis: two secreted phospholipases C (PLCs; PlcA and PlcB) and a surface protein (ActA). We found that shortly after phagocytosis, wild-type (WT) L. monocytogenes escaped from a noncanonical autophagic process that targets damaged vacuoles. During this process, the autophagy marker LC3 localized to single-membrane phagosomes independently of the ULK complex, which is required for initiation of macroautophagy. However, growth restriction of bacteria lacking PlcA, PlcB, and ActA required FIP200 and TBK1, both involved in the engulfment of microbes by xenophagy. Time-lapse video microscopy revealed that deposition of LC3 on L. monocytogenes-containing vacuoles via noncanonical autophagy had no apparent role in restricting bacterial growth and that, upon access to the host cytosol, WT L. monocytogenes utilized PLCs and ActA to avoid subsequent xenophagy. In conclusion, although noncanonical autophagy targets phagosomes, xenophagy was required to restrict the growth of L. monocytogenes, an intracellular pathogen that damages the entry vacuole.
The autophagy network includes catabolic processes (e.g., macroautophagy) that target cytoplasmic components for lysosomal degradation and is critically important to maintain cellular homeostasis (1). Defects in autophagy are associated with a wide range of pathophysiologies, including autoimmunity, neurodegeneration, infectious diseases, and cancer (1, 2). During macroautophagy, cytoplasmic components are enclosed in double-membrane vesicles (i.e., autophagosomes), which ultimately mature into autolysosomes (1). Although macroautophagy nonselectively digests parts of the cytoplasm during starvation, it also mediates the clearance of specific cellular components such as protein aggregates, damaged organelles, and intracellular microbes. Therefore, macroautophagy contributes to host immune defenses by controlling the replication of intracellular microbes (3, 4). In addition, the autophagy machinery orchestrates noncanonical processes that overlap with other cell-autonomous defense mechanisms such as the phagolysosomal pathway (5).
Intracellular bacteria can be targeted by the autophagy machinery while confined in vacuoles or free in the cytosol. Targeting can proceed through noncanonical pathways that result in lipidation of proteins of the ATG8 family (LC3s/GABARAPs) on pathogen-containing vacuoles and do not rely on the formation of autophagosomes (5–7). One of the best characterized noncanonical autophagy pathways, referred to as LC3-associated phagocytosis (LAP), is triggered during phagocytosis following the engagement of a subset of immune receptors (5). In addition to LAP, other pathways promote recruitment of the autophagy machinery and lipidation of ATG8 proteins on pathogen-containing vacuoles. For example, the ATG12–ATG5–ATG16L1 complex is recruited to and acts upon Salmonella-containing vacuoles through the ubiquitin-binding domain of ATG16L1 (6, 8). The decoration of pathogen-containing vacuoles with ATG8 proteins may restrict the growth of intracellular microorganisms by promoting phagosome maturation or the recruitment of specific antimicrobial effectors (7).
Pathogens and pathogen-containing vacuoles can be marked by “eat-me” signals that trigger a macroautophagic process termed xenophagy (3, 4). Intracellular pathogens can alter phagosomes using auxiliary secretion systems or pore-forming toxins, which cause membrane damage to endocytic/phagocytic compartments (9). These membrane breaches expose β-galactosides that normally decorate the inner leaflet of vacuoles for binding by cytosolic galectins (10). In addition, ruptured pathogen-containing vacuoles and cytosolic microbes can be targeted by ubiquitin ligases and coated with ubiquitin chains. Both galectins and ubiquitin chains mediate the recruitment of autophagy adaptors such as p62 and NDP52. These autophagy adaptors serve as bridges between the ubiquitin-associated cargo and lipidated LC3 proteins, and mediate engulfment by autophagosomal membranes. The recognition of microbes by autophagy adaptors triggers a localized macroautophagic process, which requires the activation of the protein kinase TBK1 (11).
The intracellular bacterial pathogen Listeria monocytogenes escapes from the endolysosomal pathway and grows in the cytosol of many cell types (12). In macrophages, escape from phagosomes requires the pore-forming cytolysin listeriolysin O (LLO) (13) while a phosphatidylinositol-specific phospholipase C (PlcA) and a broad-range phospholipase C (PlcB) also contribute to phagosomal escape (14). Upon exposure to the host cytosol, L. monocytogenes expresses the surface protein ActA that hijacks the host actin polymerization machinery, and allows bacteria to move intracellularly and to disseminate into neighboring cells (15).
Like many intracellular pathogens, L. monocytogenes actively circumvents host autophagy (3). First, it is targeted by the autophagy machinery during the transition from the entry vacuole to the cytosol (10, 16), a process that requires LLO (17). However, once in the cytosol, L. monocytogenes uses ActA to protect its surface from recognition by ubiquitin ligases and autophagy adaptors (18). In addition, bacterial phospholipases C (PlcA and PlcB), predominantly PlcA, interfere with autophagy by reducing the intracellular levels of phosphatidylinositol 3-phosphate (PI3P) (19, 20), a signaling molecule required for macroautophagic processes and LAP (3, 5). Importantly, results from Tattoli et al. (19) showed that PLCs may inhibit autophagy during the cytosolic phase of the intracellular life cycle of L. monocytogenes, which suggested that PLCs function beyond their role in facilitating escape from the entry vacuole. In macrophages, autophagy avoidance is essential for the intracellular growth of L. monocytogenes and requires PLCs and ActA (20). The precise roles of noncanonical autophagy and xenophagy in restricting the growth of intracellular microbes have been challenging to reconcile because intracellular pathogens evade these processes. However, in this study, by combining bacterial and host mutants, we dissect the involvement of these different autophagic pathways in controlling the growth of L. monocytogenes during macrophage infection.
