Significance
The transcription factor FoxP3 defines and controls regulatory T cells (Tregs), themselves essential components of immunoregulatory pathways. From a highly granular scanning mutagenesis, the results of our study point to very integrated functions of the protein’s domains, quite different from predictions of simple modular models. The phenotype of mutant mice carrying subtle mutations in Foxp3, which deviate from the acute lymphoproliferation and autoimmunity linked to Treg deficiency and become manifest only upon challenge, suggest that rare FOXP3 variants may contribute to a broader range of human diseases than previously recognized.
Keywords: autoimmunity, tolerance, Forkhead
Abstract
FoxP3+ regulatory T cells (Tregs) are a central element of immunological tolerance. FoxP3 is the key determining transcription factor of the Treg lineage, interacting with numerous cofactors and transcriptional targets to determine the many facets of Treg function. Its absence leads to devastating lymphoproliferation and autoimmunity in scurfy mutant mice and immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX) patients. To finely map transcriptionally active regions of the protein, with respect to disease-causing variation, we performed a systematic alanine-scan mutagenesis of FoxP3, assessing mutational impacts on DNA binding and transcriptional activation or repression. The mutations affected transcriptional activation and repression in a variegated manner involving multiple regions of the protein and varying between different transcriptional targets of FoxP3. There appeared to be different modalities for target genes related to classic immunosuppressive function vs. those related to atypical or tissue-Treg functions. Relevance to in vivo Treg biology was established by introducing some of the subtle Foxp3 mutations into the mouse germline by CRISPR-based genome editing. The resulting mice showed Treg populations in normal numbers and exhibited no overt autoimmune manifestations. However, Treg functional defects were revealed upon competition or by system stress, manifest as a strikingly heightened susceptibility to provoked colitis, and conversely by greater resistance to tumors. These observations suggest that some of the missense mutations that segregate in human populations, but do not induce IPEX manifestations, may have unappreciated consequences in other diseases.
CD4+ regulatory T cells (Tregs) are of central importance in immunological tolerance to self and in the control of inflammatory processes. They play versatile roles to balance homeostasis, regarding both immunological (autoimmunity, allergy, responses to pathogenic and commensal microbes, cancer) and nonimmunological (tissue regeneration, metabolic control) contexts (1–3). FoxP3, a winged-helix transcription factor (TF) of the Forkhead (FKH) family, is specifically expressed in Tregs, where it has pivotal roles for differentiation and function and is considered to be the defining factor of the lineage (2, 4). Treg cells have a core transcriptional signature, transcripts that are over- or under-represented relative to their naive CD4+ T cell counterparts (Tconv) (5–9). Much of this signature is controlled by FoxP3, although FoxP3 alone cannot drive the entire Treg signature (6, 7, 10–12).
Germline deletion of FoxP3 leads to Treg deficiency and to devastating multiorgan inflammation. In human immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX) patients, complete loss of FoxP3 function leads to the absence of Tregs, and there is also a spectrum of missense mutations that allow the differentiation and maintenance of some Tregs with partial function (13–15). IPEX typically begins very early in life with a typical triad of enteropathy, endocrine autoimmunity (primarily type-1 diabetes and thyroiditis), and eczematous dermatitis. Of these features, gut pathology is essentially constant, while endocrinopathies are more variable. Other manifestations occur more sporadically, such as autoimmune hepatitis, nephropathy, and cytopenias (14, 16–18). The root of this range of pathologies in IPEX patients is incompletely understood. On one hand, complete loss-of-function mutations (frameshifts, nonsense mutations, large deletions) with complete absence of FOXP3 protein are more deleterious than missense and small deletions (reviewed in ref. 15), and siblings carrying the same mutation tend to develop disease with comparable course and severity. But the range of manifestations and severity can also vary between patients with the same mutation, suggesting that modifier loci in the genetic background and/or environmental exposures modify the course of disease.
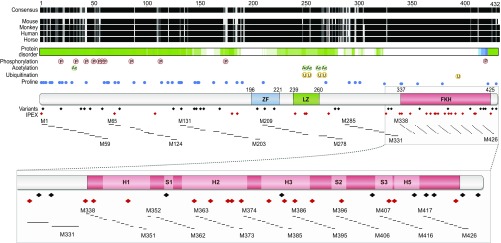
The FoxP3 protein contains several structural modules (Fig. 1): (i) a short zinc finger (ZF); (ii) a leucine zipper (LZ) domain implicated in homodimerization or heterodimer formation with other FoxP proteins (19, 20); and (iii) the family-defining FKH domain at the C terminus, which is the primary DNA-binding site but also interfaces with transcriptional coregulators (21). The structure of the FKH domain has been determined (22, 23), showing that it adopts an unusual “domain-swapped” configuration, in which two FKH domains are intertwined and can bridge two distinct DNA molecules. In contrast, the proline-rich N-terminal region appears to be an intrinsically disordered protein region (24) in computational and structural analyses (25), suggesting that it can adopt different conformations upon binding to different partners. Indeed, FoxP3 interacts with many other TFs (reviewed in ref. 2). Many of these interactions are functionally relevant and modulate specific aspects of Treg function (26–29).
Fig. 1.
FOXP3 structure, variation, and the systematic set of mutants. From the Top: conservation of FOXP3 protein across mammals; protein disorder (from D2P2 browser: d2p2.pro/); posttranslational modifications and proline positions; known protein domains; and missense variants in human FOXP3 found in IPEX patients or in population surveys. The position of alanine-scan mutants is shown in relation to these maps, where the number refers to the position of the first altered amino acid.
