Termination of replication occurs when two forks converge, an important but understudied process. In PNAS, a report from the Courcelle group examines replication termination using deep-sequencing genomic profiling of replicating cells to obtain copy number information about head-on collision of replication forks in different genetic backgrounds (1). Mutations in the SbcC-SbcD (SbcCD) and ExoI nucleases of Escherichia coli result in overreplication of DNA at the terminal replication zone where forks converge, implying that extra DNA is made upon termination and these nucleases are needed to excise the extra DNA. Furthermore, mutational studies of the RecBCD helicase/nuclease reveal that it acts at a step after SbcCD/Exo1 action to complete the processing of overreplicated DNA generated by fork convergence. Overreplication upon termination in E. coli has been reported earlier, but the DNA structures produced, and subsequent processing steps are not well understood (2–4). The report by Wendel et al. (1) demonstrates that termination of replication is a complex process orchestrated by many factors, and implies specific roles of the enzymes involved.
Termination of replication, when two replication forks meet head-on, has the potential for deleterious consequences. For example, amplifications, resections leading to deletions, and other DNA rearrangements are associated with defective replication termination (1–4). Extensive studies have outlined the events that activate origins and advance replication forks in bacteria and eukaryotes (5, 6), but little is known about the replication termination process, possibly because termination does not occur at a defined sequence, making it difficult to study.
The circular chromosome of E. coli has been an attractive model to study the termination process for two main reasons. First, E. coli has only one origin (oriC) that forms bidirectional forks that meet head-on roughly half way around the circular genome from the origin. Second, E. coli termination is restricted to a 400-kb region bordered by arrays of ter sites that let forks pass in one direction, but not the other direction, trapping forks that initiate at oriC within a 400-kb termination region between the ter site arrays (7, 8). In contrast, eukaryotic cells have numerous origins that fire at different times in different cells, confounding termination studies in eukaryotes (9).
Recent studies in E. coli indicate that replication termination requires numerous proteins (1–4). Given the myriad proteins involved in origin initiation and replication fork elongation, it may not be surprising that termination is also a multiprotein process. Indeed, accurate termination may be more important to life and death than accurate origin initiation, as the consequence of not firing an origin in eukaryotes is to simply wait for a fork from a nearby origin to duplicate the inactive origin. In contrast, termination gone awry could lead to duplications, inversions, deletions, and other genomic rearrangements. For example, mutations in E. coli SbcCD nuclease results in abnormal replication amplification in the termination region (1, 4). Furthermore, the mammalian orthologs to SbcCD, Rad50-Mre11 (10), are essential for normal development, viability, and genomic integrity (11).
Genetic studies of E. coli termination have thus far identified the involvement of RecBCD helicase/nuclease that unwinds and degrades double-strand (ds) DNA from an end, SbcCD nuclease that incises hairpins and degrades palindromic structures, ExoI, a 3′-5′ exonuclease, and the RecG branch migrating DNA translocase; DNA ligase and DNA polymerase are also assumed to be required (1–4). Mutations in the RecBCD helicase/nuclease result in various anomalies at termination. For example, RecD mutants of RecBCD, which retain helicase but not nuclease activity, overreplicate the terminal region, suggesting that fork convergence leads to overreplication that must be resected back to the doubling point (3). Conversely, mutations in RecBC, which lack both nuclease and helicase activity, result in loss of DNA in the terminal region, suggesting that RecBCD is needed to resolve and connect DNA strands and that without it the intervening DNA is exposed to nucleolytic removal (3, 4). SbcC and SbcD form a heterotetrameric nuclease that cuts at palindromes and hairpin structures (12), indicating that a hairpin or palindrome is produced during the termination process. SbcCD mutants are phenotypically dominant over RecBC mutants, indicating that SbcCD plays a role upstream of RecBCD (1). A similar phenotype is observed for ExoI mutants (1). The eukaryotic Rad50 and Mre11 orthologs of SbcCD are known to be involved in DNA resection processes (13). Mre11 and Rad50 are essential in humans, and hypomorphic mutations in these genes are associated with developmental abnormalities and predisposition to cancer (14, 15). Mutants in RecG, a branch migrating DNA translocase, also lead to overreplication in the terminal region and RecG is proposed to rearrange DNA structures to suppress aberrant processes that arise at convergent forks (4). These enzymes are involved in DNA repair, and their requirement for proper termination implies that some of the observed phenotypes in cells containing mutations in genes encoding these proteins might be explained in terms of replication termination.