Results
L. monocytogenes Escapes from an Autophagic Process That Targets Damaged Vacuoles.
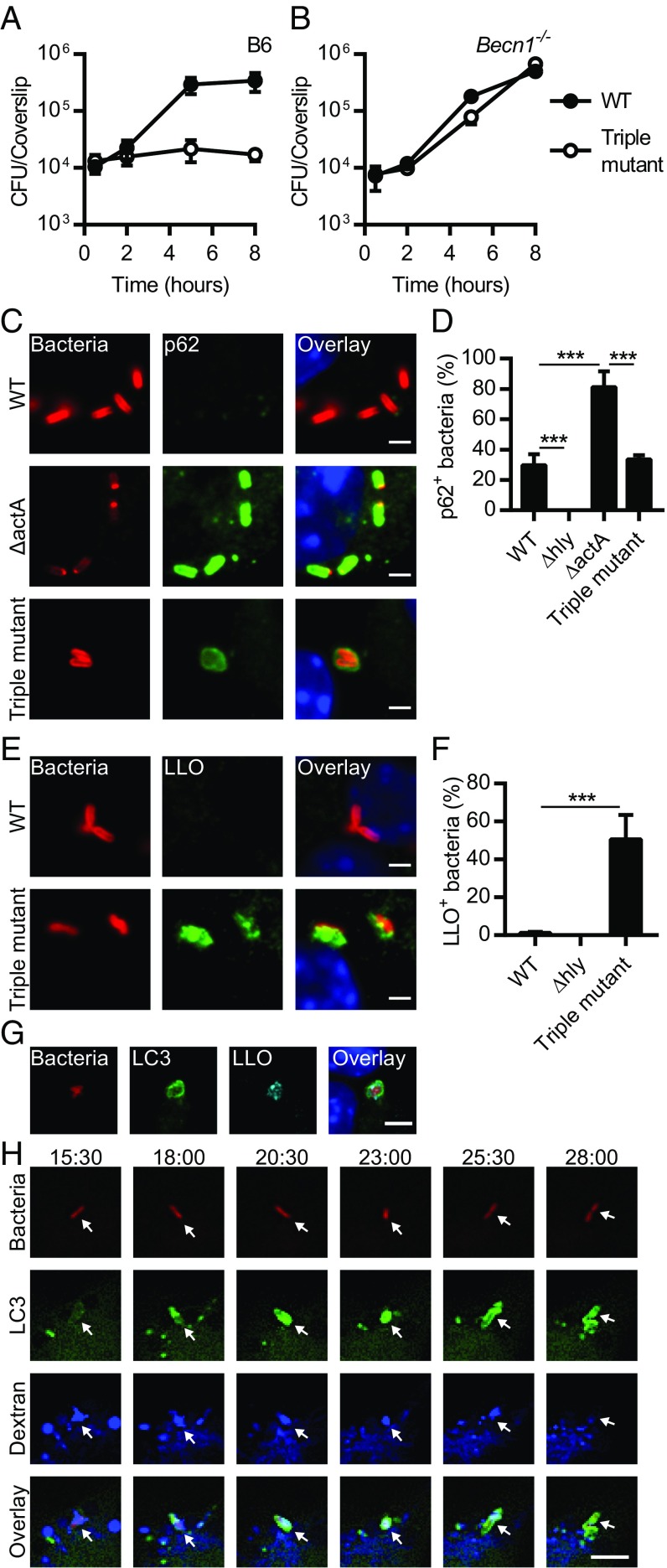
We generated a series of in-frame deletion mutants in actA, plcA, and plcB, as well as single amino acid substitution mutants in the active site of PlcA and PlcB (Table S1). Mutants lacking ActA, PlcA, and PlcB (referred to from now on as the triple mutant) had a reduced ability to grow in WT [C57BL/6 (B6)] bone marrow-derived macrophages (BMMs) (Fig. 1A). This intracellular growth defect was rescued in cells lacking the autophagy protein Beclin 1 (Fig. 1B), demonstrating that autophagy is critical for controlling the growth of the triple mutant in BMMs. Two possible scenarios were considered to explain why the triple mutant failed to grow in macrophages: (i) the triple mutant had a defect in escaping an autophagic process targeting the damaged entry vacuole, or (ii) the triple mutant entered the cytosol but was growth-restricted by xenophagy. Since ActA-minus mutants recruit p62 while targeted by xenophagy in the host cytosol (18), the colocalization of p62 with WT, Δhly, ΔactA, and the triple mutant was examined in BMMs (Fig. 1 C and D). As previously reported, ΔactA colocalized with p62 more frequently than WT bacteria (18), but, surprisingly, the triple mutant colocalized with p62 less frequently than ΔactA. In some cases, the p62-fluorescence pattern associated with the triple mutant resembled a vacuole rather than being closely associated with the bacterial surface (Fig. 1C). These results suggested that PLCs facilitated the access of L. monocytogenes to the host cytosol and promoted the recruitment of p62 to the surface of ActA-minus bacteria. Therefore, these data supported the hypothesis that p62 failed to accumulate around the triple mutant because it remained in the entry vacuole.
Fig. 1.