Genetic variation at the human FOXP3 locus includes missense variants identified in patients presenting with more or less severe IPEX, and these are preferentially represented in the FKH domain (Fig. 1). But a set of missense mutations have also been identified in systematic exome-sequencing programs among healthy donors or patients with diseases other than IPEX [e.g., aggregated in gnomAD (30)]. These non-IPEX variants are typically rare, and some are unique to one individual, but occur at a combined frequency of ∼1/300 chromosomes, mapping uniformly throughout the protein (Fig. 1). Most are likely to be completely silent, but since exome sequence aggregation efforts include a surfeit of patients with an array of diseases, their presence raises the possibility that FOXP3 missense mutations might contribute to diseases other than IPEX.
How these functional, structural, and variability aspects are integrated, and how FoxP3 operates as a transcription factor to regulate its target genes, is incompletely understood. Several studies have analyzed the functional impact of a few natural or engineered mutations of FoxP3 and identified several important positions (20, 25, 31–36). However, an integrated perspective of how FoxP3’s domains collaborate is lacking. Are there discrete, modular, regions of the protein to which specific functions can be uniquely ascribed (as would be suggested, for example, by schematic “repressor domain” representations)? Or does FoxP3 function rather as a malleable globular entity, with functional interactions being determined combinatorially by several structural elements? In an attempt to provide a wide perspective of FoxP3’s mode of operation, we performed a systematic alanine-scan of the protein. We constructed a set of 130 FoxP3 mutants and tested how this fine-grained array of mutations affects its ability to bind DNA and to regulate transcription. The results brought about a nuanced and variegated perspective on FoxP3’s structure–function relationship. Assessing the impact of some mutations by germline editing in mice suggests that variation in FoxP3 may impact diseases beyond the confines of the IPEX syndrome.
Results
Construction and Verification of the FoxP3 Alanine-Scan Library.
We constructed by site-directed mutagenesis an alanine-scan library with 130 mutations in the coding region for mouse FoxP3, the positions of which are outlined in Fig. 1. Amino acids in the essential FKH domain (P338 to P429) were replaced by alanine one-by-one, while in the less-charted N-terminal region (M1 to R337) replacements were by blocs of six alanines. Some of the alterations correspond to mutations in IPEX patients, but some also correspond to variants detected in large exome sequence projects in healthy controls or non-IPEX pathologies (Fig. 1 and Dataset S1) (30). A subset of these mutant2s was reported recently in a study that focused on the relationship between the interactions of FoxP3 with chromatin or other cofactors and its transcriptional activity (37); these results are included here for completeness.
After verification of sequence integrity, the mutants were placed in the retroviral vector MSCV-IRES-THY1.1 (with an N-terminal FLAG-tag for identification), and high-titer viruses were used to infect activated primary CD4+CD25− Tconv cells (10). This cellular setting was chosen as practical for a project on this scale, and relevant to physiological activity, since FoxP3 operates in CD4+ T cells (7, 38). Overall results are compiled in Dataset S1.
Proper expression of the mutant FoxP3 proteins was first assessed. Flow cytometric analysis of transduced cells, standardizing FoxP3-staining intensity against the cotranscribed THY1.1 reporter, showed for almost all mutants an expression similar to that of wild-type (WT) FoxP3 (Fig. 2). These FoxP3 levels reach, for cells in the higher range of the Thy1.1 reporter, that of FoxP3 in ex vivo Treg cells stained in parallel (Fig. 2, Bottom Right). A few mutants (M350, M363, M385, M389, M395, and M406) did show a significant reduction in FoxP3 expression. M350 is the position of a complete loss-of-function mutation of FoxP3 uncovered in an ENU screen (MGI:3817855). M85 prevents recognition by the anti-FoxP3 mAb used for detection. Immunoblotting of extracts from transduced cells largely confirmed the flow cytometric data and also showed that all mutant proteins were full length (Fig. S1).
Fig. 2.
Expression of the mutant FoxP3 proteins in transduced CD4+ T cells. Flow cytometry plots of FoxP3 after retroviral transduction into activated CD4+CD25− Tconv cells, displayed against the expression of the colinear Thy1.1 encoded in the same vector (red: mutants with significant loss of expression). (Inset, Bottom Right) Ex vivo Treg cells stained in the same experiment. Data are representative of three experiments.
FoxP3 localizes to the nucleus in Tregs and in transfected cells (5, 19, 39), and we verified the proper nuclear localization of the mutants by immunofluorescence microscopy (Fig. S2A). The vast majority of mutant proteins showed the correct pattern, with strictly nuclear staining, but a few showed cytoplasmic staining as well, with cell-to-cell variation in the nuclear:cytoplasmic ratios (M331, M345, M363, M385, and M389, in addition to mutants with low total FoxP3) (Fig. S2A). When positioned with reference to the 3D crystal structure of the FoxP3 FKH domain (22, 23), several of these mutations mapped to a hydrophobic pocket within the domain-swapped FoxP3 dimer structure (Fig. S2B). The M331 result was consistent with one of the nuclear localization domains previously mapped (40), but other reported motifs (19, 32) are more difficult to reconcile with our findings. Overall, these data suggest that most of the mutants in our library were properly expressed and localized, with only a minority leading to unstable or mislocalized FoxP3 (most of which were not analyzed further).
Mapping DNA-Binding Activity.
