Abstract
The present study aimed to assess the B rapidly accelerated fibrosarcoma (BRAFV600E) status in plasma from Chinese patients with melanoma, and evaluated its prognostic value following treatment with BRAF inhibitors. Mutation-specific 3D digital polymerase chain reaction (dPCR) was used to quantify BRAFV600E in circulating tumor DNA (ctDNA) in 58 patients with melanoma, prior to treatment with BRAF inhibitors. Correlations between baseline ctDNA levels and clinicopathological characteristics and clinical benefits were then statistically analyzed. The concordance and sensitivity of BRAFV600E between ctDNA and tumor tissue were 70.2% and 76%, respectively, in 58 patients with melanoma. BRAFV600E mutation in ctDNA correlated with lactate dehydrogenase concentration (P=0.04) and Eastern Cooperative Oncology Group score (P=0.04). There was no correlation between BRAFV600E of ctDNA with response, progression-free survival (PFS), or overall survival (OS) following targeted therapy. The objective response rate, PFS and OS stratified by BRAFV600E of ctDNA were 30.0% vs. 56.7%, (P=0.3), 8.1 months vs. 6.7 months, (P=0.38) and 65.6 months vs. 42.3 months (P=0.52), respectively, for undetectable and mutant types. In conclusion, 3D dPCR is appropriate for ctDNA detection and BRAFV600E in ctDNA is a non-invasive biomarker in patients with melanoma.
Keywords: melanoma, BRAFV600E in ctDNA, 3D digital PCR
Introduction
Malignant melanoma is the most aggressive skin cancer and has poor prognosis. Its incidence and mortality have increased rapidly worldwide and in Asia. In China, the estimated new cancer cases and deaths of melanoma in 2015 were 8,000 and 3,200 respectively (1). The overall survival (OS) rate at 5 years for patients with stage IV disease is <5% (2).
Somatic genetic aberrations have provided a framework for developing targeted therapy in advanced cancer. One of the most validated treatments in this area is B rapidly accelerated fibrosarcoma (BRAF) inhibition. BRAF mutations are present in 50% (3) of cases of Caucasian melanoma. The most common mutation is B rapidly accelerated fibrosarcoma (BRAFV600E) (4–6). BRAF inhibitors, such as vemurafenib and dabrafenib, lead to tumor regression in 60–80% of patients with melanoma (7). However, in Chinese patients with melanoma, the incidence of somatic mutations within the BRAF genes is 25.5% (8). Melanoma has diverse clinicopathological characteristics, especially in BRAF mutation frequencies in different ethnic groups and pathologic subtypes.
Tumor surgical or biopsy tissue is the standard material used to determine the presence of somatic mutations before the start of targeted therapy. However, mutation status is unstable and often changes (9). It is difficult to obtain tumor tissue from rebiopsies owing to discomfort and high risk and cost for patients. A liquid biopsy from blood samples, included the circulating tumor DNA (ctDNA) obtained from cell-free DNA (cfDNA), cellular tumor cells (CTCs) and others, overcome the invasive nature and heterogeneity (10). The concordance between ctDNA for BRAFV600E and BRAFV600E status in tissue has been shown to be ~70%. CtDNA for BRAFV600E has a sensitivity of 38–79% and a specificity of 40–100% (9,11–13). BRAFV600E mutated DNA was detected in CTCs of 77% of melanoma patients with recorded mutated tumor tissues (14). In colorectal cancer, a report before showed that among 23 matched CTCs and ctDNA samples, the concordance was 73.9% for BRAF mutations (15). But the disadvantage of CTCs is that the repetitive rate and the success rate of PCR in CTCs were really low in the detection of BRAF mutations (14,16). So the higher concentrations of ctDNA, which is correlated with tumor burden, can be a great alternative to rebiopsy. CtDNA can provide the genetic landscape of all cancerous lesions (primary and metastases) as well as offering the opportunity to systematically track genomic evolution which is used for evaluating response after treatment and monitoring disease recurrence. However, there is no reports associated with BRAFV600E in ctDNA and Chinese melanoma patients.
3D digital PCR (dPCR) is a third-generation dPCR technique that minimizes the use of DNA template. It can be used for true real-time quantitative detection and can detect mutation frequencies as low as 0.005–0.01% (17). In the present study we use 3D dPCR to detect ctDNA with BRAFV600E in 58 Chinese patients with melanoma and also determine whether the levels of ctDNA with BRAFV600E at baseline before BRAF inhibitor therapy correlate with treatment response and survival.
Materials and methods
Patients and sample collection
Paired tissues and blood samples from 58 patients with melanoma who were hospitalized between 2012 and 2015 in the Renal Cancer and Melanoma Department of Beijing Cancer Hospital were used in our study. Written consent was obtained from the patients or patients' parent/carer. All tissue samples were confirmed to be positive for melanoma using hematoxylin and eosin (H&E) staining. Of all the patients, the mean age is 43 years, ranged from 17 to 74 years. We obtained the following information from the patients' clinical histories, as recorded by their oncologists: disease stage, tissue mutation, Eastern Cooperative Oncology Group (ECOG) performance status, and lactate dehydrogenase (LDH) values. Blood samples were collected before BRAF inhibitor treatment and tumor response was evaluated in accordance with the Response Evaluation Criteria in Solid Tumors (RECIST version 1.1). The latest follow-up date was October 1, 2016. The present study was approved by the Medical Ethics Committee of the Beijing Cancer Hospital and Institute and was conducted according to the Declaration of Helsinki Principles.
