Significance
The histone demethylase LSD1, a critical regulator of mammalian hematopoiesis, serves as an important suppressor of endotoxic shock. Inflammation-induced deletion of LSD1 results in failure to generate all mature hematopoietic cells, and induces acute expansion of a pathological population of hyperproliferative and hyperinflammatory myeloid progenitors that cause “cytokine storm” and acute lethality. LSD1 proves to be a downstream target of Toll-like/cytokine receptors in HSCs during endotoxic shock, with its down-regulation caused by a cohort of inflammation-induced microRNAs. The resultant acute expansion of a population of pathological myeloid progenitors in bone marrow causes a septic shock phenotype. Inhibitors of these inflammation-induced microRNAs block the down-regulation of LSD1 and prevent LPS-induced mortality, suggesting a potential therapeutic approach for treatment of septic shock.
Keywords: LSD1, hematopoiesis, septic shock, hematopoietic stem cell, microRNA
Abstract
Hematopoietic stem cells (HSCs) maintain a quiescent state during homeostasis, but with acute infection, they exit the quiescent state to increase the output of immune cells, the so-called “emergency hematopoiesis.” However, HSCs’ response to severe infection during septic shock and the pathological impact remain poorly elucidated. Here, we report that the histone demethylase KDM1A/LSD1, serving as a critical regulator of mammalian hematopoiesis, is a negative regulator of the response to inflammation in HSCs during endotoxic shock typically observed during acute bacterial or viral infection. Inflammation-induced LSD1 deficiency results in an acute expansion of a pathological population of hyperproliferative and hyperinflammatory myeloid progenitors, resulting in a septic shock phenotype and acute death. Unexpectedly, in vivo administration of bacterial lipopolysaccharide (LPS) to wild-type mice results in acute suppression of LSD1 in HSCs with a septic shock phenotype that resembles that observed following induced deletion of LSD1. The suppression of LSD1 in HSCs is caused, at least in large part, by a cohort of inflammation-induced microRNAs. Significantly, reconstitution of mice with bone marrow progenitor cells expressing inhibitors of these inflammation-induced microRNAs blocked the suppression of LSD1 in vivo following acute LPS administration and prevented mortality from endotoxic shock. Our results indicate that LSD1 activators or miRNA antagonists could serve as a therapeutic approach for life-threatening septic shock characterized by dysfunction of HSCs.
Septic shock is a devastating condition and currently the leading cause of death in hospitals (1). Septic shock is a result of a systemic response to severe infection, which is commonly attributed to hyperactivation followed by exhaustion of mature hematopoietic cells such as neutrophils or macrophages (2, 3). Recently, the hematopoietic progenitors and hematopoietic stem cells (HSCs) have been found to respond to many infections to increase the output of immune cells, so-called emergency hematopoiesis (4–7). However, HSCs’ response and the pathological impact on severe infections during septic shock have not been clear. While epigenetic mechanisms clearly contribute to gene regulation in development and disease, the roles of specific epigenetic regulators in mammalian hematopoiesis and hematological disease are still largely unknown. KDM1A/LSD1 was initially described as a histone demethylase specific for histone H3 lysine 4 and 9 (H3K4 and H3K9) methylation that activates and represses many transcriptional programs (8–10). Deletion of LSD1 in mice alters the development of the pituitary gland and impairs early embryonic development (11, 12), suggesting crucial roles of LSD1 on many developmental events.
Here, we report that in addition to being a critical regulator of mammalian hematopoiesis, inflammation-induced deletion of LSD1 in adult mice leads to rapid development of a phenotype resembling septic shock and acute lethality. Poly I.poly C (pIpC)/Mx-Cre-induced LSD1 deficiency causes HSC dysregulation, promotes acute expansion of hyperproliferative and hyperinflammatory myeloid progenitors, and results in cytokine storm and multiorgan pathology. Interestingly, we observe microRNA-mediated suppression of LSD1 expression in a mouse model of endotoxin-induced septic shock that can be reversed in vivo by an anti-miRNA strategy. Our study reveals an underlying mechanism for inflammation-induced HSC dysfunction and progression to septic shock.
Results
LSD1 Deficiency Leads to Sudden Death Due to a Septic Shock-Like Phenotype.
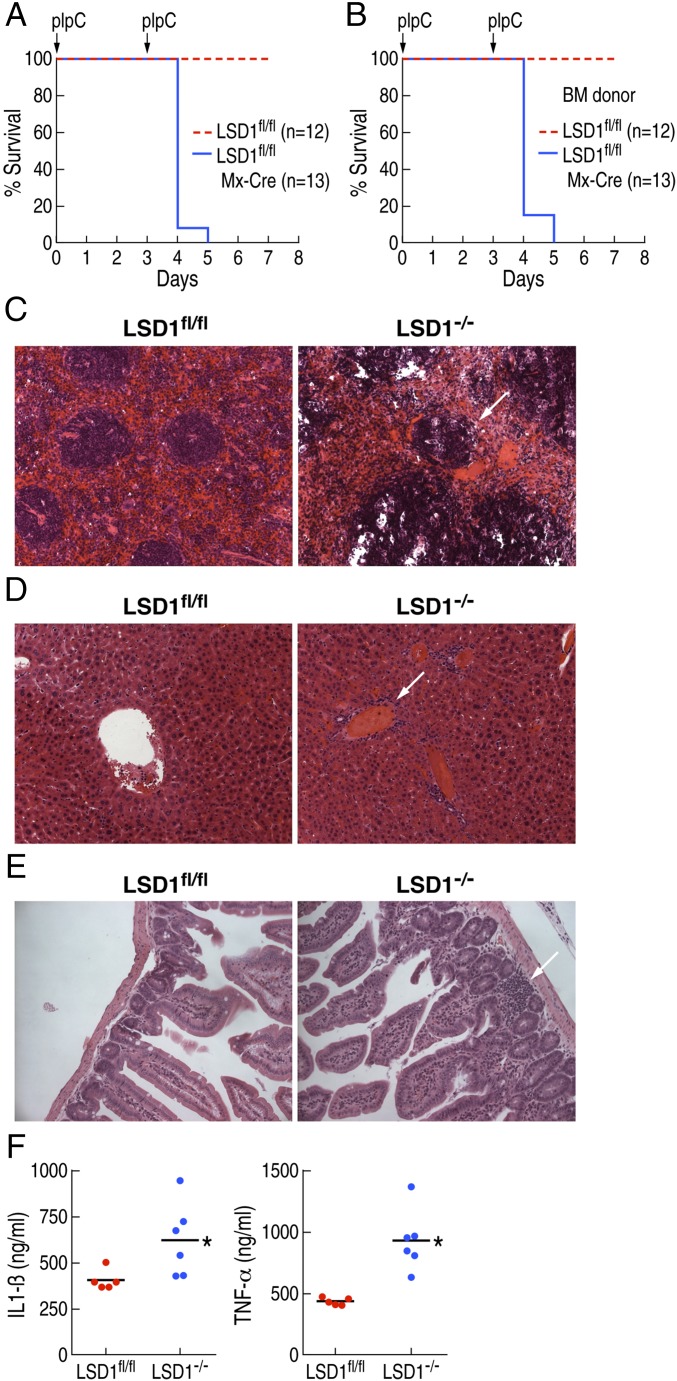
To elucidate the roles of LSD1 in mammalian hematopoiesis, LSD1 floxed mice (LSD1fl/fl) were crossed to Mx-Cre transgenic mice to delete LSD1 in a pIpC-inducible manner (11, 13). The standard protocol requires three consecutive pIpC injections (Fig. S1A). However, LSD1fl/fl:Mx-Cre mice injected with pIpC (LSD1−/−) died shortly after the second injection (Fig. 1A). Because LSD1 is broadly expressed and Mx-Cre is able to delete LSD1 from multiple organs, we performed bone marrow transplantation (BMT) from LSD1fl/fl:Mx-Cre and the control mice (LSD1fl/fl) into C57BL/6 wild-type recipient mice. The recipient mice reconstituted from the LSD1fl/fl:Mx-Cre BMT mice also died shortly after the second pIpC injection, with similar kinetics as non-BMT mice (Fig. 1B). These data suggested that the sudden death of LSD1−/− mice was caused by LSD1 deletion in bone marrow (BM) cells. Because pIpC triggers a robust innate immune reaction through Toll-like receptor 3 (TLR3), we performed BMT and induced LSD1 deletion by a single injection of pIpC to avoid a pIpC-mediated robust immune response. BMT recipient mice from LSD1fl/fl :Mx-Cre BM died after 1 wk of single pIpC injection, indicating that LSD1 deletion in BM alone was sufficient to cause the sudden death of LSD1−/− mice (Fig. S1B).