We then tested how the alanine-scan mutations affected DNA binding by FoxP3 in a solution assay with a biotinylated dimer of the canonical 5′-AAACA Forkhead Responsive Element (FKRE) motif to capture epitope-tagged protein from whole-cell extracts (Fig. 3A). Binding was detected relative to background levels observed with empty vector (EV), irrelevant recombinant protein (EBNA), or a scrambled-sequence oligonucleotide (Fig. 3B). Overall, the set of mutants showed substantial variation in the ability to bind FoxP3’s cognate motif (Fig. 3C, where results are normalized relative to binding by WT FoxP3, and Dataset S2; position highlighted on the domain structure in Fig. 3D). Several points are worth highlighting. The mutants with poor nuclear localization showed the most severely affected DNA-binding activity in vitro, which suggests that DNA binding may be important for nuclear retention of FoxP3. Aside from M350, no single mutation completely abolished binding to DNA, implying a degree of resilience. Most alterations at the very N terminus had little effect, if anything slightly enhancing binding, while those in the LZ domain were predictably deleterious. Less expected was that several mutations in the ZF also significantly decreased DNA-binding activity. Many mutations in the FKH domain had a deleterious impact, as expected from the domain’s structure, in particular those within the main DNA-contact helix (e.g., M383N383A, M386R386A, M390S390A, M397R397A) or the domain-swap coil (e.g., M340F340A, M345L345A, M348W348A, M381W381A). However, several FKH-domain mutants unexpectedly enhanced DNA binding (e.g., M369R369A, M370M370A, M377H377A, M378P378A, M380T380A). Similarly, the A384T mutation in H3 was recently reported to also increase DNA binding (36). These enhancing mutations mapped to helix H2 and to a loop immediately N-terminal to the main DNA contacts in helix H3 (Fig. 3D). This nonbinding helix has previously been shown to modulate DNA-binding specificity in other Forkhead family members and has been proposed to constitute a mode of determination and evolution of binding specificity (41, 42). In this light, the enhancing mutations may be relieving H2 structural constraints on DNA binding.
Fig. 3.
DNA-binding activity of the FoxP3 mutants. (A) Schematic of the DNA-binding assay, involving a biotinylated oligonucleotide with two FKRE motifs, and luciferase-based detection of bound FoxP3 via an N-terminal flag tag. (B) Specificity controls, comparing binding of WT FoxP3, or irrelevant EBNA protein, to FKRE2 or control (scrambled sequence) oligonucleotides (mean of two experiments ± SD); (C) Effect of each mutation on binding to the FKRE2, oligonucleotide (per A), mean ± SD of two or more experiments. Black bars indicate the mutants with atypical localization (per Fig. S2). (D) The 3D positions of the mutations with gain or loss of binding (colored shading) are shown on the structure of the FKH domain monomer (22) (structure of human FOXP3, but numbered according to mouse sequence). Amino acids the side chains of which contact DNA or the homodimerized FoxP3 are shown (yellow and orange dots, respectively). SD and P values are shown in Dataset S2.
Mapping Transactivation and Transrepression.
We then used several assays to evaluate how mutations in different regions of FoxP3 affect its impact on transcriptional targets in transduced cells. Initially, we used two short-term reporter systems in which FoxP3 has been shown to have repressive activity. One tested the inhibition of the Il2 promoter (Il2pro), which is activated by NFAT1/AP1 but repressed by NFAT1/FoxP3 complexes (21); the second assessed the inhibition of a minimal promoter by an eightfold repeat of the FKRE motif (8×FKRE) (43). Luciferase activity generated after transfection of these reporters and cell activation was strongly repressed by cotransfection of WT FoxP3 (Fig. S3A). Introduction of the FoxP3 mutant panel in these systems (Fig. 4) revealed a range of effects. First, mutation effects were generally similar for the two reporters (Fig. 4), with significant correlation (Fig. S3B), although there were differences, such as the impact of N-terminal mutations in the Il2pro but not the 8xFKRE assay. Second, the most severe effects tended to map to the FKH domain. Third, and as already observed in our narrower study (37), there was only limited correlation between DNA-binding ability and activity in these assays (Fig. S3C).
Fig. 4.
Reporter-based transcriptional activity of the FoxP3 mutants. EL4 T cells were transfected with EV, mutant, or WT FoxP3 together with the luciferase reporter plasmids described in Fig. S3 (8xFKRE, Top; Il2pro, Bottom). Data from two independent experiments are shown as mean ± SD (numeric values are in Dataset S3).