Sample collection and cfDNA extraction
Blood samples from patients with melanoma were collected in ethylene diamine tetraacetic acid (EDTA) vacutainer tubes and stored at 4°C within 24 h before isolation. After centrifugation at 2,500 rpm for 10 min and a further centrifugation at 1,200 rpm for 15 min, plasma samples were collected. CfDNA was extracted from 2 ml plasma stored at −80°C using the QIAamp Circulating Nucleic Acid kit (Qiagen China Co., Ltd.) according to the manufacturer's protocols. CfDNA was eluted in 30 µl of Buffer AVE (Qiagen China Co., Ltd.).
CfDNA detection using 3D digital polymerase chain reaction (dPCR)
The TaqMan® BRAF SNP Genotyping assay (cat. no. 4465804; Life; Thermo Fisher Scientific, Inc.) was used for dPCR detection. Each 15 µl dPCR reaction contained 0.375 µl BRAF assay, 7.5 µl QuantStudio™ 3D Digital PCR Master Mix 2X (Life; Thermo Fisher Scientific, Inc.), 6.125 µl nuclease-free water (Life, Thermo Fisher Scientific, Inc.), and 1 µl cfDNA. The dPCR reactions were then loaded onto a QuantStudio™ 3D Digital PCR Chip v2 (Life; Thermo Fisher Scientific, Inc.). The chips were preformed using QuantStudio™ 3D Digital PCR System (Life; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. The dPCR cycling was performed as follows: Initial denaturation at 96°C for 10 min, followed by 39 cycles of annealing at 60°C for 2 min, denaturation at 98°C for 30 sec, and elongation at 60°C for 2 min. The levels of cfDNA were determined using QuantStudio™ 3D AnalysisSuite™ Software (Life; Thermo Fisher Scientific, Inc.) by analyzing the numbers and proportions of positive droplets.
Statistical analyses
Statistical analysis was performed using SPSS 20.0 for Windows (IBM Corp, Armonk, NY, USA). Qualitative data are described as percentages. Quantitative data are described as means ± standard deviations. Pearson's chi-square tests, Fisher's exact tests, and Mann-Whitney U tests were used to assess correlations between the qualitative variables. Survival time was assessed using disease-free survival (DFS), progression-free survival (PFS), and OS. DFS and PFS were calculated separately as the duration from the time of initial surgery to the diagnosis of a recurrence and the time of targeted therapy to the diagnosis of progression. OS time was assessed for prognostic analysis. We used Kaplan-Meier and log-rank tests to analyze survival differences between groups. Univariate Cox regression analyses were performed to examine correlations between ctDNA levels and clinicopathologic measures and PFS. These tests were bilateral and the significance level was defined as 0.05, so that P<0.05 was considered to indicate a statistically significant difference.
Results
Concordance between BRAFV600E in ctDNA (3D dPCR) and in tumor tissue (PCR)
We used the maximum concentration of BRAFV600E in ctDNA from 4 healthy individuals (0.097–0.133 copies/ml) to establish our criteria and divided the 58 patients with melanoma into two groups: undetectable type (UT), and mutant type (MT). 43 (74.1%) of 58 plasma samples had the BRAFV600E mutation, while 13 (25.9%) did not. A total of 50 tissue samples had the BRAFV600E mutation, as assessed using PCR. The concordance and sensitivity of BRAFV600E between ctDNA and tumor tissue were 70.2% and 76%, respectively. The results are presented in Table I. Fluorescent amplitude plots of 3D dPCR in ctDNA samples and Sanger Sequencing in tumor tissues from melanoma patients are presented in Fig. 1. The plot of 3D dPCR contains the negative droplets, positive droplets for wild-type, mutant type and both. And the plot of Sanger Sequencing shows the location and variation of nucleotides in BRAF gene.
Table I.
BRAFV600E in ctDNA and tumor tissue.
| Tumor tissue | ||||
|---|---|---|---|---|
| BRAFV600E mutation | WT | MT | NA | Total |
| ctDNA | ||||
| UT | 2 | 12 | 0 | 14 |
| MT | 5 | 38 | 1 | 44 |
| Total | 7 | 50 | 1 | 58 |
Concordance and sensitivity of BRAF mutation status between ctDNA and tumor tissue were 40/57=70.2% and 38/50=76%, respectively. WT, wild-type; UT, undetectable type; MT, mutant type; NA, not available.; ctDNA, circulating tumor DNA; BRAFV600E, B rapidly accelerated fibrosarcoma.
Figure 1.
