Significance
N6-methyladnosine (m6A) represents one of the most common RNA modifications. Biochemical analyses have identified ALKBH5 as an eraser of m6A. The present study represents the first molecular characterization of the Alkbh5 knockout mouse model. Our data associate m6A erasure with mRNA length control. Specifically, proper m6A demethylation is required for correct splicing and selective degradation of longer 3′-UTR transcripts, which are abundant in mitotic and meiotic male germ cells, but these longer 3′-UTR transcripts become rapidly degraded in the haploid male germ cells. Aberrant m6A levels in spermatogenic cells are incompatible with normal spermatogenesis and male fertility.
Keywords: RNA methylation, alternative splicing, mRNA stability, 3′-UTR shortening, fertility
Abstract
N6-methyladenosine (m6A) represents one of the most common RNA modifications in eukaryotes. Specific m6A writer, eraser, and reader proteins have been identified. As an m6A eraser, ALKBH5 specifically removes m6A from target mRNAs and inactivation of Alkbh5 leads to male infertility in mice. However, the underlying molecular mechanism remains unknown. Here, we report that ALKBH5-mediated m6A erasure in the nuclei of spermatocytes and round spermatids is essential for correct splicing and the production of longer 3′-UTR mRNAs, and failure to do so leads to aberrant splicing and production of shorter transcripts with elevated levels of m6A that are rapidly degraded. Our study identified reversible m6A modification as a critical mechanism of posttranscriptional control of mRNA fate in late meiotic and haploid spermatogenic cells.
Similar to DNA and histones, mRNAs and large noncoding RNAs can also be chemically modified (1). Over 100 various chemical modifications have been discovered in RNAs (2, 3), and N6-methyladenosine (m6A) represents the most abundant (∼3–5 m6A sites per mRNA) and also the most studied in eukaryotes (4, 5). Several active components of the m6A methyltransferase complex, including METTL3, METTL14, and WTAP, have been the regarded writer proteins of m6A (6–8). Interestingly, the m6A modification is reversible in mammalian cells, and it can be removed by two m6A demethylases, ALKBH5 and FTO (9, 10). A number of RNA-binding proteins recognize m6A on RNAs and interactions between m6A sites of mRNAs and these m6A readers exert a wide variety of effects on the mRNA fate (8, 11). The functions of these m6A writers, erasers, and readers were initially established mainly in cultured cells in vitro (12). Recently, a flurry of reports has been published in which the physiological roles of these m6A regulators are demonstrated through genetic ablation in animals of various species (12–21). Inactivation of Ythdf2, an m6A reader protein, in either zebrafish (16) or mice (20) causes female infertility by affecting maternal transcript turnover and consequently disruptions of maternal-to-zygotic transition during early embryonic development. Global Mettl3 inactivation in mice has revealed critical roles of m6A in embryonic stem cell differentiation (21) and spermatogonial stem cell differentiation (13). Mutant mice lacking Ythdc2, one of the m6A reader proteins, also display a meiotic arrest phenotype in both males and females (19). FTO KO mice are fertile, but display an obesity phenotype (18). As another known m6A eraser protein, ALKBH5 has also been shown to be involved in glioblastoma (22) and to play an essential role in spermatogenesis and male fertility (23), although the underlying mechanism remains unknown.
Of great interest, these studies point to an essential role of m6A in gametogenesis and fertility control, which is in agreement with earlier studies using model organisms (24). This is not surprising given that germ cells, both male and female, all display multilayered, sophisticated posttranscriptional regulation of gene expression (25, 26). In females, a large number of maternal transcripts are synthesized in developing oocytes toward the end of folliculogenesis, and these maternal transcripts function before or shortly after zygotic genome activation and are ultimately eliminated by the two-cell stage of early embryonic development (27–29). The maternal transcripts are presynthesized in developing oocytes, but they are subjected to translational suppression so that they remain stabilized for an extended period of time until being translated shortly after fertilization (27–29). A similar phenomenon of extended delay in translation also occurs during haploid male germ cell development (also termed spermiogenesis) in the testis (25). At the beginning of spermatid elongation (step 9 in mice), nuclear condensation starts and histones are rapidly replaced by protamines (30, 31). Consequently, the transcriptional machinery is completely shut down. To provide proteins for the final seven steps (steps 9–16) of sperm assembly, mRNAs have to be premade in late pachytene spermatocytes and round spermatids before the onset of nuclear condensation in elongating spermatids. These presynthesized mRNAs have been shown to be compartmentalized into ribonuclear protein particles (RNPs) and physically segregated from the translational machinery in the cytoplasm (25, 30, 31). RNPs exist as nuage (also called intermitochondrial cements) in spermatocytes and chromatoid bodies in round spermatids (32, 33). When proteins are needed for final assembly of spermatozoa, those RNP-localized mRNAs are released and loaded onto polysomes for translation. Recent reports have revealed that RNP enrichment of mRNAs is a dynamic process, through which the overall length of 3′-UTRs in RNPs become increasingly shorter compared with polysome-enriched mRNAs from late pachytene spermatocytes to round and elongating spermatids (34). In other words, longer 3′-UTR transcripts, which are mainly transcribed during mitotic and meiotic phases of spermatogenesis, are no longer needed once spermatogenesis enters the haploid phase; thus, these longer 3′-UTR transcripts gradually come out of RNPs for the final round of translation and/or degradation. In contrast, the shorter 3′-UTR transcripts, which are mostly those required for late spermiogenesis, enter RNPs for stabilization and translational suppression and become increasingly enriched from late pachytene spermatocytes to round spermatids (34). The continuous shuffling of longer 3′-UTR mRNAs out of RNPs and of shorter 3′-UTR mRNAs into RNPs results in the overall 3′-UTR length of the entire mRNA transcriptome in elongating spermatids becoming shorter and shorter. While this global transcriptomic shortening phenomenon has been noticed for a long time (35–37), the underlying mechanism was only unveiled recently. It turns out that UPF1–3, the three mRNA quality-control proteins known to function in the nonsense mRNA decay (NMD) pathway, function to selectively degrade longer 3′-UTR transcripts, leading to relative enrichment of shorter 3′-UTR mRNAs (35, 38, 39). It is believed that shorter 3′-UTRs bind fewer regulatory factors, e.g., RNA-binding proteins and small RNAs, thus enhancing translational efficiency although the stability of these shorter transcripts may be compromised. Clearly, the posttranscriptional regulation of gene expression during late spermiogenesis features two events: (i) compartmentalization of mRNAs into RNPs for enhanced stability and suppressed translation, and (ii) global shortening of 3′-UTRs to enhance translational efficacy and quick turnover through selective degradation of longer 3′-UTR transcripts.