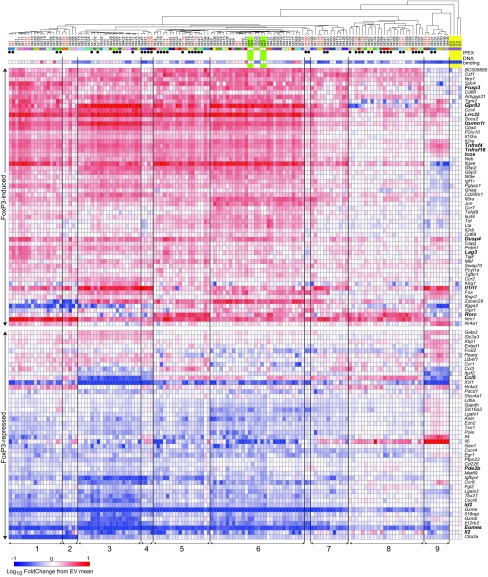
For a broader perspective, we profiled the response of a panel of FoxP3-responsive genes, using a nanostring codeset of 200 transcripts (37). Activated CD4+CD25− Tconv cells were transduced with a selected set of 70 mutants and sorted after 72 h within a constant window of Thy1.1 reporter expression to ensure a level of FoxP3 equivalent to that of ex vivo Tregs to avoid overexpression artifacts. We profiled the expression of a specific set of genes with a custom codeset for transcripts typical of the Treg signature and of tissue-Tregs and known FoxP3 targets (37). As previously reported (37), WT FoxP3 induced and repressed a set of genes relative to EV-transduced controls, including many of the Treg “usual suspects” (Icos, Lrrc32, Dusp4, Foxp3 up-regulated; Il2, Il4, Id2, Pde3b down-regulated). Complex patterns were revealed when we analyzed these induced or repressed gene sets after transduction with the selected mutants (results clustered in Fig. 5). The following observations merit bringing forth, as they have direct relevance to understanding FoxP3 function and to Tregs physiology: (i) There are general trends, several mutants being generally less active than others, captured by computing for every mutant an activation and a repression index (mean normalized expression of all FoxP3-induced or FoxP3-repressed transcripts, respectively), which are highly correlated (Fig. S4A). The activation index also correlated with the results of the reporter assay (Fig. S4B). However, we observed a diversity of effects with subtly different patterns. The heatmap (Fig. 5) groups the mutants into nine different blocs, membership being highly consistent for independent duplicate transductions of the same construct. There were even further nuances; for example, even though M14 and M276 mapped to the same bloc, they differed in the activation of Gpr83 and Lag3. Thus, varying FoxP3’s conformation or interactions with cofactors modulates diversely the effects on any one target gene. (ii) For the most part, these groupings of mutants with similar footprints did not relate to the positions of the mutations on the protein. For example, of the two mutants of bloc 4, the effects of which were quasi-identical, one mapped to the LZ and one to the FKH. This distribution suggested that the variegated effects of the mutations did not reflect local perturbations of a given interaction, but more global alterations, e.g., in multimolecular complexes. (iii) Several mutations had essentially no consequence and clustered with WT FoxP3 (bloc 6). Others appeared as gain-of-function but only for specific targets (e.g., Izumo1r, Il1rl1, and Ccl5 by bloc 3 and Rorc by bloc 5 mutations). (iv) At the other end of the spectrum (bloc 9), even the most severely affected mutants maintained some activity, including three mutants with poor or absent DNA binding (M331, M350, M406) that retained activity on some genes (e.g., Tnfrsf4/18, Eomes) and paradoxically gained induction of unexpected targets like Il4 and Il5. This remaining activity echoed a previous report of FoxP3 regulation as independent of direct DNA binding (44). However, the DNA-binding–independent activity observed here was quite specific and restricted to a small set of targets, contrary to this report (44). (v) Il1rl1 represents an interesting divergence from most other targets of FoxP3 transactivation. It encodes the receptor for the alarmin IL33 and is expressed by several tissue Treg populations (3). It was moderately activated by WT FoxP3, but markedly more so by several mutants. To capture such differences, we computed a “normalcy index” (how the expression pattern of each transcript correlated with the generic FoxP3 induction index above) (Fig. S4C). Il1rl1 was clearly an outlier, along with Rorc, Ccl5, and Il5. This observation suggests that FoxP3 may have an inherent ability to activate Il1rl1, which is normally inhibited, but can be relieved when cofactors vary in response to tissue localization or activation cues. Rorc was also an outlier among FoxP3-induced genes, but was clearly different from Il1rl1 (Fig. 5), which is of interest considering the interactions between FoxP3 and RORγ during T cell differentiation and in the control of tissue-Tregs. (vi) The Th2 cytokines Il5 and Il4 were repressed by WT FoxP3, as expected, but were actually induced by several severe FoxP3 mutations (especially the poor DNA-binding mutants of bloc 9). Il2, on the other hand, was repressed by almost all mutants, except for those of bloc 8, which slightly induced it.
Fig. 5.
Transcriptional activity of the FoxP3 mutants on endogenous genes. Activated CD4+ Tconv cells were transduced with empty vector (yellow shading) or WT (green) or mutant FoxP3; after 72 h, cells were sorted for matched expression of FoxP3 (based on colinearly expressed Thy1.1) and Nanostring-profiled in biological duplicate for a set of FoxP3-induced or -repressed genes (Right). Results are shown as FoldChange (FC) relative to the mean of EV controls. Mutants are clustered into nine blocs (Bottom; note that duplicates of the same mutant mostly cluster together). The position of known IPEX mutations (black circles, Top) and DNA-binding data are shown for reference. Mutants or genes discussed in the text are shown in red or boldface type. Numeric values are listed in Dataset S4.
Overall, these data denote specificity in the involvement of FoxP3 relative to its transcriptional targets, with a diversity that does not fit with simple domains of the protein being involved in either activation or repression, and suggests that the control of different targets involves an array of mechanisms and cofactors.
In Vivo Effects of Mild FoxP3 Mutations.