Fig. 1.
LSD1 deficiency leads to sudden death due to endotoxic shock. (A) Survival curve after pIpC injection into LSD1fl/fl: Mx-Cre (LSD1−/−) mice (n = 13) and the control (LSD1fl/fl) mice (n = 12). (B) Survival curve after pIpC injection into mice after BM transfer with LSD1fl/fl: Mx-Cre (LSD1−/−) mice (n = 13) and the control (LSD1fl/fl) mice (n = 12). (C) Histological analysis of spleens from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and the control (LSD1fl/fl) mice, revealing multiple lesions in LSD1-deficient mice. (D) Histological analysis of livers from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and the control (LSD1fl/fl) mice, revealing multiple lesions in LSD1-deficient mice. (E) Histological analysis of intestines from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and the control (LSD1fl/fl) mice, revealing multiple lesions in LSD1-deficient mice. (F) Serum level of IL-1β (Left) and TNF-α (Right) 18 h after second pIpC injection from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and control mice were determined by ELISA.
To determine the cause of sudden death observed on LSD1−/− mice, we performed histological analysis on the internal organs. We observed many lesions in the spleen, intestine, liver, kidney, and lung of LSD1−/− mice, with signs of increased inflammation (Fig. 1 C–E). This pathology is consistent with the multiple organ failure phenotype commonly observed in septic shock. Because the kinetics of sudden death, as well as the appearance and behavior of LSD1−/− mice, were consistent with a toxic shock-like syndrome, we evaluated levels of proinflammatory mediators in serum. Both interleukin (IL)-1β and tumor necrosis factor (TNF)-α were significantly elevated in LSD1−/− mice (Fig. 1F). These data suggested that the sudden death of the LSD1−/− mice was caused, at least in part, by exaggerated cytokine production upon pIpC stimulation (i.e., cytokine storm) (2, 3).
LSD1-Deficient Mice Exhibit Acute Expansion of Hyperproliferative and Hyperinflammatory Myeloid Progenitors in BM.
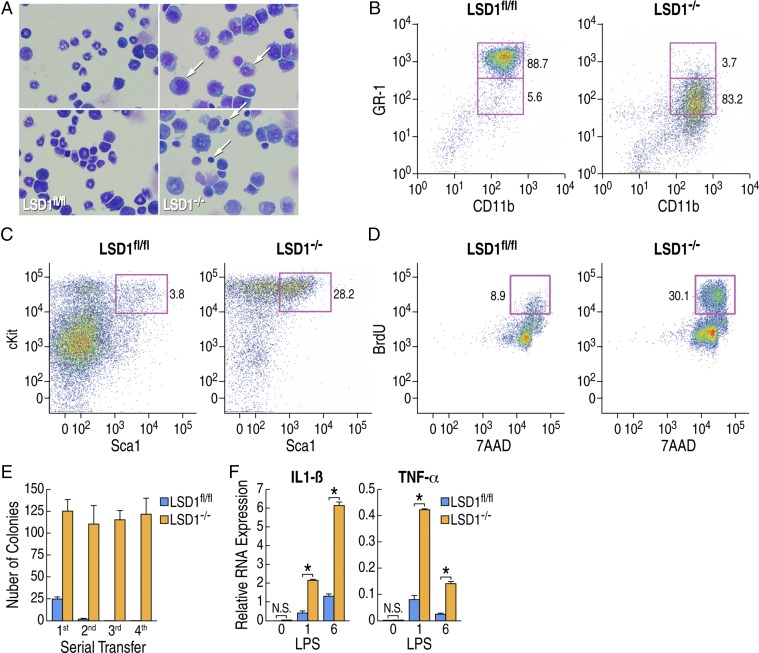
Because Mx-Cre–mediated deletion of LSD1 occurs in the HSC, we performed analysis of the hematopoietic system using BM cells. We observed a decreased number of mature granulocytes and monocytes, but an increased number of immature myeloid blast cells in these LSD1−/− mice (Fig. 2A). Next, to determine the cell populations affected in the LSD1−/− mice, BM cells were isolated and analyzed by flow cytometry based on the surface markers of BM cells. Using lineage-specific markers, we observed an aberrant CD11b+GR-1low population to be expanded in the LSD1−/− mice or BMT recipient mice (Fig. 2B and Fig. S2A). Livers and BM from LSD1−/− mice were pale and appeared anemic, indicating that LSD1−/− mice BM cells were unable to generate erythroid-lineage cells (Fig. S2B), which is consistent with previous reports (14, 15). However, these prior studies did not observe the septic shock phenotype. This is most likely due to the use of alternative methods to conditionally knock down (Tet-inducible shRNA; ref. 14) or delete (Vav-Cre; ref. 15) LSD1 that do not themselves induce an inflammatory response. Furthermore, LSD1−/− mice BM cells expressed unusual surface marker combinations such as CD11b+CD90+ (Fig. S2C) and CD11b+Sca1+ (Fig. S2D). Aberrant hematopoietic cell development was associated with the increased HSC population determined by lineage-negative Sca1+cKit+ staining (Fig. 2C). LSD1−/− mice BM cells also showed increased proliferation determined by BrdU incorporation (Fig. 2D) and could be serially transferred using methylcellulose-based colony formation assay without forming any defined lineage-specific colonies (Fig. 2E and Fig. S2E). The LSD1−/− cells formed colonies of immature cells on the methylcellulose-based colony formation assay, and became spontaneously immortalized and even grow in the absence of SCF or IL-3 (Fig. S2F). These data suggested that LSD1−/− mice BM cells showed altered development and increased proliferative capacity, similar with leukemia (16). However, the numbers of total BM cells, as well as CD11b+GR-1low, were not increased, and invasion of organs outside the BM was not observed (Fig. S2G). To test whether BM cells were responsible for the increased proinflammatory mediators, BM cells were isolated and stimulated by lipopolysaccharide (LPS) in vitro. We found that mRNA expression of both IL-1β and TNF-α after TLR4 stimulation was significantly increased in LSD1−/− BM cells (Fig. 2F). Therefore, expansion of a hyperproliferative and hyperinflammatory cell population in LSD1−/− mice BM might contribute to the exaggerated cytokine production upon pIpC stimulation (i.e., cytokine storm).