Given that m6A is predominantly enriched in 3′-UTRs close to the stop codon and numerous in vitro and in vivo studies have suggested a role of m6A in the control of mRNA dynamics (7, 8, 11), it is highly likely disruptions of spermatogenesis in mice lacking writer (Mettl3) (13), eraser (Alkbh5) (23), and reader (Ythdc2) (19) result from dysregulated mRNA fate control. As an m6A eraser, ALBKH5 has been shown to play an essential role in spermatogenesis (23). Male mice lacking Alkbh5 are completely infertile due to severe germ cell depletion and oligoasthenoteratozoospermia (OAT) (23). However, the underlying mechanism of spermatogenic disruptions in Alkbh5 KO males remains largely unknown. More importantly, data on the effects of ALKBH5 ablation on the mRNA m6A profiles in spermatogenic cells are lacking. To reveal the function of ALKBH5-dependent m6A during spermatogenesis, we purified three types of spermatogenic cell types from wild-type control and Alkbh5 KO testes and analyzed the changes in mRNA transcriptome and m6A profiles using RNA-Seq and m6A RNA immunoprecipitation sequencing (m6A-RIP-Seq), respectively. Our data suggest that proper m6A erasure is required for correct splicing of longer 3′-UTR transcripts in the nucleus, and that m6A enrichment in 3′-UTRs of mRNAs correlates with enhanced degradation in the cytoplasm. ALKBH5-dependent m6A is required for meiotic and haploid phases of spermatogenesis by controlling both splicing and stability of mRNAs.
Results
ALKBH5 Is Required for the Late Meiotic and Haploid Phases of Spermatogenesis.
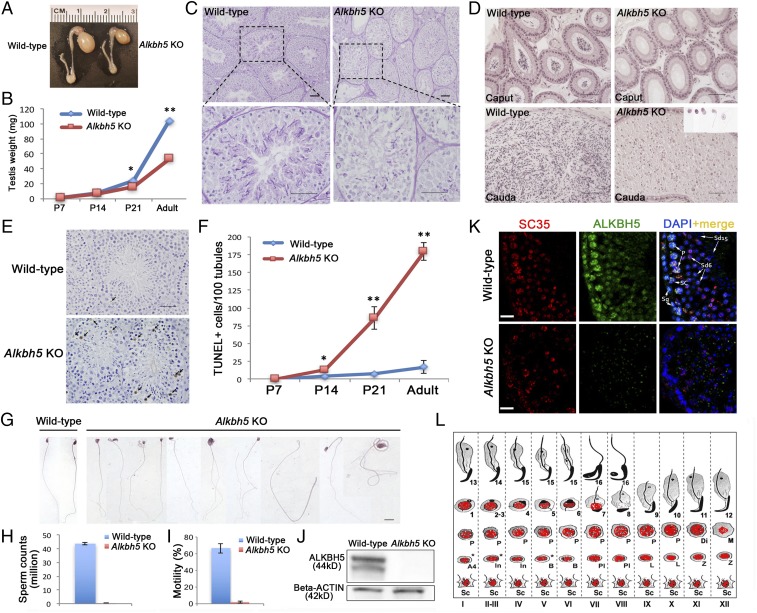
The testis expresses the highest levels of Alkbh5 mRNAs in mice, and global inactivation of Alkbh5 (herein called Alkbh5 KO mice) leads to neither discernible development defects nor adult diseases except for male infertility, suggesting an essential role of Alkbh5 in spermatogenesis and male fertility (23). To study the causes of male infertility in Alkbh5 KO mice, we first examined testicular development at both gross and histological levels (Fig. 1). Adult Alkbh5 KO testes were approximately half of the size of wild-type WT controls (Fig. 1A), and the testis weight was similar at postnatal day 14 (P14) between WT and KO mice, but the KO testes became significantly smaller compared with the WT controls at P21 and thereafter (Fig. 1B). The most advanced spermatogenic cells are midpachytene spermatocytes and round spermatids in P14 and P21 testes, respectively. Therefore, the decreased testis size between P14 and P21 and thereafter likely results from depletion of late pachytene spermatocytes and spermatids in the KO testes. Indeed, histological examination revealed that there was a delay in meiotic progress in KO testes, compared with WT controls at P14 and P21 (SI Appendix, Fig. S1). For example, midpachytene spermatocytes were present in most of the seminiferous tubules in WT testes at P14, but mostly absent in KO testes. At P21, round spermatids were detected in WT testes, but absent in the KO ones (SI Appendix, Fig. S1). The adult KO testes contained numerous vacuoles of variable sizes and the seminiferous epithelium was disorganized with much fewer spermatocytes and spermatids, suggesting active germ cell depletion (Fig. 1C and SI Appendix, Fig. S1). Supporting this notion, numerous depleted germ cells, mainly spermatids, were observed in the cauda epididymis (Fig. 1D). TUNEL analyses detected a much greater number of apoptotic germ cells in KO testes starting from P14 and the enhanced germ cell apoptosis persisted into adulthood, compared with WT controls (Fig. 1 E and F and SI Appendix, Fig. S2). Occasionally, some spermatozoa, although very few, were detected in the KO cauda epididymis; these KO sperm were all deformed, showing a wide variety of structural abnormalities (Fig. 1 G and H) without motility (Fig. 1I). Given that the male KO mice are otherwise completely healthy, Alkbh5 appears to be indispensable only for spermatogenesis and male fertility. An essential role of Alkbh5 in the testis is consistent with the fact that the testis is the organ with the highest expression levels of Alkbh5 (23).
Fig. 1.