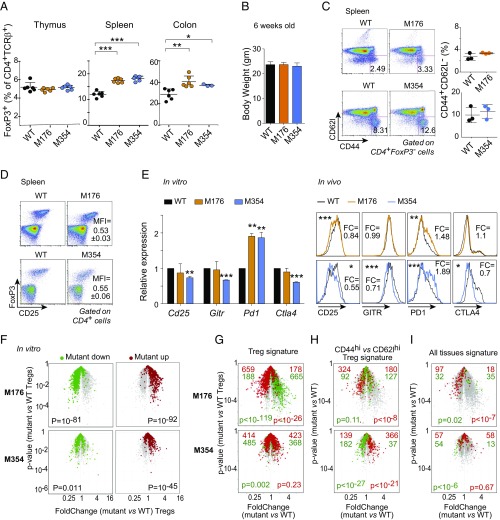
The alanine-scan mutagenesis thus brought forth a highly nuanced perspective on structure–function relationships within FoxP3, with variegated effects across the range of FoxP3 targets. Many of these mutations mapped to regions of the protein that are also affected by genetic variation in humans, some with recognized effects in the case of IPEX mutations, some unnoticed in the case of mutations uncovered in large exome-sequencing projects. Thus, it seemed important to assess the mutations’ effects in vivo at the transcriptional level to verify that the patterns observed in transduced CD4+ T cells also applied in true Tregs and at the phenotypic level to ascertain their consequence on Treg function. We chose two mutations, one with very mild effects (M176, mapping to the N-terminal region, bloc 5) and one transcriptionally more severe (M354, in the FKH domain, bloc 2). Mutant mouse lines were generated through CRISPR-based homologous recombination in fertilized oocytes (45). After screening and verification by sequencing, we obtained faithful replicas of the mutations (at 5% efficiency). Young mice homozygous for the two mutations were initially healthy and fertile with no overt phenotype. Tregs were present in essentially normal numbers and proportions in lymphoid organs and, in the colonic lamina propria, if anything slightly more abundant in the periphery (Fig. 6A). The mutant mice showed no indication of the widespread lymphoproliferation typical of scurfy or other Treg-deficient mice, histologically or from weight loss (Fig. 6B), and their Tconv cells remained mainly unactivated, judging from the CD44 and CD62l markers (Fig. 6C).
Fig. 6.
Transcriptional scars of FoxP3 mutations are recapitulated in vivo. Analysis of FoxP3 mutant mice carrying two of the above mutations. (A) Proportions of CD4+TCRb+FoxP3+ Tregs in lymphoid organs of homozygous mutant male mice or control littermates (both sets of littermate controls merged). *P < 0.05, **P < 0.005, and ***P < 0.001 (Student’s t test). (B) Body weight of 8-wk-old WT, M176, and M354 mice (average of five mice). (C) CD44 vs. CD62l expression in gated splenic Tconv from WT, M176, or M354 mice. Representative of three independent experiments. (D) FoxP3 vs. CD25 expression in CD4+ T cells; numbers: FoxP3 mean fluorescence intensity (MFI) in mutants relative to WT littermate Tregs (mean ± SD, n = 9 mice). (E) Comparison of distinguishing FoxP3 target gene expression in mutant Treg cells in vivo by flow cytometry (numbers are FoldChange relative to controls, averaged from five mice per group); for reference, mRNA expression of the same target genes in transduced CD4+ T cells (from Fig. 5) is shown at Left. *P < 0.05, **P < 0.005 and ***P < 0.001 (Student’s t test). (F) Volcano plots of microarray gene expression profiles of sorted CD4+CD25high Tregs from mutant and WT littermates (mutant/WT FoldChange vs. P value). Red and green highlights are transcripts over- or underexpressed (at FC > 1.5) when comparing mutant and WT FoxP3 in transduced CD4+ T cells in vitro (P values from a χ2 test). (G–I) Mutant vs. WT volcano plots (as in F) overlaid with (G) the canonical Treg signature (7). (H) CD44+ vs. Cd62l+ Treg signature (46). (I) All-tissue Treg signature (53); all-signature P values from a χ2 test).
On the other hand, Tregs from the mutant mice did present transcriptomic variations. First, FoxP3 levels in Tregs from both mutant lines were reduced by 40–50% relative to WT littermates (Fig. 6D). Since there was no notable difference in CD4+ T cells transduced in vitro with these mutants, when expression was driven by the vector’s retroviral promoter, these lower levels of FoxP3 suggest that the feedback that locks in Foxp3 expression may not be fully operative in the mutant Treg cells. Cell-surface markers on those Tregs (Fig. 6E, Right) mimicked changes observed in transduced cells in vitro (Fig. 6E, Left): Tregs from M354 mice displayed lower CD25, GITR, and CTLA4 but higher PD1. The M176 Tregs shared the altered CD25 and PD1 expression, but had more normal GITR and CTLA4. In gene expression profiles of Tregs from these mice, transcripts earlier seen to be overexpressed in mutant-transduced relative to WT-transduced CD4+ T cells in vitro were mostly overexpressed in mutant Tregs ex vivo, with the converse for in vitro underexpressed transcripts (Fig. 6F).
There were also marked shifts in expression of some Treg-associated signatures, but these were different in the two mutant lines. The canonical Treg signature (7) was strongly biased, but only in M176 Tregs, with underexpression of transcripts normally overexpressed in Tregs (and vice versa) (Fig. 6G). We noted an up-regulation of the signature associated with “activated Tregs” [also known as eTregs or aTregs (46)] in M354 Tregs, but the opposite in M176 Tregs (Fig. 6H). Similarly, a signature that generally distinguishes tissue from lymphoid Tregs was strongly shifted in M176, but in the opposite direction for M354 (Fig. 6I). These results indicate that the subtle transcriptional consequences of the mutations observed in vitro were also present in Tregs in vivo, each mutation differently affecting specific segments of the Treg transcriptome.
Steady-state Treg pools seemed normal in young mutant mice, but we tested their behavior in conditions of competition or challenge. First, to sensitize the Treg population analysis, we constructed radiation bone-marrow chimeras with a 50/50 mutant/WT mix of donor cells. In this context, mutant Tregs were markedly outcompeted by WT Tregs in the same mice, unlike other lineages (Fig. S5A). Mutant Tregs showed the same reduction in FoxP3 levels noted above, indicating that this is a cell-autonomous phenomenon, as was the lower CD25 in M354 (Fig. S5B).