Fig. 2.
LSD1-deficient mice have acute expansion of hyperproliferative and hyperinflammatory myeloid progenitors in BM. (A) Giemsa staining of BM cells from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and the control mice. Arrows denote cells with pyknotic nuclei in LSD1−/− conditions. (B) The representative expression of CD11b and GR-1 of BM cells from mice after BMT with LSD1fl/fl: Mx-Cre (LSD1−/−) mice and control are shown. Numbers are indicated as the percentage of indicated quadrant population among total BM cells, (C) The representative expression of Sca1 and cKit of BM cells from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and the control mice are shown. Numbers are indicated as the percentage of population among Lineage negative cells (Lin−). (D) Incorporation of BrdU and 7AAD of BM cells from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and the control are shown. Numbers are indicated as the percentage of BrdU-positive cells among total cells in the culture. (E) CD11b-positive BM cells were isolated from LSD1fl/fl: Mx-Cre (LSD1−/−) mice and the control were cultured in methylcellulose-based medium supplemented with a mixture of cytokines. Number of colonies is shown after in vitro serial transfer into the methylcellulose-based medium. Data are shown as an average of triplicates, and error bar indicated a SD. (F) Total BM cells isolated from LSD1fl/fl: Mx-Cre (LSD1−/−) mice (blue bar) and control (orange bar) were stimulated with 0.1 mg/mL LPS for the indicated time, and mRNA expression of IL-1β (Left) or TNF-α (Right) is shown. *P < 0.01.
Altered HSC Homeostasis in Mice During Endotoxic Shock.
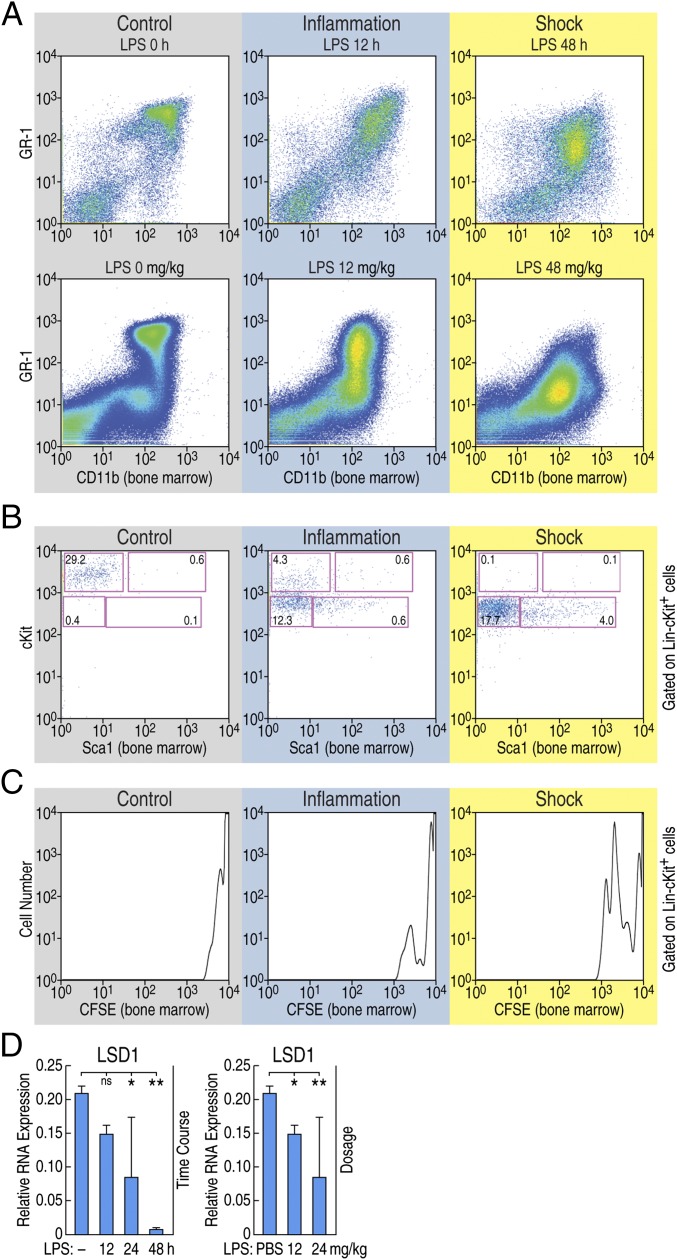
The observation that pIpC-induced deletion of LSD1 induced a septic shock phenotype, whereas pIpC treatment of wild type leads to a mild transient inflammatory response, led us investigate whether LSD1 expression is altered in vivo in response to endotoxin challenge. It was previously reported that interferons induced by sustained viral infection break HSC quiescence (4–7). These observations suggested that sustained inflammation/infection-stressed HSCs induced aberrant differentiation and maintenance of HSCs. Therefore, to test whether the breakage of HSC quiescence is also observed during experimental endotoxin shock in wild-type mice, LPS was injected into C57BL/6 in two different protocols (time and dose) to mimic the conditions of inflammation (24 mg/kg, 12 h; 12 mg/kg, 48 h) or endotoxin shock (24 mg/kg, 12 h); 24 mg/kg, 48 h) (Fig. S3A). CD11b+GR-1low cells appeared in the BM and the periphery blood observed on conditions mimicking endotoxin shock (Fig. 3A and Fig. S3B). Interestingly, under these conditions, HSCs down-regulated the surface cKit expression and increased the proliferation determined by carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling (Fig. 3 B and C). The cells observed in the periphery may not reflect the local expansion of CD11b+GR-1low cells because they do not proliferate in vitro (Fig. S3C). Rather, BM cells showed a proliferative capacity, and resultant cells might subsequently migrate into the peritoneal cavity (Fig. S3C).