Alkbh5 is essential for meiotic and haploid phases of spermatogenesis. (A) Alkbh5−/− (Alkbh5 KO) testes are much smaller than wild-type controls. (B) Significant decrease in testicular weight started at postnatal day 14 (P14) and persisted thereafter into adulthood. (C) Paraffin-embedded, periodic acid-Schiff (PAS)-stained testis sections showing robust spermatogenesis in wild-type mice, but disrupted spermatogenesis in Alkbh5 KO testes, characterized by drastically reduced number of meiotic (spermatocytes) and haploid (spermatids) male germ cells in the seminiferous epithelium and the presence of numerous vacuoles, a hallmark of active germ cell depletion. (Scale bars, 50 µm.) (D) Paraffin-embedded, HE-stained epididymal sections showing the presence of fully developed spermatozoa in wild-type caput and cauda epididymides, whereas the Alkbh5 KO caput and cauda epididymides contain no mature spermatozoa, but degenerated germ cells resembling round, elongating, or elongated spermatids, which were most likely those depleted from the seminiferous epithelium (Inset). (Scale bars, 50 µm). (E) Representative images showing TUNEL staining of apoptotic germ cells in 6-wk-old wild-type and Alkbh5 KO testes. (F) Quantitative analyses of apoptotic (TUNEL+) germ cells in developing wild-type and Alkbh5 KO testes. The number of TUNEL+ cells were counted per 100 cross-sections of seminiferous tubules. Data are presented as means ± SEM (n = 3). *P < 0.01, **P < 0.001; Student’s t test, two-tailed, homoscedasticity assumed. (G) HE-stained spermatozoa collected from wild-type and Alkbh5 KO epididymides (very rarely seen). All panels were at the same magnification. (Scale bar, 10 µm.) (H) Sperm counts in wild-type and Alkbh5 KO mice. Data are presented as means ± SEM (n = 10). (I) Sperm motility in wild-type and Alkbh5 KO mice. Data are presented as means ± SEM (n = 10). (J) Representative Western blots showing the detection of ALKBH5 protein in wild-type testes and the absence of ALKBH5 in Alkbh5 KO testes. β-Actin was used as a loading control. (K) Immunofluorescent localization of ALKBH5 (green) and SC35 (marker for nuclear speckles) in stage VI seminiferous tubules of wild-type and Alkbh5 KO testes. ALKBH5 is localized to the nuclei of Sertoli cells (SCs), spermatogonia (Sg), spermatocytes with higher levels in pachytene (P) spermatocytes, and step 6 round spermatids (Sd6). Step 15 spermatids (sd15) are devoid of ALKBH5. ALKBH5 is only partially colocalized with SC35. (Scale bars, 20 µm.) (L) Schematic illustration showing ALKBH5 localization in the murine seminiferous epithelia. A4, type A4 spermatogonia; B, type B spermatogonia; Di, diplotene spermatocytes; In, intermediate spermatogonia; L, leptotene spermatocytes; M, dividing spermatocytes; P, pachytene spermatocytes; PL, preleptotene spermatocytes; Sc, Sertoli cells; Z, zygotene spermatocytes; and 1–16, steps 1–16 spermatids.
To determine the stage-specific role of ALKBH5, we examined the testicular localization of ALKBH5 using immunofluorescence. Specificity of the anti-ALKBH5 antibody used was validated by Western blots, in which specific bands were detected in WT, but absent in the KO testes (Fig. 1J). ALKBH5 was localized in the nuclei of almost all testicular cell types except elongating and elongated spermatids (steps 9–16) (Fig. 1 K and L and SI Appendix, Fig. S3). Among spermatogenic cells, ALKBH5 appeared to be the most abundant in spermatocytes; medium levels were detected in spermatogonia and lower levels in round spermatids (Fig. 1 K and L and SI Appendix, Fig. S3). Consistent with an earlier report (23), ALKBH5 was partially colocalized with SC35, a marker for nuclear speckles (Fig. 1 K and L and SI Appendix, Fig. S3). The localization pattern of ALKBH5 is consistent with the testicular phenotype observed, i.e., depletion of pachytene spermatocytes and round spermatids, in which the ALKBH5 is abundantly expressed. The phenotype of Alkbh5 KO male mice suggests an essential role of ALKBH5 in the meiotic and haploid phases of spermatogenesis.
m6A Tends to Mark the 3′-UTRs of Longer mRNAs That Are Destined to Be Degraded During Spermiogenesis.
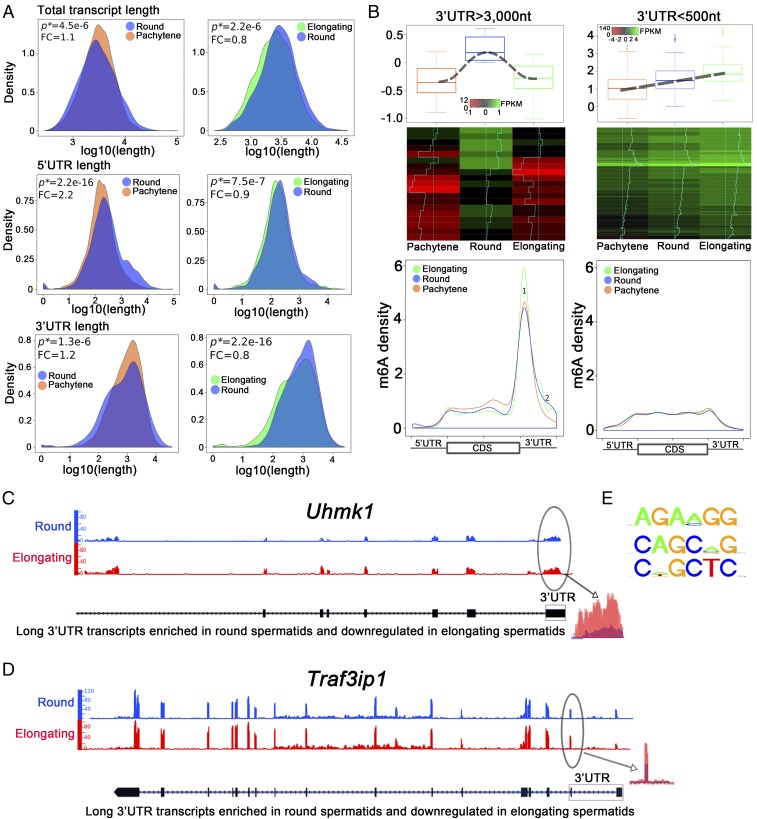
The biggest change in the overall transcriptome between the late meiotic and haploid phases of spermatogenesis lies in the global shortening of mRNAs (34, 35). The global transcriptomic shortening allows for efficient translation and quick mRNA/protein turnover during late spermiogenesis (34, 35), and was once believed to be achieved through increased production of transcripts with shorter 3′-UTRs via alternative polyadenylation (36, 37). However, recent data suggest that enhanced degradation of longer mRNAs (usually with longer 3′-UTRs) by the UPF1–3-mediated, noncanonical NMD pathway contributes significantly to the global transcriptomic shortening process (35, 38–41). We purified pachytene spermatocytes, round and elongating spermatids from the WT adult mouse testes, and performed RNA-Seq. As expected, a global shortening trend of all transcripts was observed from round and elongating spermatids (Fig. 2A). As reported in previous studies, larger transcripts, which are mainly those with longer 3′-UTRs expressed in spermatocytes and round spermatids, are down-regulated through degradation in elongating/elongated spermatids; consequently, the shorter mRNAs, which are mostly needed for sperm assembly in the final several steps of spermiogenesis, get increasingly enriched in elongating/elongated spermatids during late spermiogenesis (34, 35). In this study, we found that ∼60% of the longer 3′-UTR (>1,500 nt) transcripts enriched in round spermatids were significantly down-regulated in the elongating spermatids, and levels of ∼70% of the shorter 3′-UTR (<500 nt) transcripts were steadily increased. Given that m6A has been implicated in the control of mRNA stability (7, 8, 11), we explored whether m6A is involved in the degradation of those longer 3′-UTR transcripts during spermiogenesis.