L. monocytogenes escapes from an autophagic process that targets damaged vacuoles. Growth kinetics of WT L. monocytogenes and the triple mutant lacking ActA, PlcA, and PlcB in B6 (A) and Becn1−/− (B) BMMs (n = 2–4). (C) Representative micrographs of BMMs infected for 2 h with WT, ΔactA, and the triple mutant. Infected cells were stained for L. monocytogenes (red), p62 (green), and DNA (blue). (D) Colocalization of p62 with WT, Δhly, ΔactA, and the triple mutant in BMMs infected for 2 h. Relevant statistically significant differences are indicated [***P < 0.001 (ANOVA with Tukey’s post hoc test); n = 5]. (E) Representative micrographs of BMMs infected with WT and the triple mutant for 2 h and stained with L. monocytogenes (red), LLO (green), and DNA (blue). (F) Colocalization of LLO with WT, Δhly, and the triple mutant in BMMs infected for 2 h. Relevant statistically significant differences are indicated [***P < 0.001 (ANOVA with Tukey’s post hoc test); n = 3]. (G) Representative micrographs of GFP-LC3 BMMs infected with the triple mutant for 2 h and stained for L. monocytogenes (red), GFP-LC3 (green), LLO (cyan), and DNA (blue). (H) Selected Z-stacked micrographs from a time-lapse microscopy experiment performed with GFP-LC3 (green) BMMs infected with the triple mutant expressing mCherry (red) in presence of fluorescent dextran (blue). Arrowheads indicate the position of bacteria in the different channels and at different time points. Times are indicated (min:s) above the top row. Results are expressed as means and SDs. (Scale bars: C and E, 2 µm; G and H, 5 µm.)
To test the hypothesis that the triple mutant was targeted by an autophagic process in a vacuole, the association between the membrane-binding cytolysin LLO and L. monocytogenes was evaluated during macrophage infection. The triple mutant colocalized with LLO structures reminiscent of vacuoles or membrane remnants (Fig. 1E). Importantly, the degree of colocalization was significantly increased in macrophages infected with the triple mutant in comparison with macrophages infected with WT bacteria (Fig. 1F). In addition, the triple mutant that associated with LC3 was often also positive for LLO (Fig. 1G) (70.1 ± 11.2%; n = 3), suggesting that LC3 is recruited to phagosomes perforated by the cytolysin. This relationship between LC3 recruitment and vacuole damage was further established using time-lapse video microscopy and GFP-LC3 BMMs infected in the presence of fluorescent dextran molecules, which diffuse into the cytosol following vacuole disruption (21). LC3 recruitment to triple mutant-containing vacuoles correlated with the loss of dextran signal (Fig. 1H and Movie S1). These results showed that the triple mutant was targeted by an autophagic process in damaged vacuoles.
L. monocytogenes Is Targeted by Noncanonical Autophagy.
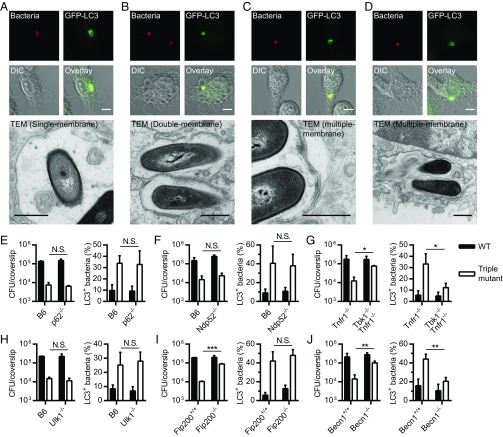
Cytosolic bacteria and bacteria-containing vacuoles can be targeted by xenophagy and trapped in double-membrane vacuoles (3, 4). Alternatively, ATG8 proteins can be lipidated on single-membrane vacuoles through a noncanonical process (5–7). To gain additional insight into the autophagic pathway(s) that targets L. monocytogenes during macrophage infection, the ultrastructure of the LC3+ vacuoles associated with the triple mutant was studied using correlative light-electron microscopy (CLEM), as previously described (16, 22). At 1 h postinfection (p.i.), the ultrastructure of LC3+ triple mutant-containing vacuoles was heterogeneous, with bacteria in single-, double-, and multiple-membrane vacuoles (Fig. 2 A–D). Similar results were obtained at 2 h p.i. These results showed that the triple mutant was targeted by multiple autophagic processes during infection.
Fig. 2.
L. monocytogenes is targeted by noncanonical autophagy. (A–D) CLEM of GFP-LC3 BMMs infected with the triple mutant expressing mCherry for 1 h. Micrographs showing bacteria (red), GFP-LC3 (green), differential interference contrast (DIC) images, and transmission electron microscopy (TEM) images are shown. (Scale bars: fluorescence/DIC micrographs, 5 µm; TEM micrographs, 500 nm.) CFUs (at 5 h p.i.) and LC3 colocalization (at 2 h p.i.) in p62−/− (E), Ndp52−/− (F), Tbk1−/− Tnfr1−/− (G), Ulk1−/− (H), Fip200−/− (I), and Becn1−/− (J) BMMs infected with WT or the triple mutant. Tnfr1−/− (tumor necrosis factor receptor 1, TNFR1) BMMs are the control cells for the Tbk1−/− Tnfr1−/− BMMs. Results are expressed as means and SDs [N.S., nonsignificant; *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired t test); n = 2–4].
To study pathways involved in the autophagic targeting of L. monocytogenes, intracellular growth and LC3 colocalization of WT and the triple mutant were monitored in BMMs lacking genes important for specific autophagic processes. Intracellular growth and LC3 colocalization were not altered in BMM mutants lacking the autophagy adaptors p62 and NDP52 (Fig. 2 E and F). These data suggested that xenophagy was not involved in the targeting of L. monocytogenes or that autophagy adaptors act redundantly, as suggested for mitophagy (23). TBK1 is involved in cargo engulfment and autophagosome maturation during xenophagy (11). Interestingly, the intracellular growth defect of the triple mutant was rescued and the colocalization of LC3 with the triple mutant was decreased in BMMs lacking TBK1 (Fig. 2G). These results suggested that xenophagy restricts the growth of the triple mutant by a process that might involve several autophagy adaptors.