Fig. 3.
Altered HSC homeostasis in mice following endotoxin challenge. (A) The representative expression of CD11b and GR-1 on BM cells during animal models of sepsis is shown. After indicated time of 24 mg/kg LPS injection (Upper) or 48 h after indicated LPS dosage (Lower) are shown. (B) The representative expression of Sca1 and cKit on BM cells is shown. Numbers are indicated of percentage of the cell in the lineage negative, cKit, and Sca1 gated population. (C) Total BM cells were isolated from sepsis-induced mice and cultured for 24 h in vitro, and expression of CFSE gated on lineage negative cKit-positive cells is shown. (D) Wild-type animals were injected with LPS for indicated time (Left) or dosage (Right), and expression of LSD1 in BM cells was determined by RT-qPCR. *P < 0.01; **P < 0.001; ns, not significant.
Cytokine storm is a term commonly employed to describe the overproduction of proinflammatory mediators during endotoxin shock and sepsis (2, 3). Since cytokine storm can be a fatal incident during sepsis, the establishment of tolerance is a critical mechanism to avoid septic death (17, 18). To test the tolerance induction, BM cells from inflammation and endotoxin shock-induced animals were isolated and stimulated with LPS again in vitro. Tolerance was not induced under endotoxin shock conditions, because the BM cells still responded and up-regulated iNOS and TNF-α mRNA (Fig. S3D). As Nurr1 and LSD1 were shown to be recruited by NF-κB p65 and mediate transcriptional repression of NF-κB target genes in glia cells (10), we next investigated whether LSD1 and Nurr1 also localized to promoters directing expression of proinflammatory mediators in the BM cells. Chromatin immunoprecipitation (ChIP) assays were performed for iNOS and TNF-α promoters using BM cells isolated from LPS-treated animals. Nurr1 recruitment was observed under both of inflammation and endotoxin shock. However, we failed to detect LSD1 recruitment to the promoter regions of iNOS and TNF-α under endotoxin shock conditions (Fig. S3E). Therefore, the expression of LSD1 was tested in the BM cells isolated from wild-type mice induced by inflammatory signals to have “septic shock.” We found that the expression of LSD1 in the BM cells was strongly suppressed under these conditions of endotoxin shock (Fig. 3D). These data suggested a dramatic inflammation/sepsis-induced suppression of LSD1 in BM cells, indicating that LSD1 deficiency occurs in vivo during endotoxic shock.
miRNA-Mediated Down-Regulation of LSD1 During Sepsis.
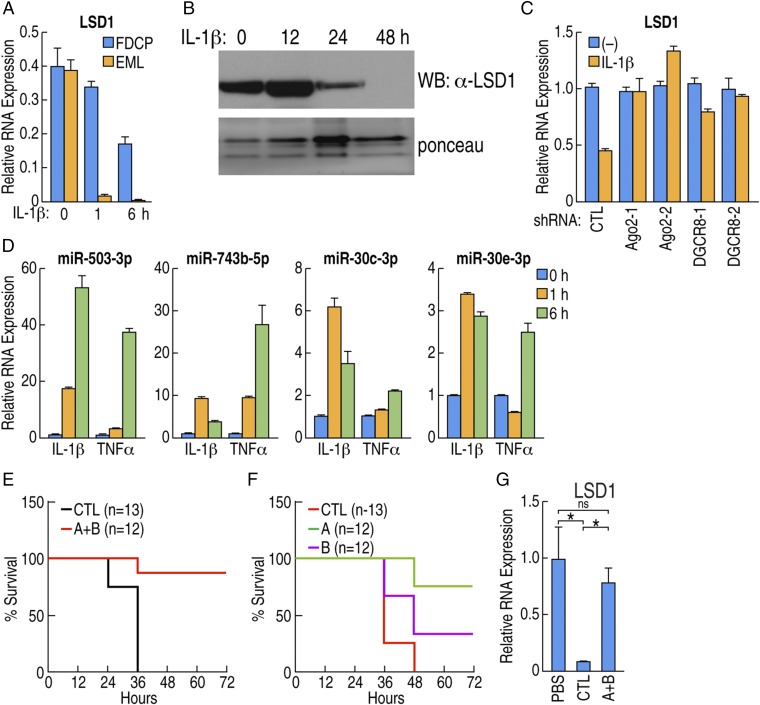
Because depletion of LSD1 caused loss of homeostasis in HSCs, we next studied the molecular mechanism underlying the suppression of LSD1 in HSCs. First, we tested whether LSD1 suppression could be mimicked in vitro using murine HSC-like cell lines. Since cytokine storm/endotoxin shock broke HSC homeostasis, we treated HSC-like cell lines with proinflammatory cytokines and then examining the expression of LSD1. IL-1β stimulation significantly down-regulated the expression of LSD1 mRNA and protein (Fig. 4 A and B). Because mRNA down-regulation was observed before the protein down-regulation, we focused on the mechanism that induced the down-regulation of LSD1 mRNA. We first tested whether the LSD1 locus was transcriptionally repressed during the sepsis condition. The LSD1 promoter (−2 kb upstream to transcription start site) was subcloned into a luciferase-reporter vector and tested for promoter activity. We found that LSD1 promoter activity was slightly increased on cytokine stimulation, rather than transcriptionally silenced (Fig. S4A). No significant change was observed in the H3K27me3 mark at the LSD1 promoter, generally considered as a mark of transcriptional repression (19), suggesting suppression of LSD1 is due to posttranscriptional regulation.
Fig. 4.
miRNA-mediated down-regulation of LSD1 in response to inflammatory cytokines. (A) FDCPmix cell (blue bar) or EML cell (orange bar) were treated with IL-1β. LSD1 mRNA levels are shown, measured by RT-qPCR. (B) FDCPmix cells were treated with IL-1β, and LSD1 expression was determined by Western blot; Ponceau S staining as loading control. (C) FDCPmix cells were transduced with lentiviral vectors carrying two independent shRNAs against argonout2 (Ago2) or DGCR8, and mRNA expression of LSD1 after IL-1β treatment was measured by RT-qPCR. (D) FDCPmix cells were stimulated by IL-1β or TNF-α for the indicated time, and the expression level of indicated miRNA was determined by qPCR. (E) Wild-type mice were reconstituted with BM cells transduced with pooled lentiviral vectors carrying antisense for indicated miRNAs (n = 12, group A+B, see text for grouping) or scramble control (n = 12). After reconstitution, mice were injected with 24 mg/kg LPS to induce sepsis, and the survival curve is shown. (F) Wild-type mice were reconstituted with BM cells transduced with pooled lentiviral vectors carrying antisense against miRNAs (scramble control, n = 12, group A, n = 12 or group B, n = 12). After reconstitution, mice were injected with 24 mg/kg LPS, and survival curve is shown. (G) Expression of LSD1 was determined in animals described in E. BM cells were isolated 36 h after LPS injection, and LSD1 expression is shown as an average of four animals. *P < 0.01; ns, not significant.