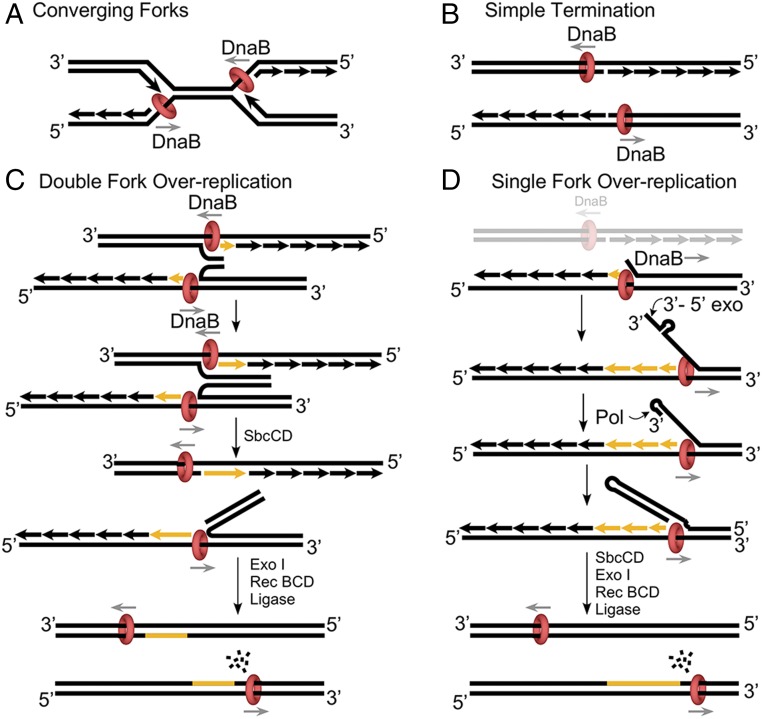
Replication fork convergence in E. coli is illustrated in Fig. 1A. DnaB helicase encircles the lagging strand single-strand (ss) DNA and, upon colliding with the opposite fork, the DnaB ring faces a leading strand 3′ ss/ds junction. In vitro, DnaB, like other replicative helicase rings, requires a forked structure to unwind DNA, and upon encountering a flush 3′ ss/ds structure, DnaB typically slides onto the dsDNA rather than unwinding (16). Hence, a simplistic view of termination would have the two helicases pass one another without effect on the completed chromosomes (Fig. 1B). However, DnaB can sometimes unwind at a flush junction (17), and overreplication has been observed in in vitro studies of plasmid replication (18). Hence, DnaB is proposed to invade the replicated leading strand of a converging fork at some frequency.
Fig. 1.
Possible consequences of fork convergence at termination of E. coli replication. (A) Two replication forks with DnaB helicase encircling the lagging strand collide. (B) The two DnaB helicases may slipover the flush 3′ ss/ds ends of the leading stands of the opposite fork. (C) The two DnaB helicases may invade both leading strand ss/ds ends to form complementary 3′ ssDNAs that pair. SbcCD could then cleave the palindrome-like sequence, followed by RecBC/ExoI processing. (D) One DnaB invades a leading strand of the opposite fork. The displaced 3′ ssDNA is susceptible to a 3′-5′exonuclese, forming a primed junction for polymerase extension back to DnaB. The resulting hairpin/palindrome is processed by SbcCD and RecBCD as in C.
The model proposed by Wendel et al. (1) is illustrated in Fig. 1C, in which overreplication initiates when both DnaB helicases of converging forks invade the head-on leading strand of the opposite fork. The two displaced 3′ single-strands are complementary and in close proximity, and are proposed to pair and form a growing duplex of overreplicated DNA that connects the two daughter chromosomes. The process will necessarily stop upon running into the particular array of ter sites that normally allow passage of forks coming from the origin, but will block advance of forks emanating from the termination region. SbcCD nuclease is proposed to incise the palindrome-like structure of the overreplicated DNA, and then ExoI and RecBCD can trim the overreplicated DNA to form a precise junction for ligation and two exact copies of the parental chromosome.
A slight variation on this model may occur when only one DnaB of two colliding forks invades the duplex, producing just one 3′ single strand (Fig. 1D). In this model, excision of the 3′ terminus by ExoI, or another nuclease, may digest the ssDNA until reaching a frequent indirect repeat, the size of a restriction enzyme site, yielding a hairpin 3′ ss/ds junction that primes polymerase extension (e.g., as described in ref. 19). The replication fork would be stopped within the terminal region by ter sites, as described above for Fig. 1C. Incision of the hairpin/palindrome of the overreplicated DNA segment by SbcCD, and processing by RecBCD, would enable accurate ligation and complete the termination event. In light of these models, one may speculate that ter sites function to stop forks initiated by termination processes in one direction, and stop forks initiated at origins in the other direction.