During xenophagy, the formation of autophagosomal membranes is initiated by the ULK complex consisting of ULK1 or ULK2, ATG13, FIP200, and ATG101 (24). However, this complex is dispensable for lipidation of ATG8 proteins on single-membrane vacuoles (5, 6). The intracellular growth and LC3 colocalization of WT and the triple mutant were not affected in Ulk1−/− BMMs (Fig. 2H), suggesting that LC3 is directly lipidated to L. monocytogenes-containing vacuoles during infection. However, loss of function of ULK1 can be compensated by ULK2 in mammalian cells (24), so it was possible that L. monocytogenes was still targeted by a macroautophagic process in Ulk1−/− BMMs. The intracellular growth and LC3 colocalization of WT and the triple mutant were then evaluated in Fip200−/− BMMs (Fig. 2I), since FIP200 is essential for the function of the ULK complex (24). Conversely, while the intracellular growth defect of the triple mutant was rescued in Fip200−/− BMMs, the triple mutant still colocalized at a high level with LC3. This showed that while FIP200 (and the ULK complex) was not required for the deposition of LC3 on Listeria-containing vacuoles, FIP200 was involved in restricting the growth of the triple mutant. Control experiments using Becn1−/− BMMs were performed as Beclin 1 is involved in both macroautophagy and LAP (Fig. 2J), and confirmed that Beclin 1 is required for the targeting of the triple mutant by LC3. Overall, the results of these experiments indicated that LC3 was recruited to L. monocytogenes-containing vacuoles by a noncanonical autophagy pathway, but that growth restriction required xenophagy.
Multiple Autophagic Processes Target Intracellular Bacteria Sequentially.
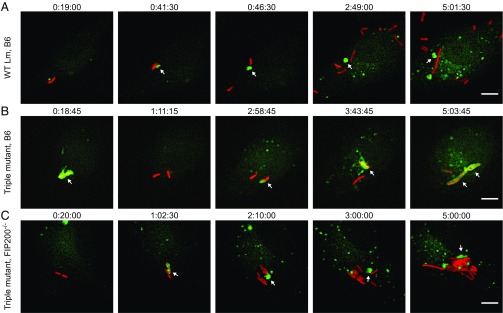
We suspected that the triple mutant was targeted sequentially, first by noncanonical autophagy and then by xenophagy. To test this hypothesis, time-lapse video microscopy was performed using GFP-LC3 BMMs infected with L. monocytogenes. Results from control experiments revealed that LC3 was frequently recruited to the entry vacuole of WT bacteria in WT BMMs (Fig. 3A and Movies S2 and S3). However, colocalization between LC3 and WT bacteria was weak and transient since WT bacteria escaped from the entry vacuoles, and proliferated in the host cytosol (Fig. 3A and Movies S2 and S3). Notably, LC3+ membranes were removed from WT bacteria and formed aggregates that persisted in the host cytosol for hours (Fig. 3A and Movies S2 and S3). In contrast, the triple mutant was often observed to be repeatedly targeted by distinct LC3 recruitment events in WT (Fig. 3B and Movies S4 and S5) and control FIP200+/+ (Movies S6 and S7) BMMs. For example, in Movie S4 (Fig. 3B), one can clearly see robust LC3 recruitment to the triple mutant at the early onset of the infection, then loss of signal and reappearance of LC3 in the surrounding of bacteria. This suggested that the damaged vacuole associated with the triple mutant was strongly targeted by LC3 and then possibly degraded before retargeting of cytosolic bacteria by xenophagy. Reappearance of LC3 was rarely observed during infection with WT bacteria. Taken together, these results suggested that both WT and the triple mutant were targeted by noncanonical autophagy shortly after phagocytosis, but that the triple mutant was subsequently growth-restricted by xenophagy. In accordance with this hypothesis, sequential targeting of the triple mutant was not observed in FIP200−/− BMMs (Fig. 3C and Movies S8 and S9). Although LC3 was recruited to L. monocytogenes-containing vacuoles in these macrophages, the triple mutant escaped and grew in the host cytosol (Fig. 3C and Movies S8 and S9). Interestingly, LC3+ membrane aggregates often localized to triple mutants following escape into the cytosol of FIP200−/− BMMs (Fig. 3C and Movies S8 and S9), and, in some cases, were observed to transiently coat entire microcolonies (Movie S9). Overall, these results demonstrated that noncanonical autophagy and xenophagy sequentially target intracellular bacteria during macrophage infection.
Fig. 3.
Multiple autophagic processes sequentially target intracellular bacteria. Selected Z-stacked micrographs from time-lapse microscopy experiments of GFP-LC3 (green) BMMs infected with mCherry L. monocytogenes strains (red). (A) GFP-LC3 WT BMMs infected with WT bacteria. Arrowheads show a LC3+ vacuole that was removed from a bacterium and formed a membrane aggregate in the host cytosol. (B) GFP-LC3 WT BMMs infected with the triple mutant. Arrowheads show bacteria that colocalized with LC3. Note that the triple mutant colocalized with LC3 multiple distinct times. (C) GFP-LC3 Fip200−/− BMMs infected with the triple mutant. Arrowheads show a LC3+ vacuole that was removed from bacteria and formed a membrane aggregate in the host cytosol. Times are indicated (h:min:s) above each micrograph. (Scale bars: 5 µm.)