We therefore investigated the possibility of microRNA (miRNA)-mediated suppression of LSD1 mRNA (20). FDCPmix cells were transduced with lentiviral vectors carrying two independent shRNAs against Argonaute 2 (Ago2-1, Ago2-2) and DiGeorge critical region 8 (Dgcr8-1 and Dgcr8-2) to knock down miRNA processing machinery (21–23). We found that knockdown of Ago2 and Dgcr8 significantly blocked the down-regulation of LSD1 in FDCPmix cells (Fig. 4C). The contribution of the 3′UTR of LSD1 was tested by luciferase assay in FDCPmix cells. IL-1β and TNF-α stimulation significantly down-regulated LSD1-3′UTR reporter activity, consistent with a miRNA-mediated down-regulation mechanism regulating LSD1 mRNA expression (Fig. S4B).
To identify the candidate miRNAs that might down-regulate LSD1 mRNA in HSCs, miR-seq was performed in FDCPmix cells treated with IL-1β and TNF-α (Fig. S4 C and D). Candidate miRNAs that would possibly bind to the LSD1 3′UTR were selected by using multiple algorithms, focusing on the miRNAs exhibiting significant up-regulation by proinflammatory cytokines. Induction of the most prominent candidate miRNAs, miR-503-3p, miR-743b-5p, miR-30c-3p, and miR-30e-3p upon IL-1β and TNF-α stimulation, was confirmed by qPCR (Fig. 4D). Based on these data, potential seed regions of candidate miRNAs were mutated, LSD1 cDNAs harboring these deletions were infected into FDCPmix cells, and tagged-LSD1 protein levels upon IL-1β stimulation was determined. Several mutations of the seed region including miR-30e-3p, miR-137-5p, miR-503-3p, and miR-743b-5p were able to block the down-regulation (Fig. S4E). Finally, to confirm whether these miRNAs were responsible for the down-regulation of LSD1 during sepsis, HSCs from wild-type mice were isolated and transduced with lentiviral vectors to express the antisense miRNAs (miRZiP) to block the candidate miRNAs. Reconstituted mice were injected with LPS, and the survival was tested. Control mice infected with scramble miRZip were induced sepsis and died within 36 h. In contrast, mice transduced with pooled miRZip against miR-503-3p, miR-743b-5p, and miR-200a-5p (group A), and miR-30b-3p, 30c-3p, 30d-3p, and 30e-3p (group B) were protected from septic death and from aberrant HSC development (Fig. 4E). Next, pooled miRZip was divided into individual groups A and B. Mice reconstituted with scramble control died after 48 h of sepsis induction; in contrast, most of the group A reconstituted animals survived more than 72 h (Fig. 4F). Inhibition of down-regulation of LSD1 was confirmed from BM cells from experimental mice, with no such effects in the mice treated with the scramble shRNA control (Fig. S4F). Therefore, inhibitors of inflammation-induced miRNAs may serve as a novel approach to block LSD1 suppression upon severe infection and, hence, promote survival during septic shock.
LSD1 Inhibitors Alter HSC Development in ex Vivo BM Culture.
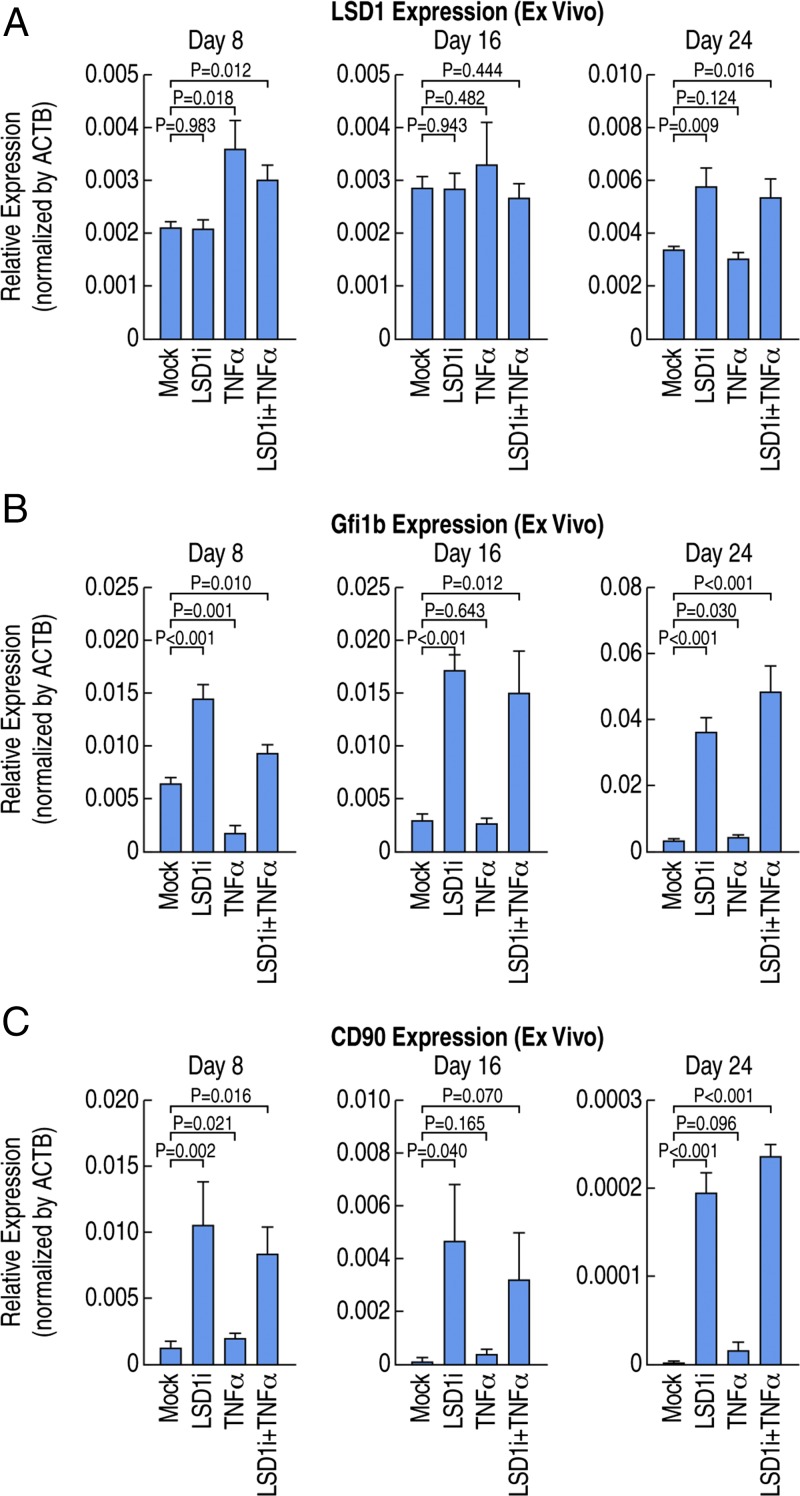
Recent publications suggest the inactivation of LSD1 might be effective for the treatment of acute myelogenous leukemia (AML) caused by MLL-AF9 or PML-RARα chromosomal translocation (24–26), and other types of cancers (27, 28). However, our data suggest that inhibition of LSD1 activity in the context of inflammation/infection potentially impairs the quiescence of HSCs and, therefore, might even cause a leukemic/septic outcome. To determine whether LSD1 inhibitors can target HSCs, we performed BM culture ex vivo in the presence of LSD1 inhibitors or mock controls. We observed increased expression of Gfi1b upon treatment of LSD1 inhibitors while there were no significant changes of expression of LSD1 (Fig. 5 A and B), similar with the LSD1 knockout phenotype and consistent with the role of LSD1 in HSC regulation. In addition, we observed increased expression of CD90 (Fig. 5C), a surrogate marker of HSCs, suggesting LSD1 inhibitors can induce expansion of a pathological population of HSCs in BM culture ex vivo.