Fig. 2.
m6A marks the longer 3′-UTR transcripts that are destined to be eliminated during spermiogenesis (from round to elongating/elongated spermatids). (A) Density plots showing that the total transcript, 5′-UTR, and 3′-UTR lengths all decreased when round spermatids developed into elongating spermatids although such a trend is less obvious during late meiotic and early haploid phases of spermatogenesis (i.e., from pachytene spermatocytes to round spermatids). Total transcript, 5′-UTR, and 3′-UTR lengths were determined based on RNA-Seq data using SpliceR. P values of statistical significance (P*) and fold changes (FC) are shown. (B) m6A sites and levels in longer vs. shorter 3′-UTR transcripts in pachytene spermatocytes and round and elongating spermatids. Two types of transcripts were analyzed: (i) Thirty longer 3′-UTR (>3,000 nt) mRNAs that were mainly synthesized in round spermatids, but were drastically down-regulated when round spermatids developed into elongating spermatids. (ii) Two hundred shorter 3′-UTR (<500 nt) mRNAs, whose levels continuously increased from round to elongating spermatids. These transcripts are most likely those required for the final several steps of sperm assembly and are subjected to delayed translation in pachytene spermatocytes and round spermatids. Density of m6A reads detected in m6A RIP-Seq datasets were plotted against the total mRNA length. Longer 3′-UTR mRNAs contain much higher levels of m6A, which is mainly enriched in 3′-UTRs proximal to the stop codon. In contrast, levels of m6A in shorter 3′-UTR transcripts are much lower and no significant enrichment was noticed. Note that m6A levels of the longer 3′-UTR mRNAs were noticeably higher in elongating spermatids than in pachytene spermatocytes and round spermatids (peak 1). (C and D) Two example genes (Uhmk1 and Traf3ip1) showed higher m6A levels in the 3′-UTRs close to the stop codon in elongating spermatids than in round spermatids. (E) Common motifs detected surrounding the m6A sites.
To map m6A sites in mRNAs, we established and optimized a m6A RIP-Seq protocol based on previous reports (42, 43). We also adopted the latest method reported for bioinformatic analyses of m6A RIP-Seq data (16). We validated our method by performing m6A-RIP-Seq using HEK293 cells and our method detected ∼70% of the m6A sites reported previously using high-resolution m6A CLIP-SEq (42, 43) (SI Appendix, Fig. S4). To explore the relationship between 3′-UTR length and m6A levels and sites, we selected the top 30 transcripts with the longest 3′-UTRs (>3,000 nt) that were enriched in round spermatids, but drastically down-regulated in elongating spermatids and compared them with 200 randomly chosen, shorter 3′-UTR (<500 nt) transcripts showing increasing abundance from round to elongating spermatids (Fig. 2B). Interestingly, we found that the longer 3′-UTR (>3,000 nt) transcripts contained much higher levels of m6A in their 3′-UTRs close to the stop codon, compared with the shorter 3′-UTR (<500 nt) mRNAs (Fig. 2B). These data suggest that m6A predominantly marks the longer 3′-UTR transcripts that are down-regulated in elongating spermatids during spermatogenesis. It is noteworthy that m6A levels in those long 3′-UTR transcripts appeared to be higher in elongating spermatids than in pachytene spermatocytes and round spermatids (Fig. 2B, peak 1, Lower Left). Indeed, two representative genes (Uhmk1 and Traf3ip) showed much higher m6A levels in elongating spermatids than in round spermatids (Fig. 2 C and D). Given that m6A is mainly present in the 3′-UTRs of longer transcripts (Fig. 2B), the increased m6A levels may serve as a signal for decreased stability in longer 3′-UTR transcripts when round spermatids develop into elongating and elongated spermatids. Consistent with published data (8, 42, 43), we also found the conserved m6A motifs [G/A/U] [G > A]m6AC[U > A > C] (Fig. 2E). Together, these data suggest that m6A tends to mark the transcripts with longer 3′-UTRs that are destined to be degraded when spermiogenesis progresses from round to elongating spermatids.
m6A Erasure Controls Correct Splicing of Long 3′-UTR Transcripts in Spermatocytes and Round Spermatids.
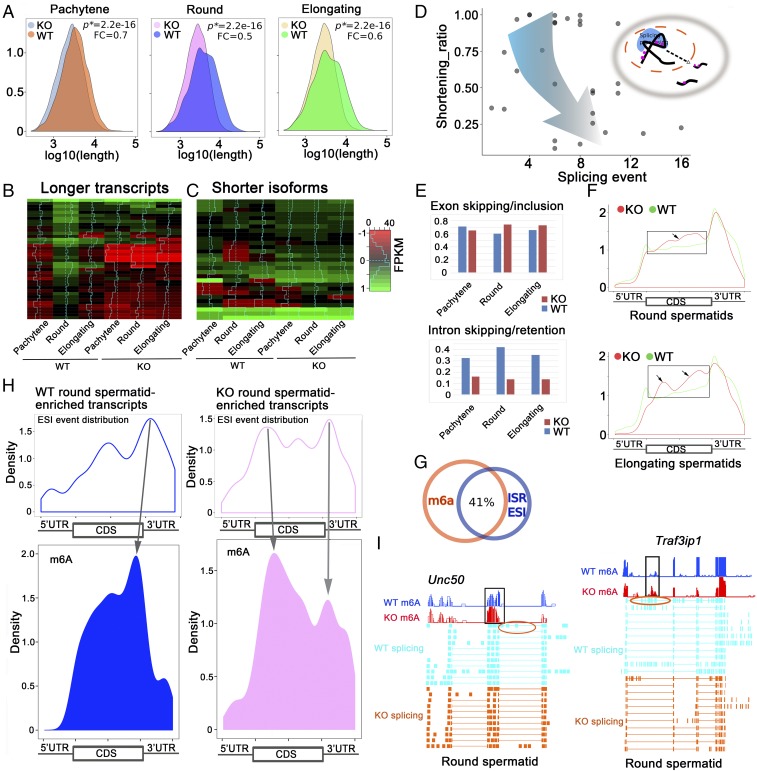
We performed RNA-Seq and m6A-RIP-Seq using pachytene spermatocytes, round, and elongating spermatids purified from Alkbh5 KO and WT adult testes. A significant decrease in transcript length was observed in all three KO spermatogenic cell types, compared with the WT controls (Fig. 3A). We chose the same 30 longer 3′-UTR (>3,000 nt) transcripts, which are mainly synthesized in round spermatids and degraded in elongating spermatids under physiological conditions (Fig. 2B), and analyzed their expression levels. Interestingly, almost all 30 longer 3′-UTR mRNAs analyzed were significantly down-regulated in the KO cells (Fig. 3B). Meanwhile, we noticed that the shorter isoforms of these 30 long transcripts were all up-regulated (Fig. 3C). Gene ontology (GO) term enrichment analyses revealed that these longer 3′-UTR mRNAs were almost all involved in the regulation of spermatogenesis (SI Appendix, Fig. S5). Where did these up-regulated shorter 3′-UTR transcripts come from? To answer this question, we plotted the shortening ratios (defined as average length of the shorter isoform/length of the longest transcript) against the total splicing events [combined counts of intron skipping/retention, exon skipping/inclusion (ESI) and alternative 5′/3′-UTR splicing, etc.], and found an interesting trend: the shorter the transcripts became, the more splicing events were detected (Fig. 3D), suggesting that the up-regulated shorter transcripts in the three Alkbh5 KO spermatogenic cell types are most likely derived from enhanced splicing events of those longer transcripts normally expressed in WT spermatogenic cells. We then analyzed the total splicing events in all of the transcripts detected in the three spermatogenic cell types purified from KO and WT testes. ESI appeared to be up-regulated and intron skipping/retention (ISR) was drastically down-regulated in Alkbh5 KO compared with WT controls (Fig. 3E), implicating enhanced splicing events in the absence of ALKBH5 in these three spermatogenic cell types.