L. monocytogenes Avoids Growth-Restricting Xenophagy in the Macrophage Cytosol.
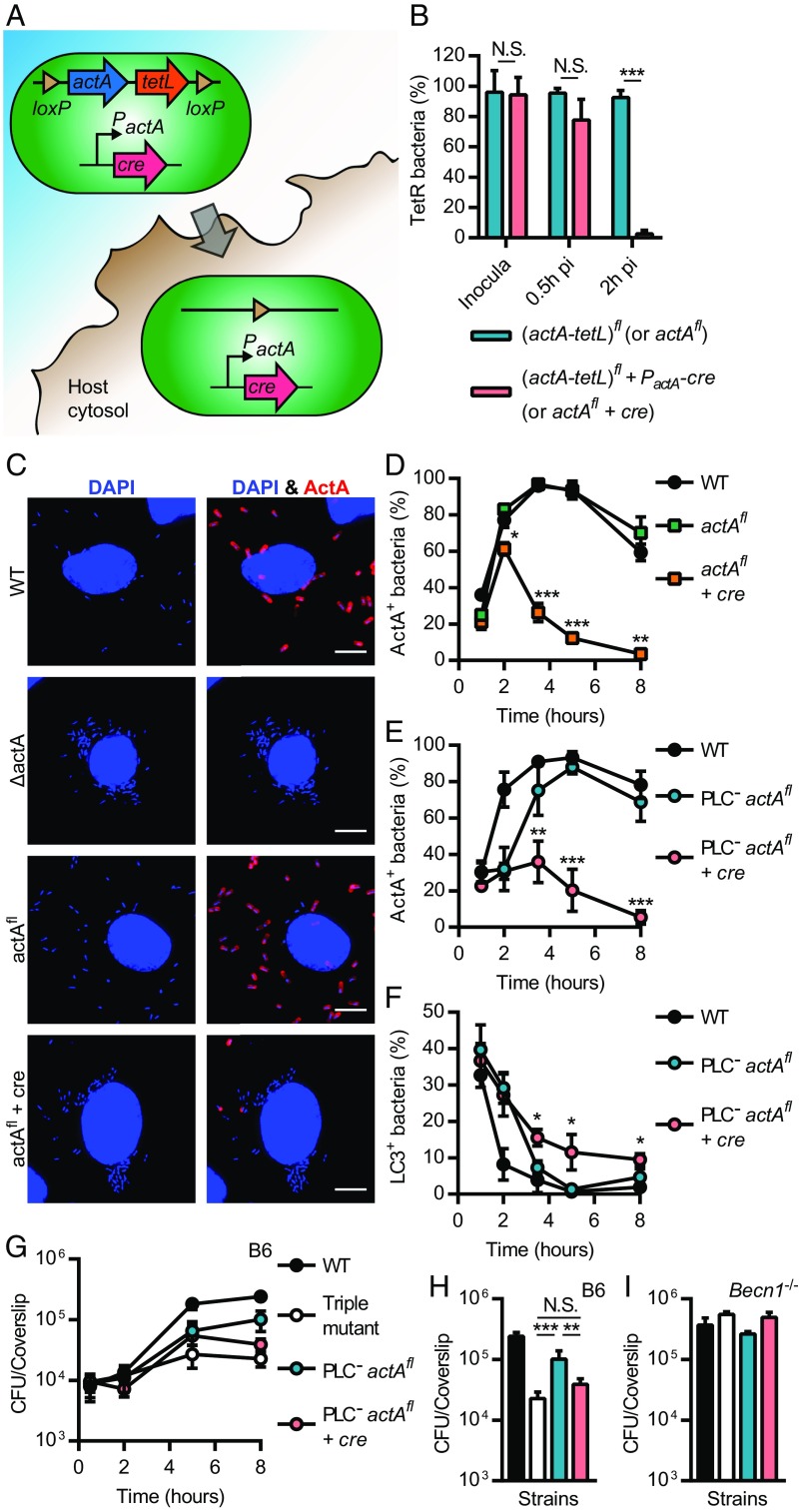
L. monocytogenes is clearly targeted in damaged vacuoles by an autophagy-related process early during infection, but these studies do not address whether bacteria in the host cytosol are also subject to growth restriction by the autophagy machinery once free of host membranes. In addition to the role of PLCs in escaping the entry vacuole, we hypothesized that ActA and PLCs act in concert in the host cytosol to interfere with growth-restricting xenophagy. To test this hypothesis, we constructed a double PLC mutant that retained a functional actA gene, which would be deleted upon entry into the host cytosol, thereby generating a triple mutant later during infection. This strain was engineered using a Cre-lox system activated by PactA, a promoter robustly up-regulated intracellularly (Fig. 4A and Supporting Information), as previously described (25). A tetracycline resistance gene (tetL) was also inserted between the loxP sites to monitor deletion during infection. A control WT strain that deletes actA upon access to the host cytosol was also constructed. These bacteria escaped from the entry vacuole and then rapidly deleted actA (Fig. 4 B–E). The deletion of actA from the genome of the PLC-minus strain resulted in an increased colocalization with LC3 at later infection time points in comparison with the PLC-minus strain (i.e., PLC− actAfl) (Fig. 4F). Deletion of actA in the PLC-minus strain did not impact bacterial growth for the first 5 h of infection, but resulted in a decrease in colony-forming units (CFUs) of 2.6 ± 0.8-fold by 8 h p.i (Fig. 4 G and H). This growth defect was not observed in Becn1−/− BMMs (Fig. 4I). These data demonstrated that ActA was involved in xenophagy avoidance and promoted growth in the macrophage cytosol following escape from the entry vacuole.
Fig. 4.