Fig. 5.
LSD1 inhibitors alter HSC development in ex vivo BM culture. (A) Expression of LSD1 in ex vivo cultured BM cells, revealing no change or slightly up-regulated LSD1 expression upon LSD1 inhibitor treatment. (B) Expression of Gfi1b in ex vivo cultured BM cells, revealing statistically significant up-regulation of Gfi1b expression upon LSD1 inhibitor treatment. (C) Expression of CD90 in ex vivo cultured BM cells, revealing statistically significant up-regulation of CD90 expression upon LSD1 inhibitor treatment.
LSD1 Regulates HSC Gene Expression.
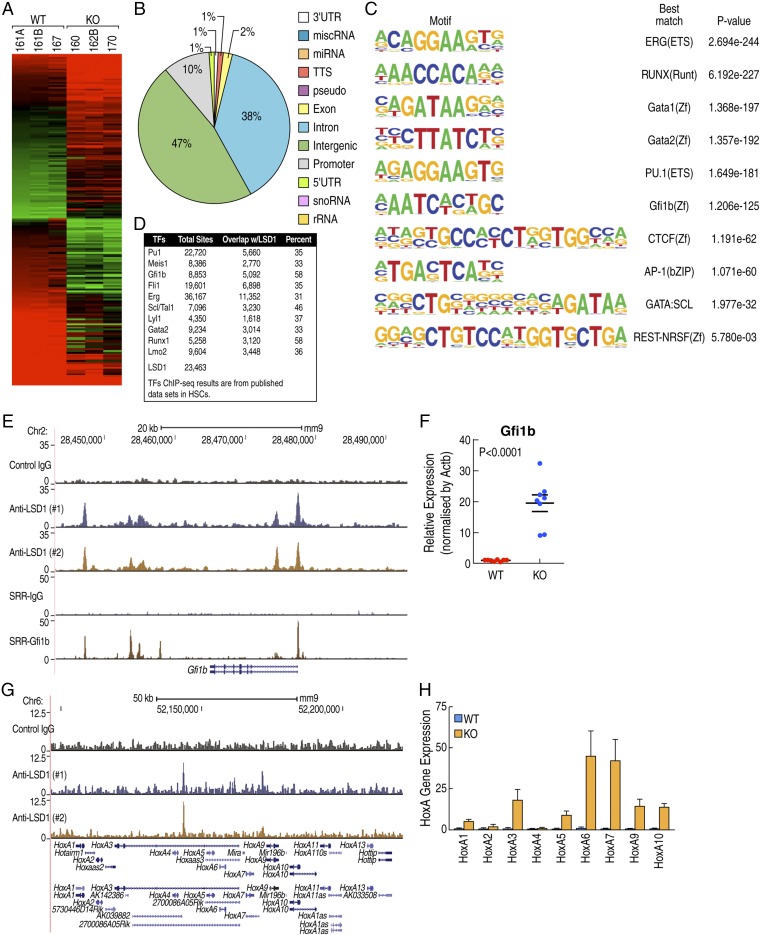
To investigate the transcriptional programs regulated by LSD1, we performed transcriptional profiling experiments comparing purified CD11b+ BM progenitor cells from LSD1fl/fl and LSD1−/− mice, identifying 518 genes up-regulated and 623 genes down-regulated in LSD1−/− mice (Fig. 6A). To determine the direct gene targets of LSD1, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) experiments. We identified 23,463 LSD1 binding sites in a murine HSC line (EML C1) using the validated anti-LSD1 antibody. Overlapping of LSD1-regulated genes with LSD1-binding sites revealed 464 of 518 (90%) genes up-regulated in LSD1−/− cells exhibited LSD1 binding sites within 250 kb, while 383 of 623 (61%) genes down-regulated in LSD1−/− cells exhibited LSD1 binding sites within 250 kb, consistent with direct regulatory roles of LSD1 at these loci. The majority of LSD1 binding sites are located in the intergenic or intronic region, in agreement with previous reports of distal enhancer binding of LSD1 (Fig. 6B). Motif analysis of LSD1 binding sites revealed enrichment of Ets, Runx, Gata, Gfi1b, CTCF, and AP-1 motifs (Fig. 6C). Intriguingly, we found that LSD1 binding sites are overlapping with binding sites of PU.1, Meis1, Gfi1b, or Runx1 on HPC7 cells (Fig. 6D), a different murine HSC line. Coenrichment of these motifs suggests that LSD1 is a coregulator of the corresponding transcription factors, important for HSC function. For example, we found that LSD1 was recruited to multiple sites on the Gfi1b locus, overlapping with Gfi1b binding sites (Fig. 6E). Gfi1b expression was significantly up-regulated in LSD1−/− BM (Fig. 6F), suggesting that LSD1 is a critical regulator of an Gfi1b autoregulatory loop. In addition, we observed increased level of H3K4 dimethylation and decreased level of H3K27 trimethylation on Gfi1b locus (Fig. S5C). These findings are consistent with a previous report that LSD1 is a critical regulator of Gfi1b by modulations of histone modifications, in addition, revealing a self-regulation loop by Gfi1b/LSD1 on Gf1b locus. Consistent with the observed leukemia-like phenotype, we found that LSD1 was recruited to the HoxA locus and that multiple HoxA genes were up-regulated in LSD1−/− BM cells (Fig. 6 G and H), suggesting LSD1 is a direct regulator of HoxA gene expression. These data suggest that LSD1 is an essential coregulator of multiple transcriptional factors required for the maintenance of HSC homeostasis and this normal regulatory strategy becomes critically interrupted during endotoxic shock.
Fig. 6.