Fig. 3.
Proper m6A erasure is required for the production of longer 3′-UTR mRNAs in pachytene spermatocytes and spermatids. (A) Density plots showing that the total transcript length was significantly decreased in Alkbh5 KO (KO) pachytene spermatocytes and round and elongating spermatids compared with corresponding wild-type (WT) spermatogenic cells. P values of statistical significance (P*) and fold changes (FC) are shown. (B) Heat maps showing the 30 longer 3′-UTR (>3,000 nt) transcripts predominantly expressed in round spermatids in wild-type testes were significantly down-regulated in all three spermatogenic cell types in Alkbh5 KO testes. (C) Heat maps showing that the shorter 3′-UTR isoforms of these 30 longer transcripts were all up-regulated in three spermatogenic cell types in Alkbh5 KO testes compared with wild-type (WT) controls. (D) Dot plot showing the relationship between shortened transcripts and splicing events in the three Alkbh5 KO spermatogenic cell types (pachytene spermatocytes and round and elongating spermatids) analyzed. Shortening ratios (defined as the length of shorter isoform/the length of the longest transcript) were plotted against the number of total splicing events detected based on the RNA-Seq data. The shorter the transcript isoforms become, the more splicing events they tend to have, suggesting that those shorter transcript isoforms were derived from enhanced splicing of those longer transcripts in the three Alkbh5 KO spermatogenic cell types. (E) Histograms showing splicing events, including exon skipping/inclusion and intron skipping/retention, in the three spermatogenic cell types in WT and Alkbh5 KO testes. (F) Comparison of m6A density in transcripts enriched in Alkbh5 KO round (Upper) and elongating (Lower) spermatids with >3 exon skipping/inclusion (ESI) events and those enriched in wild-type (WT) round and elongating spermatids without ESI events. Note that frames indicate elevated m6A levels in the CDS region of the transcripts. (G) Venn diagram showing ∼41% of the m6A sites overlap with the sites with splicing events (±200 nt distance), including exon skipping/inclusion (ESI) and intron skipping/retention (ISR). (H) Density plots showing correlations between splicing and m6A sites in wild-type (WT) and Alkbh5 KO (KO) round spermatid-enriched transcripts. The transcripts enriched in KO round spermatids appear to contain more splicing sites proximal to the m6A sites, compared with those enriched in WT round spermatids, suggesting enhanced splicing events due to m6A accumulation in the KO cells. (I) Examples of two mRNAs (Unc50 and Traf3ip1) up-regulated in Alkbh5 KO round spermatids showing exon skipping events (circled) near the m6A accumulation sites.
To further reveal the relationship between m6A levels and ESI events, we compared the m6A density between the transcripts that not only were up-regulated in Alkbh5 KO round and elongating spermatids, but also contained >3 ESI events with those enriched in WT round and elongating spermatids without ESI events (Fig. 3F). Significantly higher m6A levels were observed in coding sequences (CDSs) in Alkbh5 KO round and elongating spermatids, compared with WT cells (Fig. 3F), further supporting a role of m6A in the control of splicing. More intriguingly, ∼41% of the m6A sites overlapped with ESI/ISR sites (Fig. 3G), and m6A appeared to be enriched near the sites where the aberrant splicing events took place (Fig. 3H). Indeed, two transcripts (Unc50 and Traf3ip1) showed that the dominant m6A retention sites were close to the sites with exon skipping events in Alkbh5 KO round spermatids (Fig. 3I). These data suggest that m6A sites may guide the splicing machinery during pre-mRNA processing in the nucleus, as proposed in previous studies (6, 10, 14). This notion is also supported by the nuclear localization of ALKBH5 in pachytene spermatocytes and round spermatids during spermatogenesis (Fig. 1 K and L and SI Appendix, Fig. S3). qPCR analyses confirmed that these longer 3′-UTR transcripts of these two genes were indeed drastically down-regulated in Alkbh5 KO round and elongating spermatids compared with WT controls (SI Appendix, Fig. S6). Together, our data strongly suggest that proper erasure of m6A from the pre-mRNAs by ALKBH5 is critical for the production of longer 3′-UTR transcripts in pachytene spermatocytes and round spermatids in WT testes, and failure to do so leads to aberrant splicing and consequently the production of aberrant shorter transcript isoforms in the Alkbh5 KO testes.
Fate of Aberrantly Spliced Transcripts Due to m6A Erasure Failure.