L. monocytogenes avoids growth-restricting xenophagy in the macrophage cytosol. (A) A Cre-lox system that deletes actA (and tetL) upon access to the host cytosol was engineered. The gene encoding the Cre recombinase was cloned downstream of the actA promoter, which is robustly up-regulated intracellularly. Upon access to the host cytosol, both actA and cre are expressed until Cre mediates recombination of loxP sites and deletion of the (actA-tetL) cassette (hereafter referred to as actAfl). (B) Proportion of tetracycline resistant (TetR) CFU in inocula and samples recovered from BMMs infected with actAfl and actAfl + PactA-cre (referred to as actAfl + cre) in the PlcAH86A PlcBH69G (referred to as PlcAB−) background [N.S., nonsignificant; ***P < 0.001 (unpaired t test); n = 2–5]. (C) Micrographs of BMMs infected for 5 h with WT, ΔactA, and ΔactA bacteria carrying actAfl or actAfl + cre. Infected cells were strained for DNA (blue) and ActA (red). (D and E) Kinetics of ActA-positive bacteria in BMMs infected with WT and actAfl +/− cre (D), or WT and PlcAB− actAfl +/− cre (E). (F) Colocalization kinetics of GFP-LC3 with L. monocytogenes in BMMs infected with WT and PlcAB− actAfl +/− cre. (D–F) Significant differences between actAfl and actAfl +/− cre strains are indicated for each time point [*P < 0.05; **P < 0.01; ***P < 0.001 (ANOVA with Tukey’s post hoc test performed for each time point); n = 2–3]. (G) Growth kinetics for WT, the triple mutant, and PlcAB− actAfl +/− cre in BMMs. CFU recovered from the 8 h time point are represented in H. Relevant statistically significant differences are indicated [N.S., nonsignificant; **P < 0.01; ***P < 0.001 (ANOVA with Tukey’s post hoc test); n = 4]. (I) CFU recovered from Becn1−/− BMMs infected for 8 h with WT, the triple mutant, and PlcAB− actAfl +/− cre [no significant differences (ANOVA with Tukey’s post hoc test); n = 2]. Results are expressed as means and SDs. (Scale bars: 5 µm.)
Discussion
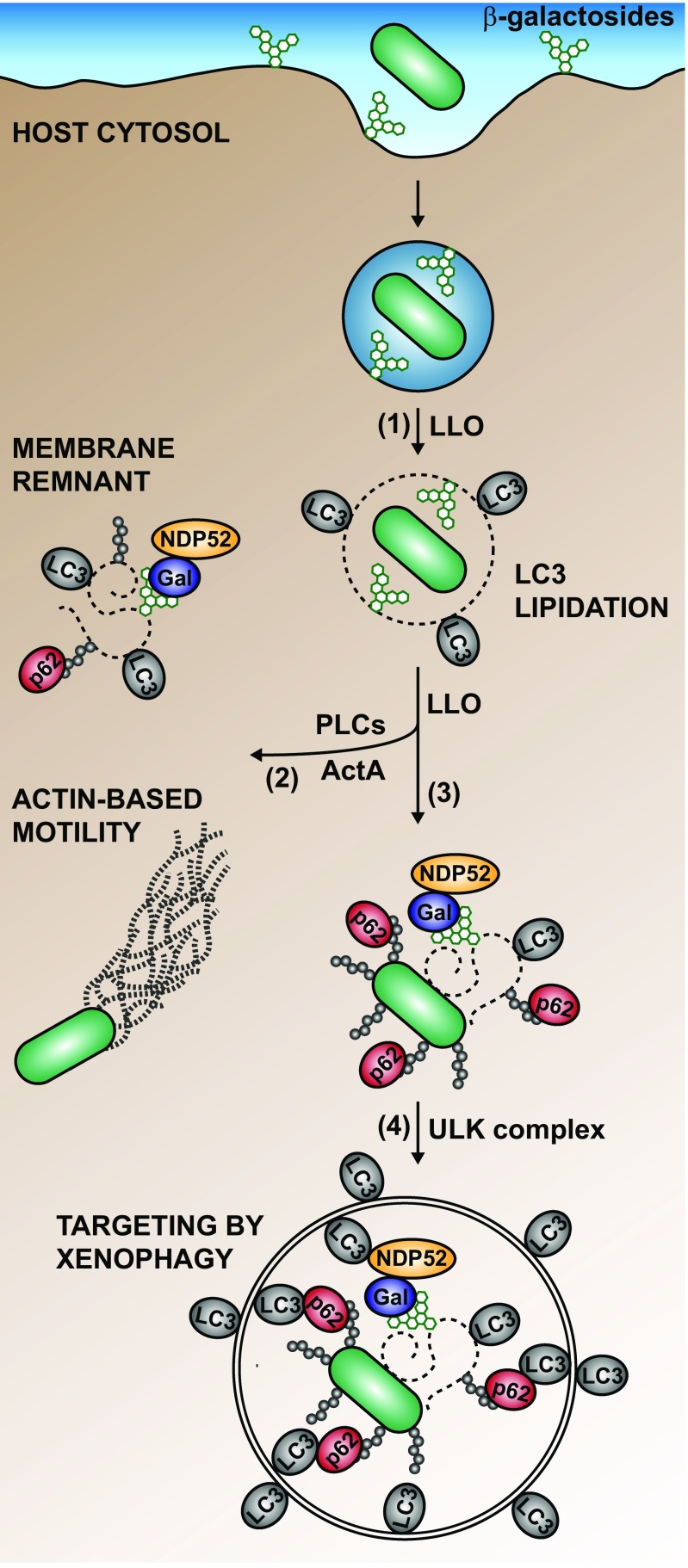
The results of this study showed that distinct autophagic processes sequentially target pathogenic bacteria during infection (Fig. 5) and that the presence of LC3 is not indicative of functional xenophagy. Although, before this study, it was appreciated that the autophagy machinery acts on intracellular microbes through multiple distinct pathways (3, 4), it was not clear whether these pathways are independent or function in a coordinated fashion. Our data support a model in which autophagic processes sequentially act on pathogen-containing vacuoles and cytosolic microbes, constituting a multilayered network of cell-autonomous defense. These processes are expected to range in importance depending on the nature of the internalized microbe. For example, while nonpathogenic microbes may be neutralized by the phagolysosomal pathway without involvement from the autophagy machinery, microbial pathogens are more likely to be targeted by one or several autophagic processes depending on their intracellular lifestyle (e.g., intravacuolar vs. cytosolic) and their pathogenic strategies.
Fig. 5.