RecA mutants show normal replication termination (3), and thus RecA-mediated recombination events, such as double-strand break repair or D-loop–based replication restart are not involved in termination. However, mutations in RecBCD, and SbcCD/ExoI that result in loss of cell viability, are largely rescued by RecA (1). Hence, in these mutant backgrounds, RecA restores cell viability, but the terminal region of the chromosome in these cells contains abnormalities (1).
One may question the percentage of cells that resolve converging forks in a simple fashion (Fig. 1B), relative to converging forks that overreplicate DNA (e.g., as in Fig. 1 C and D). The fact that RecA is not involved in termination, and that mutants in either RecBC, RecD, or SbcCD/ExoI clearly loose viability (in the absence of RecA), together with the finding of deletions and amplifications of DNA in the terminal region of these mutants, imply that most termination events are not simple. These observations suggest that termination is often complex and that simple fork passage/ligation may be the exception, not the rule.
It is interesting to contemplate the termination process in eukaryotes, especially given numerous origins, and thus the high frequency with which termination must occur for each cell cycle. Eukaryotes differ from bacteria in their core replication proteins, which show no common ancestor in evolution (20). In particular, eukaryotes contain distinct DNA polymerases (i.e., polymerases delta and epsilon) for each daughter strand, and the CMG (Cdc45, Mcm2-7, GINS) helicase encircles the leading strand, not the lagging strand (5, 6, 21). Perhaps the problems of fork convergence in eukaryotes might be somehow mitigated by these large evolutionary changes, and could even have been a driving force for evolving such a different strategy of chromosome replication relative to bacteria. Deeper studies of replication termination in both bacteria and eukaryotes will certainly be an important and fascinating area for the future.
Acknowledgments
The authors are grateful for support from the NIH (Grant GM115809) and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
See companion article on page 349.
References
- 1.Wendel BM, Cole JM, Courcelle CT, Courcelle J. SbcC-SbcD and Exol process convergent forks to complete chromosome replication. Proc Natl Acad Sci USA. 2017;115:349–354. doi: 10.1073/pnas.1715960114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courcelle J, Wendel BM, Livingstone DD, Courcelle CT. RecBCD is required to complete chromosomal replication: Implications for double-strand break frequencies and repair mechanisms. DNA Repair (Amst) 2015;32:86–95. doi: 10.1016/j.dnarep.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wendel BM, Courcelle CT, Courcelle J. Completion of DNA replication in Escherichia coli. Proc Natl Acad Sci USA. 2014;111:16454–16459. doi: 10.1073/pnas.1415025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG. Avoiding chromosome pathology when replication forks collide. Nature. 2013;500:608–611. doi: 10.1038/nature12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SP, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill TM. Arrest of bacterial DNA replication. Annu Rev Microbiol. 1992;46:603–633. doi: 10.1146/annurev.mi.46.100192.003131. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T, Hidaka M, Horiuchi T. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J. 1989;8:2435–2441. doi: 10.1002/j.1460-2075.1989.tb08374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission yeast. Cell. 2009;136:852–864. doi: 10.1016/j.cell.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 11.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51:195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connelly JC, de Leau ES, Leach DR. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 1999;27:1039–1046. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng SK, Yin Y, Petes TD, Symington LS. Mre11-Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol Cell. 2015;60:500–508. doi: 10.1016/j.molcel.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo G, et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan DL, O’Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 17.Shin JH, Kelman Z. DNA unwinding assay using streptavidin-bound oligonucleotides. BMC Mol Biol. 2006;7:43. doi: 10.1186/1471-2199-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiasa H, Marians KJ. Tus prevents overreplication of oriC plasmid DNA. J Biol Chem. 1994;269:26959–26968. [PubMed] [Google Scholar]
- 19.Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao NY, O’Donnell ME. Evolution of replication machines. Crit Rev Biochem Mol Biol. 2016;51:135–149. doi: 10.3109/10409238.2015.1125845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel TA, Burgers PMJ. Arranging eukaryotic nuclear DNA polymerases for replication: Specific interactions with accessory proteins arrange Pols α, δ, and ε in the replisome for leading-strand and lagging-strand DNA replication. BioEssays. 2017;39:8. doi: 10.1002/bies.201700070. [DOI] [PMC free article] [PubMed] [Google Scholar]