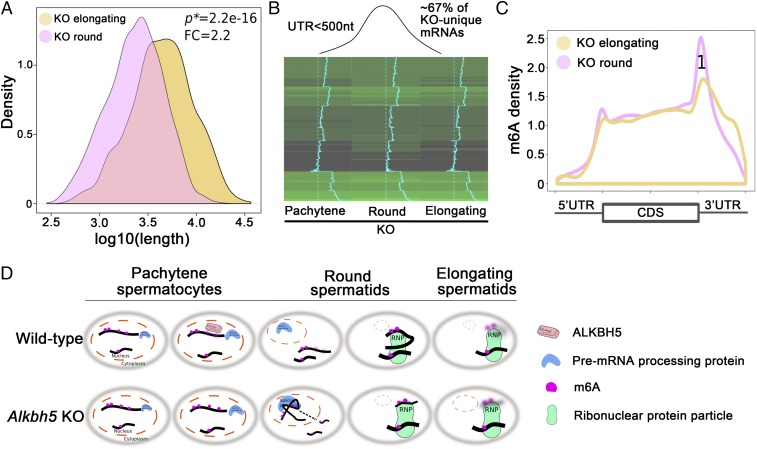
From round to elongating spermatids, the overall transcript, 3′-UTR, and 5′-UTR lengths all further decreased in wild-type mice (Fig. 2A). However, an opposite trend was observed in Alkbh5 KO spermatogenic cells (Fig. 4A), i.e., the overall mRNA length significantly increased, instead of decreased (Fig. 2A), from round to elongating spermatids. Given that the KO spermatogenic cells lack longer 3′-UTR transcripts due to m6A-induced aberrant splicing (Fig. 3), we examined the fate of ∼600 shorter 3′-UTR (<500 nt) transcripts derived from aberrant splicing and found that ∼67% of the shorter 3′-UTR transcripts that were up-regulated in KO round spermatids became down-regulated in KO elongating spermatids (Fig. 4B). GO term enrichment analyses identified that these dysregulated, shorter transcripts in the KO spermatids were mostly derived from genes involved in RNA splicing, cilium development, and spermatogenesis (SI Appendix, Fig. S7). It is interesting to see that genes encoding splicing factors (e.g., Sfswap, U2af2, Srsf1, Khdrbs3, and Snrnp70) were aberrantly spliced in the absence of ALKBH5 (SI Appendix, Table S1) because dysregulation of these splicing factors inevitably would lead to more splicing errors in their target genes, thus amplifying the initial adverse effects and generating a vicious cycle of aberrant splicing. Aberrantly spliced transcripts due to m6A erasure failure in pachytene spermatocytes and round spermatids quickly degraded in elongating spermatids (Fig. 4B). Many of the implicated genes, e.g., Foxj1 and Dnaaf3, are involved in ciliogenesis (SI Appendix, Table S2) and are known to be important for sperm flagellar development (44, 45). Among 53 dysregulated spermatogenesis-related genes (SI Appendix, Table S3), many play an essential role in spermiogenesis. For example, Crem encodes a transcriptional factor serving as a master regulator of the spermatid differentiation program (46). Prm2 encodes protamine 2, which works together with other protamine proteins, as well as transition proteins, to properly pack the chromatin to achieve a high degree of nuclear condensation in spermatozoa (47). The OAT phenotype observed in our Alkbh5 KO testes likely represents combined effects of all dysregulated genes on spermiogenesis.
Fig. 4.
Fate of the aberrantly spliced short transcripts in Alkbh5 KO (KO) round spermatids. (A) Density plot showing the average length of total transcripts increased from round to elongating spermatids in Alkbh5 KO testes; this expression pattern is opposite to the shortening trend in wild-type controls, as shown in Fig. 2A (Upper Right). P values of statistical significance (P*) and fold changes (FC) are shown. (B) Heat map showing ∼2/3 of the shorter 3′-UTR (<500 nt) transcripts up-regulated in Alkbh5 KO round spermatids were quickly down-regulated when round spermatids develop into elongating spermatids. Given that the up-regulated shorter 3′-UTR transcripts in Alkbh5 KO round spermatids were mostly those aberrantly spliced from the longer transcripts in the wild-type cells, this result suggests that these KO cell-specific shorter 3′-UTR transcripts are not stable in the Alkbh5 KO cells. (C) Elevated levels of m6A in the 3′-UTR close the stop codon in Alkbh5 KO cell-unique shorter 3′-UTR transcripts. Note that this m6A pattern is typical to the longer, but not the shorter 3′-UTR transcripts in WT round and elongating spermatids, as shown in Fig. 2B, Lower Right. (D) Schematic presentation showing the physiological functions of ALKBH5-dependent m6A erasure in late meiotic (pachytene spermatocytes) and haploid (round and elongating spermatids) phases of spermatogenesis in the wild-type testes, and the molecular consequences of m6A erasure failure due to Alkbh5 inactivation.
Large-scale degradation of those short transcripts may result from the relative increase in the overall length of the transcripts from round to elongating spermatids in the KO testes (Fig. 4A). By mapping the m6A sites in those rapidly degraded short transcripts in KO round spermatids, we observed much higher m6A levels at their 3′-UTR (Fig. 4C), and this m6A pattern is very different from that of short 3′-UTR transcripts in WT round and elongating spermatids (Fig. 2B). The elevated m6A levels in the short 3′-UTR transcripts in the KO testes likely represent m6A carried over from their precursors, i.e., those longer 3′-UTR mRNAs in WT spermatogenic cells, but aberrantly spliced in the Alkbh5 KO cells. This notion is consistent with the finding that the degradation of longer 3′-UTR transcripts in WT spermatogenic cells contains higher m6A levels at the 3′-UTRs compared with shorter 3′-UTR mRNAs (Fig. 2B). Together, these data suggest that the KO-unique shorter transcripts, which represent those spliced from the WT longer mRNAs and marked with elevated levels of m6A, are not stable and become quickly degraded when round spermatids develop into elongating spermatids in Alkbh5 KO testes.
Discussion
Although m6A was first reported >40 y ago, a comprehensive mapping of m6A in total mRNAs was only accomplished recently using anti-m6A antibody-based m6A RIP-Seq or m6A CLIP-SEq (13, 16, 18, 19, 42, 43). Over 12,000 m6A sites have been identified in mRNAs encoded by >7,000 genes in mammalian cells. In general, m6A is often enriched in 3′-UTRs close to the stop codon, where factors regulating alternative polyadenylation and splicing, subcytoplasmic compartmentalization, and stability (e.g., RNA-binding proteins and small RNAs) tend to bind (42, 43). This suggests that m6A most likely plays a role in posttranscriptional regulation. Indeed, by manipulating genes encoding m6A writers, erasers, and readers in cultured cells in vitro and in the whole animals in vivo (e.g., Drosophila, zebrafish, and mice), recent studies have demonstrated that m6A in mRNAs appear to be involved in the control of alternative splicing, translational efficiency, and stability (6–8). In the present study, we studied the molecular consequences of m6A erasure failure in spermatogenic cells in murine testes. As an m6A erasure, ALKBH5 has been shown to be nuclear (22, 23). In the testis, ALKBH5 is predominantly nuclear and partially colocalized with SC35, a DNA speckle marker, suggesting that ALKBH5 most likely acts on pre-mRNAs during splicing events in the nucleus. Indeed, our data do support a role of m6A in the control of correct splicing. First, elevated m6A levels are associated with altered splicing events because transcripts enriched in Alkbh5 KO pachytene spermatocytes and round spermatids display increased splicing events (Fig. 3 E and F). Second, m6A sites are often proximal to the sites of splicing events (Fig. 3 G and H). Third, genes encoding splicing factors appear to be preferentially affected by m6A retention due to Alkbh5 inactivation (SI Appendix, Table S1). Dysregulated splicing factors, in turn, can further cause aberrant splicing in their target genes. Taken together, ALKBH5-dependent m6A erasure appears to be able to protect longer transcripts from aberrant splicing in the nuclei of pachytene spermatocytes and round spermatids.