LSD1 regulates HSC gene expression. (A) Gene-expression profiling analysis reveals LSD1-regulated genes. (B) LSD1 genome occupancy in EML C1 cells is enriched in distal regions. (C) Top 10 enriched motif on LSD1 binding sites in EML C1 cells. (D) LSD1 binding sites are overlapping with multiple transcriptional factor binding sites. (E) LSD1 and Gfi1b cooccupy the Gfi1b locus; genome browser shot of LSD1 and Gfi1b ChIP-seq experiments. (F) LSD1 represses Gfi1b gene expression in vivo; expression level of Gfi1b is determined by qPCR using purified CD11b-positive BM cells from LSD1−/− and control mice. (G) LSD1 occupies HoxA gene clusters; genome browser shot of LSD1 ChIP-seq experiments. (H) LSD1 represses HoxA gene expression in vivo; expression level of HoxAs is determined by qPCR using purified CD11b-positive BM cells from LSD1−/− and control mice.
Discussion
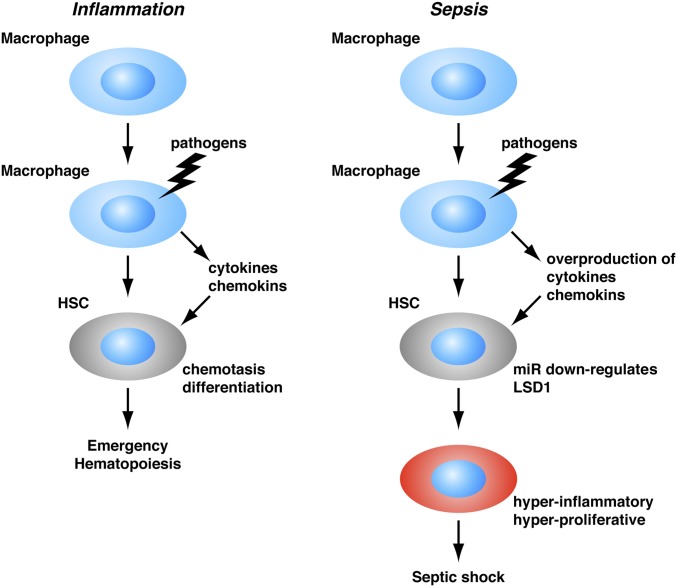
Septic shock is a devastating condition and currently the leading cause of death in hospitals. Septic shock is a result of a systemic response to infection, which is commonly attributed to dysfunction of mature hematopoietic cells such as neutrophils or macrophages. Traditional treatment for septic shock is focused to block endogenous mediators of the host inflammation response, such as cytokines. However, those treatments are usually not very effective. Therefore, novel targets or treatment strategies are actively explored. In this study, we found that a population of myeloid progenitors (CD11b+/Gr1low) is generated in response to acute infection, which has some markers of HSCs such as Sca1 and CD90, which has not been reported before. Those pathological populations of myeloid progenitors are hyperproliferative and hyperinflammatory, and may contribute to the cytokine storm and septic shock. Interestingly, inhibitors for Brd4, which have been reported to be an effective treatment for leukemia by promoting leukemia cell differentiation (29), can prevent death from septic shock in mouse models (30). Inhibitors of TOP1, commonly used for cancer therapy to block cancer cell proliferation, prove to be effective in mouse models of septic shock treatment as well (31), suggesting there are potential links between dysfunction of hematopoietic cell proliferation and differentiation and septic shock progression. Therefore, our findings of the roles of hematopoietic progenitors and/or HSCs in endotoxic shock development suggest the possibility that LSD1 activators, miRNA antagonists, or HSC-targeting strategies such as Brd4 or TOP1 inhibitors might be effective treatment strategies (Fig. 7).
Fig. 7.
Model: Sepsis-induced miRNAs suppress expression of LSD1 and induce acute expansion of hyperproliferative and hyperinflammatory HSC, leading to septic shock.
Our findings are consistent with previous studies demonstrating critical roles of LSD1 in regulation of hematopoiesis (14, 15). However, these prior studies did not observe the shock phenotype. This is most likely due to their use of alternative methods to conditionally knock down (Tet-inducible shRNA; ref. 14) or delete (Vav-Cre; ref. 15) LSD1 from BM progenitor cells that do not themselves induce an inflammatory response. Recently, infection-induced HSC activation has been reported as a mechanism of emergency hematopoiesis with biased myeloid differentiation. However, the underlying molecular mechanism is largely unknown. We found that a few inflammation mediators such as TNF-α and IL-1β, typically produced during acute bacterial or viral infections can suppress LSD1 expression through a subset of inflammation-induced miRNAs in HSCs and LSD1 deficiency is sufficient to induce a biased myeloid proliferation and differentiation program in mouse models. Therefore, suppression of LSD1 expression in HSC by miRNAs may underlie the biased myeloid differentiation and proliferation observed on emergency hematopoiesis upon acute infection. In addition, LSD1-deficient mice have defects in lymphocytes development. It is likely that suppression of the LSD1 expression in HSCs contributes to late stage immune paralysis phenotype observed in sepsis patients due to dysfunctions other than myeloid lineage, such as lymphocytes.
LSD1 inhibitors have been found to induce leukemia cells differentiation, suggesting a therapeutic strategy in cancer treatment. However, because we found that LSD1 deficiency in adult mice can lead to sudden death with acute expansion of a population of HSC-like cells, this raises a cautionary note, particularly as LSD1 inhibitors can interfere with hematopoiesis in ex vivo BM cultures in mice. Therefore, the safety of application of LSD1 inhibitors on cancer treatment should be rigorously reevaluated, especially under inflammation conditions. However, activators of LSD1, as well as inhibitors of miRNAs that regulate LSD1, might also represent potential therapeutic targets for sepsis and leukemia.
Materials and Methods
Animal.
LSD1fl/fl and Mx-Cre mice were described previously (11, 13). C57BL/6 wild-type mice were purchased from Charles River. For BM transfer experiments, wild-type animals were lethally irradiated (1,100 rad) and 1 × 106 total BM cells were injected intravenously. For HSC reconstitution, 1 × 106 infected cells were injected into recipient mice. Sepsis was induced as described in Fig. S3A, animals were observed every 6 h, and moribund animals were killed. All animal housing and experiments were approved by the Institutional Animal Care and Use Committee at the University of California, San Diego (UCSD).
Cell Culture.
EML C1 cell was kindly provided by Schickwann Tsai, University of Utah, Salt Lake City, and maintained by following the protocol (32). FDCPmix cell was kindly provided by Venkateshwar Reddy, Scripps Research Institute, La Jolla, CA, and maintained with IMDM (Life Technologies) supplemented with 20% horse serum (HyClone), 1% penicillin/streptomycin, and 10 ng/mL mouse IL-3 (Cell Signaling) (33).
BM ex Vivo Culture.