Second, we aged the mice. Older M354 mice failed to thrive after ∼35 wk and uniformly lost weight, while M176 mice stayed healthy (Fig. 7A). Even though Treg numbers and frequency remained normal in aged mice (Fig. 7B), CD4+ Tconv cells shifted to an activated CD44hiCD62Llo phenotype, as did CD8+ T cells, particularly in M354 (Fig. 7C). Widespread inflammation was detected in the skin of aged M354 mice, but not in any other tissues usually affected in fully deficient scurfy mice, pointing to a specific autoimmune or inflammatory attack (Fig. 7D). Third, we challenged M176 and M354 mice in the trinitrobenzenesulfonic acid (TNBS)-induced colitis model. At the normal dose, mutant mice of both lines succumbed within a few days of recall (Fig. 7E). At a lower dose of TNBS, which elicited only limited colitis in WT littermates, both M176 and M354 showed severe disease (Fig. 7F).
Fig. 7.
Physiological consequences of nonscurfy FoxP3 mutations. (A) Late body weight of WT, M176, and M354 mice up to 60 wk. (B) CD4+TCRb+FoxP3+ Treg proportions in 40-wk-old WT, M176, and M354 mice. (C) CD44 vs. CD62l expression in CD8+ or Tconv (Bottom) from 40-wk-old WT and M176 or M354 mice (plots representative of three experiments). (D) Tissue histopathology from the same aged mice (representative pictures from three independent experiments). (E) Survival rate of WT and mutant mice upon challenge with the standard 200-μg dose of TNBS (results pooled from two independent experiments). (F) Representative histology, clinical score, and weight loss after challenge with low dose (40 μg) of TNBS; each point is an individual mouse, pooled from three independent experiments. (G) Growth of MC38 tumors transplanted s.c. into mutant mice or WT littermates (results combined from three independent experiments; each line represents an individual mouse). (H) Representative cytometry plots for CD4+TCRβ+FoxP3+ Tregs in tumor tissue. (I) Proportion of TNFα+ or IFNγ+TNFα+ cells among tumor-infiltrating CD8+ T (Left), Tconv (Middle), and Treg cells (Right). Data are from three independent experiments; P values are from a paired Student’s t test (*P < 0.05, **P < 0.005, and ***P < 0.001).
As a further evaluation of Treg function, we tested the susceptibility of the FoxP3 mutant mice to tumor growth by s.c. injection of MC38 colon tumor cells. M176 and M354 mice showed strikingly delayed tumor progression (Fig. 7G), suggesting a more effective antitumor response. Tregs normally form the majority of infiltrating CD4+ T cells in MC38 tumors, but these Tregs were far less abundant in mutant hosts, especially for M354 (Fig. 7H). This deficiency was accompanied by heightened production of TNFα and IFNγ by tumor-infiltrating CD4+ Tconv and CD8+ T cells (Fig. 7I), especially in M354 hosts, confirming that the functional Treg deficiency allowed stronger activation of T-effector pathways in the tumor microenvironment. This phenotype is similar to the poor Treg function described for Nrp1-deficient Tregs in the tumor environment (47). Overall, these missense mutations in FoxP3 led to Tregs with reduced fitness and function, the deficit of which became manifest only upon challenge. These subtle defects markedly influenced immune response in disease contexts, but in a manner that bore little or no relation to known pathology of FoxP3 deficiency.
Discussion
The level of resolution provided by this set of 130 FoxP3 mutants sheds a light on the operation of this transcription factor. The complexity of FoxP3 structure–function relationships operated at two levels. First, the mutations diversely affected the transactivation of different FoxP3 target blocs, some with subtle difference within the general axis of activity, others very distinctly. Second, these functions could not be ascribed to simple modular architectures, but implicated the whole molecule to some extent. These variegated mutational effects translated into in vivo phenotypes distinct from the usual IPEX/scurfy pathology.
Our recent mutagenesis study, involving a much smaller number of alterations, showed that FoxP3’s transactivation ability generally correlated with the potential to bind with multimolecular complexes that include RelA and Ikzf2 (37). In keeping with this notion, we found here that a generic “activation index” can effectively summarize the ability of the mutants to activate FoxP3 targets (and inversely for the overall repression index). However, the results were also more nuanced than these dominant effects. First, the response of individual genes showed subtle differences, reflected in the finely resolved ability of mutants of any one bloc to affect a particular target (e.g., Il2ra and Tnfrsf18 responded similarly, but distinctly from Dusp4 or Lag3). Second, some targets showed radically different patterns, objectivized by the “normalcy index” of Fig. S4C, which brought out the very different sensitivity to mutations of Il5, Rorc, or Il1rl1. Interestingly, the expression of the latter two is found predominantly in tissue-Tregs (3), implying that the adaptation of FoxP3 to functioning in these particular environments harnesses functional facets different from those characteristic of its more usual function in lymphoid tissues.
Transactivation and transrepression of particular targets involved several structural regions identifiable by sequence composition, with convergent mutational effects of mutations in either the N-terminal moiety or the FKH domain. This spread was observed with profiling as well as reporter assays. There was no evidence for a simple repressor domain or for any region with a dominant activating role. This integrated view, in which the entire molecule takes part in different functions, is at odds with the simpler interpretations of modular TF structure (48), in which well-demarcated domains are ascribed distinct functions, which can be shuffled evolutionarily (in fairness, however, the modular model had already been shown to be an oversimplification; see, e.g., ref. 49). In the same vein, our study of FoxP3 interactions with transcriptional cofactors also found that the regions of FoxP3 that conditioned the interactions with specific cofactors encompassed multiple domains (37). As a consequence, we cannot ascribe the different transcriptomes of M176 and M354 Treg cells to perturbed interactions between FoxP3 and any given cofactor. These results are consistent with the notion of FoxP3 assembled into multicomponent “molecular machines” for transcriptional control, such that many facets of the protein contribute to assembly of one such complex.