The most abundant expression of ALKBH5 in spermatocytes suggests a critical role in meiotic progression. Indeed, a delay in spermatocyte development occurs in the KO testes at P14; dysregulation of many genes known to be critical for meiotic progression, e.g., Sycp1, Sycp2, and Marf1 (SI Appendix, Table S3) due to failure in m6A demethylation in Alkbh5 KO spermatocytes could contribute to the meiotic defects observed. Levels of ALKBH5 are much reduced in round spermatids and there is no expression in elongating/elongated spermatids. This expression pattern is consistent with the role of m6A in the control of enhanced degradation of longer transcripts (with higher levels of m6A) and enrichment of shorter mRNAs (containing lower levels of m6A) when spermiogenesis progresses from round to elongating/elongated spermatids.
In KO elongating spermatids, the overall length of mRNAs appears to be increased from round to elongating spermatids, which is in contrast to the continuous decrease in transcript length from WT round to elongating spermatids. Failure of m6A erasure in the Alkbh5 KO pachytene spermatocytes and round spermatids causes retention of more m6A in pre-mRNAs for longer transcripts than in those for shorter 3′-UTR mRNAs; consequently, the longer 3′-UTR transcripts are affected more severely due to enhanced splicing events. These aberrantly spliced, KO spermatogenic cell-unique short transcripts are likely much shorter than normal shorter transcripts in WT cells, thus leading to a relative increase in length from round to elongating spermatids in the Alkbh5 KO testes. These KO spermatogenic cell-unique shorter mRNAs contain more m6A compared with normal shorter isoforms and thus, are subjected to quick degradation in KO elongating spermatids.
Based on the dynamic changes in m6A and the transcriptomic profiles in the Alkbh5 KO spermatogenic cells, we propose here a biphasic action of m6A, including (i) nuclear action by affecting splicing of the longer transcripts and (ii) cytoplasmic activity by marking transcripts with m6A for degradation (Fig. 4D). In WT pachytene spermatocytes and round spermatids, m6A is effectively erased by ALKBH5 to ensure correct splicing of longer 3′-UTR transcripts. Despite m6A erasure, the longer 3′-UTR transcripts tend to contain much greater levels of m6A compared with shorter mRNAs, and the former become less stable in elongating spermatids. The degradation of longer transcripts leads to enrichment of shorter transcripts during late spermiogenesis. In KO spermatogenic cells, failure to reduce m6A levels in longer pre-mRNAs results in enhanced splicing and the production of shorter transcripts. These aberrant short transcripts are different from those normally expressed shorter transcripts in that these are derived from longer mRNAs normally expressed in WT spermatogenic cells, thus retaining much greater levels of m6A, which may serve as the degradation signals in elongating spermatids. Therefore, ALKBH5-dependent m6A plays a critical role in the production of longer mRNAs and their subsequent degradation. The fact that spermatogonia are normal in Alkbh5 KO testes suggests that ALKBH5-dependent m6A has no significant effects on the mitotic phase of spermatogenesis. In contrast, Ythdc2 KO mice display spermatogonial differentiation defects, suggesting different m6A pathways are in operation in mitotic (spermatogonia) vs. meiotic (spermatocytes) spermatogenic cells. Interestingly, inactivation of the writer METTL3 in mice causes a much earlier meiotic arrest (in zygotene spermatocytes) compared with Alkbh5 KO mice (depletion of pachytene spermatocytes and spermatids) (13, 23). The phenotypic differences likely reflect the cell- and stage-specific roles of m6A during spermatogenesis.
In summary, we have demonstrated that ALKBH5-dependent m6A mainly controls mRNA fate in spermiogenesis. Appropriate erasure of m6A from pre-mRNAs is required for correct splicing and production of longer 3′-UTR transcripts in spermatocytes and round spermatids, and elevated levels of m6A in 3′-UTRs tend to induce degradation in elongating spermatids.
Materials and Methods
Animal Use, Histology, PAS Staining, and TUNEL Analyses.
Animal use protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nevada, Reno (Protocol 00494), and are in accordance with the Guide for the Care and Use of Experimental Animals established by NIH (48). The male Alkbh5 KO mice used in this study were described previously (23). Other details are provided in SI Appendix, SI Materials and Methods.
Gross Morphology and Sperm Analysis.
Testis and the whole epididymis were dissected from morphological and sperm analyses. Details are provided in SI Appendix, SI Materials and Methods.
Immunofluorescence Staining and Confocal Microscopy.
Testicular cryosections were used for the analyses. Details are provided in SI Appendix, SI Materials and Methods.
Western Blots.
Total protein lysates were extracted from whole testis for the analyses. Details are provided in SI Appendix, SI Materials and Methods.
Purification of Spermatogenic Cells.
Pachytene spermatocytes, round and elongating/elongated spermatids were purified from adult mouse testes using the STA-PUT method (34). Details are provided in SI Appendix, SI Materials and Methods.
RNA Extraction.
RNA was extracted from HEK293 cells using the mirVana miRNA Isolation Kit (AM1560; Thermo Fisher), according to the manufacturer’s instructions. Extracted RNA quantification was done using the Qubit RNA High Sensitivity Assay Kit (Q32855, Invitrogen) measured on the Qubit 2.0 Fluorometer (Invitrogen).
m6A RNA Immunoprecipitation.
Rabbit anti-m6A antibody (ab151230, Abcam) or normal rabbit IgG (10500C, Invitrogen) (6 μg each) were used for the analyses. All immunoprecipitations were performed in triplicate. The representative results of the procedures are shown in SI Appendix, Fig. S8. Other details are provided in SI Appendix, SI Materials and Methods.
RNA Library Construction.
Immunoprecipitated RNA (1 ng) and nonimmunoprecipitated RNA (300 ng) were constructed into next-generation sequencing libraries (Illumina) using the KAPA Stranded RNA-Seq Library Preparation Kit (KK8400) according to the manufacturer’s instructions. Details are provided in SI Appendix, SI Materials and Methods.