BM cells were isolated according to standard protocols. BM ex vivo culture was established according to standard protocols in the presence of cytokines. LSD1 inhibitors (C76 from Bioscience) or mock control were added to the culture medium every 2 d. Cells were split every 8 d.
Antibodies and Reagents.
Polyinosinic:polycytidylic acid (pIpC or poly I:C) was obtained from GE Healthcare Life Science and used as described in Fig. S1A. LPS 0111:B4 was purchased from Sigma and used as described in Fig. S3A. LPS was maintained with IMDM (Life Technologies).
For flow cytometry analysis, the following antibodies were purchased from eBioscience: anti-CD11b (M1/70), anti-GR-1 (RB6-8C5), anti-cKit (2B8), anti-Sca1 (D7), anti-CD90 (53-2.1), anti-TER-119 (TER-119), anti-CD45R (RA3-6B2). BrdU staining kit was obtained from BD Bioscience, and CFSE was purchased from Life Technologies and used by following manufacturer’s protocols. Stained cells were acquired by LSRII, and data were analyzed by FlowJo software.
Mouse IL-1β and TNF-α ELISA kit were purchased from eBioscience. HSC colony assay was performed with MethoCult kit purchased from STEMCELL Technologies.
Western blotting was performed as described before using anti-LSD1 (Cell Signaling for Western blotting; Abcam for ChIP); anti-Nurr1, anti-Nur77, and anti-Nor1 (Santa Cruz); and anti-actin (EMD Millipore). pGIPZ lentivirus vectors carrying shRNA and miR-Zip vectors for antisense miRNAs were purchased from Thermo Scientific and System Biosciences, respectively, and viral vector production was performed following the manufacturer’s protocol.
microRNA Isolation and qPCR.
miRNA isolation was performed using PureLink miRNA isolation kit and reverse transcription, and qPCR was performed using NCode Express SyberGreenER kit. qPCR data were normalized against U6. All kits were purchased from Life Technologies. RNA isolation and qPCR were described before (10).
ChIP-Seq.
ChIP assays were performed according to the previously described protocol (34). For LSD1 ChIP-seq experiments, EML C1 cells were cross-linked using 2 mM disuccinimidyl glutarate (ProteoChem c1104-100mg) for 45 min before cross-linking in the presence of 1% formaldehyde for 10 min. ChIP experiments were performed using the following antibody: LSD1 ab17721 (Abcam).
miR-Seq.
miR-seq was performed according to a protocol described previously (35). Briefly, total RNA was isolated with or without treatments. Small RNAs were isolated using PAGE purification. A 5′ adapter ligation/purification step was followed by a 3′ adapter ligation/purification step. The sequencing libraries were prepared by reverse transcription and PCR amplification. The libraries were then sequenced using Solexa plate form.
Statistics.
All data are presented as mean ± SD. P value was calculated using Prism 6 software, and P < 0.01 was considered significant.
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. No randomization and blinding were employed.
Data distribution was assumed to be normal, but this was not formally tested.
There was correction for multiple comparisons.
No animals or data points were excluded from analyses.
The P value, degree of freedom, and t value are calculated using online tools from www.graphpad.com/quickcalcs/ttest1.cfm.
Bioinformatics.
For the ChIP-seq experiments, the FASTQ files were aligned to the Mus musculus genome build mm9 using bowtie2 (36) with default parameters. Peaks were called on two replicates separately with matched input DNA controls using HOMER with parameters “-style factor -size 200 -tbp 1 -minDist 200 -P 0.1 -LP 0.1” that specified a relaxed P value threshold. These replicate peaksets were then processed with IDR 2.0.3 (37) with parameters “–rank 5 -i 0.05” parameters to yield a single set of high-confidence peaks. H3K4me2 peaks were considered overlapping LSD1 sites if their coordinates overlapped by a single base pair or more. The H3K4me2 tag densities around LSD1 binding sites were computed using HOMER’s “annotatePeaks.pl” command with parameters “-hist 10 -ghist -norm 0.”
Supplementary Material
Acknowledgments
We thank Dr. S. Tsai at University of Utah for EML C1 cells; Dr. V. Reddy at the Scripps Research Institute for FDCPmix cells; Dr. X. Zhou at Washington University for miRNA profiling analysis; Drs. J. G. Collier, M. Lam, D. Gosselin, X. Zhu, J. Zhang, K. Ohgi, and H. Taylor at UCSD for help with experiments; Dr. I. Garcia-Bassets for discussion, comments, suggestions, and critical reading of the manuscript; Ms. R. Pardee for proofreading of the manuscript; and Ms. J. Hightower for help with figure preparation. This research was supported by: NIH/National Cancer Institute (NCI) Grant CA173903 (to C.K.G.) and NIH/NCI Grant CA21371 (to M.G.R.); NIH/National Institute of Diabetes and Digestive and Kidney Diseases Grants DK018477 and DK039949 (to M.G.R.); and NIH/National Heart, Lung, and Blood Institute Grant HL065445 (to M.G.R.). M.G.R. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718759114/-/DCSupplemental.
References
- 1.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 2.D’Elia RV, Harrison K, Oyston PC, Lukaszewski RA, Clark GC. Targeting the “cytokine storm” for therapeutic benefit. Clin Vaccine Immunol. 2013;20:319–327. doi: 10.1128/CVI.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tisoncik JR, et al. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldridge MT, King KY, Goodell MA. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 2011;32:57–65. doi: 10.1016/j.it.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, et al. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 10.Saijo K, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 13.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 14.Sprüssel A, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26:2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 15.Kerenyi MA, et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TK, Gore SD, Zeidan AM. Epigenetic therapy in acute myeloid leukemia: Current and future directions. Semin Hematol. 2015;52:172–183. doi: 10.1053/j.seminhematol.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: New mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 19.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 23.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Harris WJ, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 25.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mould DP, McGonagle AE, Wiseman DH, Williams EL, Jordan AM. Reversible inhibitors of LSD1 as therapeutic agents in acute myeloid leukemia: Clinical significance and progress to date. Med Res Rev. 2015;35:586–618. doi: 10.1002/med.21334. [DOI] [PubMed] [Google Scholar]
- 27.Mohammad HP, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28:57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Zheng YC, et al. A systematic review of histone lysine-specific demethylase 1 and its inhibitors. Med Res Rev. 2015;35:1032–1071. doi: 10.1002/med.21350. [DOI] [PubMed] [Google Scholar]
- 29.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rialdi A, et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science. 2016;352:aad7993. doi: 10.1126/science.aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 33.Spooncer E, Heyworth CM, Dunn A, Dexter TM. Self-renewal and differentiation of interleukin-3-dependent multipotent stem cells are modulated by stromal cells and serum factors. Differentiation. 1986;31:111–118. doi: 10.1111/j.1432-0436.1986.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, et al. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci. 2015;18:1256–1264. doi: 10.1038/nn.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5:1752–1779. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.