Model for the interaction of L. monocytogenes with the host autophagy machinery during macrophage infection. L. monocytogenes is targeted sequentially by distinct autophagic processes during its intracellular life cycle. (1) LLO triggers lipidation of LC3 on the Listeria-containing vacuole through a process independent of the ULK complex. (2) Bacteria that expressed ActA and PLCs interfere with autophagy and proliferate in the host cytosol. (3) During escape from the phagolysosomal pathway, L. monocytogenes might associate with a ruptured vacuole or membrane remnants, which are marked by galectins (Gal) and ubiquitin (Ub) chains, and recruit autophagy adaptors (e.g., p62 and NDP52). Once in the cytosol, L. monocytogenes can also directly associate with Ub and autophagy adaptors (e.g., p62). (4) Autophagy adaptors mediate the engulfment of bacteria through xenophagy, a process that requires the ULK complex. Not represented on this model is the possibility that L. monocytogene-containing autophagosomes are damaged by LLO and retargeted by the autophagy machinery.
For at least some microbial pathogens, there is evidence that lipidation of ATG8 proteins on pathogen-containing vacuoles is triggered by LAP (5, 16, 26), a process initiated by the engagement of immune receptors. However, permeabilization of the pathogen-containing vacuole might be required for autophagy induction during infection by L. monocytogenes (16, 17, 20), Mycobacterium marinum (27), Mycobacterium tuberculosis (28), and Salmonella enterica serovar Typhimurium (S. Typhimurium) (29). This suggests that the engagement of immune receptors is not necessarily sufficient to trigger lipidation of ATG8 proteins on phagosomes. For example, the production of reactive oxygen species during LAP initiates a cascade of events that may lead to destabilization of the phagosome membrane (30). Therefore, it is possible that vacuole destabilization is the unifying mechanism leading to deposition of ATG8 proteins on pathogen-containing vacuoles.
Although it was first suggested that LAP mediates antimicrobial activity by promoting phagosome maturation, a recent study showed that phagosome maturation is not always affected by autophagy proteins (31). It is possible that the autophagy machinery promotes repair of damaged endosomal/phagosomal membranes (32) or that lipidation of ATG8 proteins on pathogen-containing vacuoles is inherently required for the targeting of membranous structures by xenophagy (6). Perhaps, lipidation of ATG8 proteins might have a broader function in marking and licensing pathogen-containing vacuoles to interact with other antimicrobial effectors such as IFN-regulated GTPases (7).
The results of this study suggest that intracellular pathogens need to avoid xenophagy to grow in the macrophage cytosol. Surprisingly, FIP200, but not ULK1, was necessary for restricting the growth of the L. monocytogenes triple mutant (Fig. 2). It is possible that ULK2 plays a privileged role in triggering a subset of autophagic processes important for immunity, although it was suggested that ULK1 and ULK2 have compensatory functions (24). As shown for S. Typhimurium (11), this study confirmed that the initiation of a localized xenophagic response involved the protein kinase TBK1, which promotes macroautophagy by phosphorylating autophagy adaptors (11, 33, 34). Our finding that single mutants lacking p62 and NDP52 still have functional xenophagy against L. monocytogenes contrasts with results obtained with macrophages lacking TBK1. Taken together, this suggests that p62 and NDP52 act redundantly or that another autophagy adaptor has a dominant role in the targeting of L. monocytogenes. It is tempting to speculate that different autophagy adaptors are recruited to damaged pathogen-containing vacuoles and cytosolic microbes, and participate in discrete xenophagic pathways. In this scenario, it is conceivable that cytosolic pathogens, such as L. monocytogenes, need to circumvent both of these pathways to successfully grow in macrophages.
Current models of xenophagy were influenced by our knowledge of macroautophagy, but it is becoming increasingly recognized that these processes are not identical. This study supports a model in which the autophagy machinery coordinates a series of distinct responses to intracellular pathogens. One interesting notion is that the sequential nature of these processes allows coupling to different immune pathways in a manner that calibrates the host response to a specific microbial threat. Future research should aim to define these distinct xenophagic processes and to characterize their unique features, which are likely to provide insights that are relevant to microbial pathogenesis and immunity, and may lead to the development of specific therapeutics targeting infectious diseases without affecting the housekeeping functions of macroautophagy.
Materials and Methods
L. monocytogenes and Escherichia coli strains used in this study are described in Supporting Information and listed in Tables S1 and S2. BMMs were prepared and cultured as previously described (20). Detailed information on analysis, growth conditions, infection assays, CLEM, immunofluorescence microscopy, plasmids and strains construction, and time-lapse microscopy can be found in Supporting Information. Relevant primers are listed in Table S3. This study was performed in accordance with the guidelines in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (35), and protocols were approved by the Animal Care and Use Committee of the University of California, Berkeley.
Supplementary Material
Acknowledgments
We thank Yukio Ishii (University of Tsukuba, Japan) for generously providing us with mouse femurs, J.D. Sauer (University of Wisconsin–Madison) for bacterial strains carrying the fluorescent protein mCherry, and Alexander Louie [University of California (UC) Berkeley], Jen-Yi Lee [Cancer Research Lab (CRL) Molecular Imaging Center, UC Berkeley] and Reena Zalpuri (Electron Microscope Lab, UC Berkeley) for technical assistance and helpful advice. This work was supported by National Institutes of Health Grants 1P01 AI063302 (to J.S.C. and D.A.P.), 1R01 AI027655 (to D.A.P.), and R01 AI46406 (to D.R.G.), and a grant from Aduro Biotech to the UC Berkeley Immunotherapy and Vaccine Research Initiative (IVRI) (to D.A.P.). G.M. was supported by fellowships from Fonds de Recherche du Québec–Nature et Technologies (166871), Fonds de Recherche du Québec–Santé (29406 and 33790), and the Natural Sciences and Engineering Research Council of Canada (PDF-453906-2014). B.N.N. was supported by National Science Foundation Graduate Research Fellowship DGE 1106400.