RNA-Seq Data Analyses.
Quality control of RNA-Seq data is shown in SI Appendix, Fig. S9. Trimmomatic was used to remove adaptor sequences and low-quality reads from the sequencing data (49). To identify all of the transcripts, we used Tophat2 and Cufflinks to assemble the sequencing reads based on the University of California, Santa Cruz MM9 mouse genome (50). The differential expression analysis was performed by Cuffdiff (50). The global statistics and quality controls are presented in SI Appendix, Fig. S1. The UTR and alternative splicing analyses were performed using the SpliceR pipeline (51). The sequences without identified coding frames were extracted and subjected to coding potential calculating (52). The sequencing depth and alignment ratio were listed in SI Appendix, Fig. S10. We performed the data mining using our in-house R script.
Data Normalization.
Fragments per kilobase of exon per million reads mapped counts are scaled in Cuffdiff analyses via the median of the geometric means of fragment counts across all libraries, as described in ref. 53. The principle was identical to the one used by DESEq (54).
m6a RIP-Seq Data Analyses.
Trimmomatic was used to remove adaptor sequences and low-quality reads from the sequencing data (48). Other details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the NIH (Grants HD071736 and HD085506 to W.Y.) and the Templeton Foundation (PID: 50183 to W.Y.). RNA-Seq and bioinformatics were conducted in the Single Cell Genomics Core of the University of Nevada, Reno School of Medicine, which was supported, in part, by the NIH COBRE Grant 1P30GM110767.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Both RNA-Seq and m6A RIP-Seq datasets have been deposited in the Sequence Read Archive (SRA) database, www.ncbi.nlm.nih.gov/sra (accession no. PRJNA420607).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717794115/-/DCSupplemental.
References
- 1.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 2.Cantara WA, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machnicka MA, et al. MODOMICS: A database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rottman F, Shatkin AJ, Perry RP. Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs: Possible implications for processing. Cell. 1974;3:197–199. doi: 10.1016/0092-8674(74)90131-7. [DOI] [PubMed] [Google Scholar]
- 5.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 6.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 9.A Alemu E, He C, Klungland A. ALKBHs-facilitated RNA modifications and de-modifications. DNA Repair (Amst) 2016;44:87–91. doi: 10.1016/j.dnarep.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11:669–672. doi: 10.4161/rna.28829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N, Pan T. N6-methyladenosine–Encoded epitranscriptomics. Nat Struct Mol Biol. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 13.Xu K, et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke S, et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haussmann IU, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 16.Zhao BS, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li A, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu PJ, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova I, et al. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67:1059–1067.e4. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 eExpression and cell proliferation program. Cancer Cell. 2017;31:591–606 e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klungland A, Dahl JA, Greggains G, Fedorcsak P, Filipczyk A. Reversible RNA modifications in meiosis and pluripotency. Nat Methods. 2016;14:18–22. doi: 10.1038/nmeth.4111. [DOI] [PubMed] [Google Scholar]
- 25.Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Semin Cell Dev Biol. 1998;9:483–489. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 26.Clarke HJ. Post-transcriptional control of gene expression during mouse oogenesis. Results Probl Cell Differ. 2012;55:1–21. doi: 10.1007/978-3-642-30406-4_1. [DOI] [PubMed] [Google Scholar]
- 27.Farley BM, Ryder SP. Regulation of maternal mRNAs in early development. Crit Rev Biochem Mol Biol. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- 28.Reyes JM, Ross PJ. Cytoplasmic polyadenylation in mammalian oocyte maturation. Wiley Interdiscip Rev RNA. 2016;7:71–89. doi: 10.1002/wrna.1316. [DOI] [PubMed] [Google Scholar]
- 29.Susor A, Jansova D, Anger M, Kubelka M. Translation in the mammalian oocyte in space and time. Cell Tissue Res. 2016;363:69–84. doi: 10.1007/s00441-015-2269-6. [DOI] [PubMed] [Google Scholar]
- 30.Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anat Embryol (Berl) 2001;203:323–334. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- 31.Bettegowda A, Wilkinson MF. Transcription and post-transcriptional regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1637–1651. doi: 10.1098/rstb.2009.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Idler RK, Yan W. Control of messenger RNA fate by RNA-binding proteins: An emphasis on mammalian spermatogenesis. J Androl. 2012;33:309–337. doi: 10.2164/jandrol.111.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. MicroRNAs control mRNA fate by compartmentalization based on 3′ UTR length in male germ cells. Genome Biol. 2017;18:105. doi: 10.1186/s13059-017-1243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao J, et al. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3'UTR transcripts. PLoS Genet. 2016;12:e1005863. doi: 10.1371/journal.pgen.1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D, et al. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacDonald CC, McMahon KW. Tissue-specific mechanisms of alternative polyadenylation: Testis, brain, and beyond. Wiley Interdiscip Rev RNA. 2010;1:494–501. doi: 10.1002/wrna.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacDonald CC, Grozdanov PN. Nonsense in the testis: Multiple roles for nonsense-mediated decay revealed in male reproduction. Biol Reprod. 2017;96:939–947. doi: 10.1093/biolre/iox033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fanourgakis G, Lesche M, Akpinar M, Dahl A, Jessberger R. Chromatoid body protein TDRD6 supports long 3′ UTR triggered nonsense mediated mRNA decay. PLoS Genet. 2016;12:e1005857. doi: 10.1371/journal.pgen.1005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mühlemann O. Spermatogenesis studies reveal a distinct nonsense-mediated mRNA decay (NMD) mechanism for mRNAs with long 3'UTRs. PLoS Genet. 2016;12:e1005979. doi: 10.1371/journal.pgen.1005979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones SH, Wilkinson M. RNA decay, evolution, and the testis. RNA Biol. 2017;14:146–155. doi: 10.1080/15476286.2016.1265199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 44.Mitchison HM, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012;44:381–389, S1–2. doi: 10.1038/ng.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: Transcriptional networks regulating ciliogenesis. Development. 2014;141:1427–1441. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- 46.Foulkes NS, Schlotter F, Pévet P, Sassone-Corsi P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature. 1993;362:264–267. doi: 10.1038/362264a0. [DOI] [PubMed] [Google Scholar]
- 47.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: What is the link? Hum Reprod Update. 2007;13:313–327. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- 48.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed Natl Acad Press; Washington, DC: 2011. [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitting-Seerup K, Porse BT, Sandelin A, Waage J. spliceR: An R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinformatics. 2014;15:81. doi: 10.1186/1471-2105-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong L, et al. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li P, Piao Y, Shon HS, Ryu KH. Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-seq data. BMC Bioinformatics. 2015;16:347. doi: 10.1186/s12859-015-0778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.