The variegation of mutational effects across the spectrum of FoxP3 targets and across the phenotypes of M176 and M354 mice opens perspectives on pathologies associated with FoxP3 deficiency in humans. First, the phenotype of mutants with more severe mutations suggests that the high IgE levels of many IPEX patients may be connected to the paradoxical induction of Il5 and Il4 observed here, in addition to the defective control of Th2 cells. More generally, the range of transcriptional effects observed here is consistent with the wide heterogeneity of severity and symptoms that result from missense mutations in IPEX patients (13–15). Some IPEX mutations with milder phenotypes (F324L, R347H, V408M) affect the same position as some of our less dramatic mutants. Similarly, a recent report also showed a partial scurfy phenotype after introduction of the IPEX A384T mutation (36). Even more divergent, however, is the absence of an overt phenotype in the M176 and M354 mutant mice in which Treg defects, clearly apparent by transcriptional analysis, were revealed only in a competitive context in response to inflammatory stress or after aging. Had these defects appeared spontaneously, they would likely not have been ascribed to Foxp3. This observation suggests the possibility that humans with comparable rare missense variants in FOXP3 may exist. These variants would not be recognized because of the marked departure from the IPEX syndrome, but may contribute to exacerbated susceptibility to insults in the gut or to isolated skin pathology such as aged M354 mice. Indeed, a survey of rare missense mutations of FOXP3 in the Inflammatory Bowel Disease Exomes Browser (https://ibd.broadinstitute.org) shows 13 such mutations in irritable bowel disease patients. One might question why FOXP3 does not appear in genome-wide association study data; the answer may be in the rare nature of the mutations, which are likely subject to rapid purifying selection, which cannot be detected by association studies that track more frequently distributed variants.
In conclusion, this refined structure–function dissection has brought a very different perspective on FoxP3 and its integration into flexible molecular machines and the particular dysfunction of which may affect human disease in unexpected ways.
Materials and Methods
All experimental procedures are described in detail in SI Materials and Methods.
Mice.
The C57BL/6J mice (Jackson Laboratory) and mutants were bred in an specific pathogen-free (SPF) facility at Harvard Medical School (IACUC protocol 02954). The M176 and 354 mutations were introduced by Cas9-targeted mutagenesis with oligonucleotide-directed resealing and direct injection into mouse zygotes (45). For BM radiation chimeras, recombination-activating genes (RAG)-deficient recipients were injected 6 h after 6 Gy irradiation with a 50/50 mix of bone marrow (BM) cells from WT B6.CD45.1 congenic and M176 or M354 (CD45.2) donors and analyzed 10 wk later. Experiments were performed under protocol IS00001257 approved by the Harvard Medical School Institutional Animal Care and Use Committee.
Antibodies and Plasmids.
N-terminal FLAG-tagged FoxP3 was cloned into the MSCV-IRES-THY1.1 retroviral vector, and alanine replacements were generated by site-directed mutagenesis; the entirety of the coding sequence was verified by Sanger sequencing.
Retroviral Infection and Expression Analysis.
CD4+CD25− Tconv were isolated by negative magnetic selection and activated with anti-CD3/CD28 beads for 36 h before infection. Cells were spin-infected for 2 h and flow-sorted 72 h later within a window of Thy1.1 expression determined to correspond to normal levels of FoxP3 in Tregs.
DNA-Binding Assay.
Nuclear extract from EV, WT, or mutant transduced CD4+ T cells was induced with 25-bp double-stranded biotinylated oligo with two copies of canonical FoxP3-binding motif. FoxP3 binding was measured with an Episeeker DNA-protein–binding assay kit (ab117139; Abcam).
Luciferase Reporter Assay.
EV, WT, or each mutant FoxP3 plasmid was cotransfected with luciferase reporter plasmids driven by 8XFKRE (50) or IL2 promoter with pRL-tk renilla vector in EL4 T cells. Forty-eight hours after transfection, cells were stimulated with phorbol myristate acetate/ionomycin for 2 h, and reporter activity was measured.
Tumor and Colitis Challenge.
Mice (6 wk old) were sensitized with 50 μL of 1% TNBS (4:1 acetone:olive oil solution) on their backs. A week later, colitis was induced by intrarectal administration of 100 μg (standard dose) or 40 μg (low dose) of TNBS per gram of mouse in 50% ethanol in anesthetized mice. Mice were injected s.c. with 1 × 105 MC38 colon adenocarcinoma cells. Tumor size was measured every 2 d with a caliper.
Expression Profiling.
RNA was prepared and used for expression profiling on Affymetrix ST1.0 microarrays per ImmGen SOP (51). Cell lysates were used directly for profiling by Nanostring nCounter [custom Treg codeset (37)]. Data were processed and normalized using Nanostring or Affymetrix software, per refs. 51 and 52.
Supplementary Material
Acknowledgments
We thank Dr. E. Sefik for help with the mutant evaluation; K. Hattori, C. Araneo, and A. Rhoads for help with mice, cell sorting, and the Helmsley Inflammatory Bowel Disease Exomes Program; and the groups that provided exome variant data for comparison (listed at https://ibd.broadinstitute.org/about). This work was supported by NIH Grant AI116834 and a Sponsored Research Agreement from GSK, and by National Research Foundation Fellowship 357-2011-1C00084 (to H.-K.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE104344 and GSE104345).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718599115/-/DCSupplemental.