Footnotes
Conflict of interest statement: D.A.P. has a consulting relationship with and a financial interest in Aduro Biotech. Both he and the company stand to benefit from the commercialization of the results of this research. G.M. and Ralph R. Isberg are coauthors on a 2017 review article.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716055115/-/DCSupplemental.
References
- 1.Galluzzi L, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang J, Brumell JH. Bacteria-autophagy interplay: A battle for survival. Nat Rev Microbiol. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Randow F, Youle RJ. Self and nonself: How autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez J, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Kageyama S, et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell. 2011;22:2290–2300. doi: 10.1091/mbc.E10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell G, Isberg RR. Innate immunity to intracellular pathogens: Balancing microbial elimination and inflammation. Cell Host Microbe. 2017;22:166–175. doi: 10.1016/j.chom.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurston TL, et al. Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy. EMBO J. 2016;35:1779–1792. doi: 10.15252/embj.201694491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnupf P, Portnoy DA. Listeriolysin O: A phagosome-specific lysin. Microbes Infect. 2007;9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Smith GA, et al. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambrechts A, Gevaert K, Cossart P, Vandekerckhove J, Van Troys M. Listeria comet tails: The actin-based motility machinery at work. Trends Cell Biol. 2008;18:220–227. doi: 10.1016/j.tcb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Lam GY, Cemma M, Muise AM, Higgins DE, Brumell JH. Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy. 2013;9:985–995. doi: 10.4161/auto.24406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer-Morse N, et al. Listeriolysin O is necessary and sufficient to induce autophagy during Listeria monocytogenes infection. PLoS One. 2010;5:e8610. doi: 10.1371/journal.pone.0008610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshikawa Y, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 19.Tattoli I, et al. Listeria phospholipases subvert host autophagic defenses by stalling pre-autophagosomal structures. EMBO J. 2013;32:3066–3078. doi: 10.1038/emboj.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell G, et al. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect Immun. 2015;83:2175–2184. doi: 10.1128/IAI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaughnessy LM, Hoppe AD, Christensen KA, Swanson JA. Membrane perforations inhibit lysosome fusion by altering pH and calcium in Listeria monocytogenes vacuoles. Cell Microbiol. 2006;8:781–792. doi: 10.1111/j.1462-5822.2005.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell-Valois FX, Sachse M, Sansonetti PJ, Parsot C. Escape of actively secreting Shigella flexneri from ATG8/LC3-positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. MBio. 2015;6:e02567-14. doi: 10.1128/mBio.02567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alers S, Löffler AS, Wesselborg S, Stork B. The incredible ULKs. Cell Commun Signal. 2012;10:7. doi: 10.1186/1478-811X-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reniere ML, et al. Glutathione activates virulence gene expression of an intracellular pathogen. Nature. 2015;517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubber A, et al. Bacterial secretion system skews the fate of Legionella-containing vacuoles towards LC3-associated phagocytosis. Sci Rep. 2017;7:44795. doi: 10.1038/srep44795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerena MC, Colombo MI. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell Microbiol. 2011;13:814–835. doi: 10.1111/j.1462-5822.2011.01581.x. [DOI] [PubMed] [Google Scholar]
- 28.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 30.Boyle KB, Randow F. Rubicon swaps autophagy for LAP. Nat Cell Biol. 2015;17:843–845. doi: 10.1038/ncb3197. [DOI] [PubMed] [Google Scholar]
- 31.Cemma M, Grinstein S, Brumell JH. Autophagy proteins are not universally required for phagosome maturation. Autophagy. 2016;12:1440–1446. doi: 10.1080/15548627.2016.1191724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreibich S, et al. Autophagy proteins promote repair of endosomal membranes damaged by the Salmonella type three secretion system 1. Cell Host Microbe. 2015;18:527–537. doi: 10.1016/j.chom.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 34.Richter B, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Research Council . Guide for the Care and Use of Laboratory Animals. Natl Acad Press; Washington, DC: 1996. [Google Scholar]
- 36.Bécavin C, et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio. 2014;5:e00969-14. doi: 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones S, Portnoy DA. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect Immun. 1994;62:5608–5613. doi: 10.1128/iai.62.12.5608-5613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohtsuka S, et al. SQSTM1/p62/A170 regulates the severity of Legionella pneumophila pneumonia by modulating inflammasome activity. Eur J Immunol. 2014;44:1084–1092. doi: 10.1002/eji.201344091. [DOI] [PubMed] [Google Scholar]
- 39.Lauer P, et al. Constitutive activation of the PrfA regulon enhances the potency of vaccines based on live-attenuated and killed but metabolically active Listeria monocytogenes strains. Infect Immun. 2008;76:3742–3753. doi: 10.1128/IAI.00390-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- 41.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camilli A, Tilney LG, Portnoy DA. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whiteley AT, et al. c-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Mol Microbiol. 2017;104:212–233. doi: 10.1111/mmi.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bannam T, Goldfine H. Mutagenesis of active-site histidines of Listeria monocytogenes phosphatidylinositol-specific phospholipase C: Effects on enzyme activity and biological function. Infect Immun. 1999;67:182–186. doi: 10.1128/iai.67.1.182-186.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zückert WR, Marquis H, Goldfine H. Modulation of enzymatic activity and biological function of Listeria monocytogenes broad-range phospholipase C by amino acid substitutions and by replacement with the Bacillus cereus ortholog. Infect Immun. 1998;66:4823–4831. doi: 10.1128/iai.66.10.4823-4831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marquis H, Doshi V, Portnoy DA. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vincent WJ, Freisinger CM, Lam PY, Huttenlocher A, Sauer JD. Macrophages mediate flagellin induced inflammasome activation and host defense in zebrafish. Cell Microbiol. 2016;18:591–604. doi: 10.1111/cmi.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.