References
- 1.Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: Mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panduro M, Benoist C, Mathis D. Tissue Tregs. Annu Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegler SF. FOXP3: Of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 5.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Sugimoto N, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 7.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Ferraro A, et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci USA. 2014;111:E1111–E1120. doi: 10.1073/pnas.1401343111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arvey A, et al. Genetic and epigenetic variation in the lineage specification of regulatory T cells. Elife. 2015;4:e07571. doi: 10.7554/eLife.07571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu W, et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang W, et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekiya T, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 13.Ramsdell F, Ziegler SF. FOXP3 and scurfy: How it all began. Nat Rev Immunol. 2014;14:343–349. doi: 10.1038/nri3650. [DOI] [PubMed] [Google Scholar]
- 14.Verbsky JW, Chatila TA. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) and IPEX-related disorders: An evolving web of heritable autoimmune diseases. Curr Opin Pediatr. 2013;25:708–714. doi: 10.1097/MOP.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.d’Hennezel E, Bin Dhuban K, Torgerson T, Piccirillo CA. The immunogenetics of immune dysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J Med Genet. 2012;49:291–302. doi: 10.1136/jmedgenet-2012-100759. [DOI] [PubMed] [Google Scholar]
- 16.Powell BR, Buist NR, Stenzel P. An X-linked syndrome of diarrhea, polyendocrinopathy, and fatal infection in infancy. J Pediatr. 1982;100:731–737. doi: 10.1016/s0022-3476(82)80573-8. [DOI] [PubMed] [Google Scholar]
- 17.Wildin RS, Freitas A. IPEX and FOXP3: Clinical and research perspectives. J Autoimmun. 2005;25(Suppl):56–62. doi: 10.1016/j.jaut.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Barzaghi F, Passerini L, Bacchetta R. Immune dysregulation, polyendocrinopathy, enteropathy, x-linked syndrome: A paradigm of immunodeficiency with autoimmunity. Front Immunol. 2012;3:211. doi: 10.3389/fimmu.2012.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopes JE, et al. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol. 2006;177:3133–3142. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- 20.Li B, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 22.Bandukwala HS, et al. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011;34:479–491. doi: 10.1016/j.immuni.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, et al. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 2015;43:1268–1282. doi: 10.1093/nar/gku1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen KG, Nissen JK, Betz AG. Comparative genomics reveals key gain-of-function events in Foxp3 during regulatory T cell evolution. Front Immunol. 2012;3:113. doi: 10.3389/fimmu.2012.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darce J, et al. An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity. 2012;36:731–741. doi: 10.1016/j.immuni.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bettini ML, et al. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36:717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bacchetta R, et al. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest. 2006;116:1713–1722. doi: 10.1172/JCI25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magg T, Mannert J, Ellwart JW, Schmid I, Albert MH. Subcellular localization of FOXP3 in human regulatory and nonregulatory T cells. Eur J Immunol. 2012;42:1627–1638. doi: 10.1002/eji.201141838. [DOI] [PubMed] [Google Scholar]
- 33.Song X, et al. Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep. 2012;1:665–675. doi: 10.1016/j.celrep.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gambineri E, et al. Clinical and molecular profile of a new series of patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome: Inconsistent correlation between forkhead box protein 3 expression and disease severity. J Allergy Clin Immunol. 2008;122:1105–1112.e1. doi: 10.1016/j.jaci.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Wang L, Han R, Beier UH, Hancock WW. Two lysines in the forkhead domain of foxp3 are key to T regulatory cell function. PLoS One. 2012;7:e29035. doi: 10.1371/journal.pone.0029035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayatsu N, et al. Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity. 2017;47:268–283.e9. doi: 10.1016/j.immuni.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Kwon HK, Chen HM, Mathis D, Benoist C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat Immunol. 2017;18:1238–1248. doi: 10.1038/ni.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arvey A, et al. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2014;15:580–587. doi: 10.1038/ni.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J Biol Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 40.Hancock WW, Ozkaynak E. Three distinct domains contribute to nuclear transport of murine Foxp3. PLoS One. 2009;4:e7890. doi: 10.1371/journal.pone.0007890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc Natl Acad Sci USA. 2013;110:12349–12354. doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biggs WH, III, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie X, et al. The regulatory T cell lineage factor Foxp3 regulates gene expression through several distinct mechanisms mostly independent of direct DNA binding. PLoS Genet. 2015;11:e1005251. doi: 10.1371/journal.pgen.1005251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nat Immunol. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Overacre-Delgoffe AE, et al. Interferon-γ drives Treg fragility to promote anti-tumor immunity. Cell. 2017;169:1130–1141.e11. doi: 10.1016/j.cell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frankel AD, Kim PS. Modular structure of transcription factors: Implications for gene regulation. Cell. 1991;65:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- 49.Cutler G, Perry KM, Tjian R. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol Cell Biol. 1998;18:2252–2261. doi: 10.1128/mcb.18.4.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci USA. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heng TS, Painter MW. Immunological Genome Project Consortium The immunological genome project: Networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 52.Ye CJ, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sefik E, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koh KP, Sundrud MS, Rao A. Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3. PLoS One. 2009;4:e8109. doi: 10.1371/journal.pone.0008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shay T, et al. ImmGen Consortium Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci USA. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.