A negative sample from a melanoma patient of ctDNA in 3D dPCR (A) and the other positive for this mutation (B). A negative sample in tumor tissue by Sanger Sequencing (C) and the other positive for this mutation (D). In plot of 3D dPCR, yellow cluster corresponds to negative droplets, red cluster corresponds to droplets positive for wild-type, blue cluster corresponds to droplets positive for mutant BRAFV600E, and green cluster corresponds to droplets both positive for wild-type and mutant BRAFV600E. In plot of Sanger Sequencing, the nucleotide refers to 1799T >A and the amino acid refers to V600E. dPCR, digital PCR; BRAFV600E, B rapidly accelerated fibrosarcoma.
Clinical correlations between BRAFV600E in ctDNA and clinical characteristics of patients
The presence of the BRAFV600E mutation in ctDNA was correlated with LDH concentration (P=0.04) and ECOG performance status (P=0.04). The levels of BRAFV600E mutation in ctDNA levels were markedly higher in patients with high LDH levels compared to those with normal LDH levels [UT: ctDNA median, 0.08 copies/ml (0–0.094 copies/ml); LDH median, 197 IU/l (125–269 IU/l); MT: ctDNA median, 0.37 copies/ml (0.149–456.9 copies/ml); LDH median, 206 IU/l (129–1371 IU/l)]. Worse ECOG performance status was associated with higher BRAFV600E mutation levels in plasma. There were no significant associations between BRAFV600E mutation in ctDNA and other clinical characteristics, such as sex, age, subtype, stage, thickness, or ulcer. The details of our findings are presented in Table II.
Table II.
Correlation between BRAFV600E in ctDNA and characteristics of patients.
| BRAFV600E mutation of ctDNA | |||
|---|---|---|---|
| Clinical charateristics | UT (n=15,%) | MT (n=43,%) | P-value |
| Sex | |||
| Male | 5 (26.3) | 14 (73.7) | 1.00 |
| Female | 10 (25.6) | 29 (74.4) | |
| Age, years | |||
| >60 | 1 (14.3) | 6 (85.7) | 0.46 |
| ≤60 | 14 (27.5) | 37 (72.5) | |
| Subtype | |||
| Acral + Mucosal | 3 (20) | 12 (80) | 0.68 |
| +Uveal | |||
| CSD + non-CSD | 10 (30.3) | 23 (69.7) | |
| Unknown | 2 (25) | 8 (75) | |
| Stage | |||
| I–III | 8 (29.6) | 19 (70.4) | 0.54 |
| IV | 7 (22.6) | 24 (77.4) | |
| Thickness, mm | |||
| <4 | 3 (37.5) | 5 (62.5) | 0.76 |
| ≥4 | 3 (27.3) | 8 (72.7) | |
| Ulcer | |||
| With | 5 (29.4) | 12 (70.6) | 1.00 |
| Without | 6 (26.1) | 17 (73.9) | |
| LDH | |||
| <240 IU/L | 12 (36.4) | 21 (63.6) | 0.04 |
| ≥240 IU/L | 1 (6.7) | 14 (93.3) | |
| ECOG | |||
| 0 | 12 (42.9) | 16 (57.1) | 0.04 |
| 1 | 3 (12) | 23 (88) | |
| 2 | 0 (0) | 4 (100) | |
UT, undetectable type; MT, mutant type; CSD, chronic sun-induced damage; LDH, lactate dehydrogenase; ECOG, Eastern Cooperative Oncology Group; BRAFV600E, B rapidly accelerated fibrosarcoma; ctDNA, circulating tumor DNA.
Clinical correlations between BRAFV600E in ctDNA and clinical benefits after targeted therapy
A total of 30 out of 58 patients with melanoma were treated with BRAF inhibitors. This accounted for 20% (6/30), 66.7% (20/30), and 13.3% (4/30) of patients with first-, second-, and third-line targeted therapy, respectively. The median follow-up time was 34.9 months (range, 30.3–39.6 months). There was no correlation between BRAFV600E in ctDNA and response, PFS, or OS after targeted therapy with BRAF inhibitors. The subset of patients undergoing targeted therapy included 13 patients with partial responses (PR), 13 with stable disease (SD), and 1 progressive disease (PD), as assessed using RECIST criteria. We also had 3 unassessable patients (Table III). Objective response rate (ORR), PFS, and OS stratified by BRAFV600E in ctDNA were 30.0% vs. 56.7% (P=0.3) (Table III), 8.1 months vs. 6.7 months (P=0.38) (Fig. 2), and 65.6 months vs. 42.3 months (P=0.52), respectively for the UT and MT groups. Quantitative and qualitative data of response are presented in Fig. 3 and Table III, respectively.
Table III.
Associations between baseline ctDNA concentrations and response to targeted treatment.
| BRAFV600E mutation | PR | SD | PD | Total | P-value |
|---|---|---|---|---|---|
| UT | 3 | 6 | 0 | 9 | 0.30 |
| MT | 10 | 7 | 1 | 18 | |
| Total | 13 | 13 | 1 | 27 |
Qualitative data were statistically analyzed using Mann-Whitney U tests. UT, undetectable type; MT, mutant type; PR, partial responses; SD, stable disease; PD, progressive disease.
Figure 2.
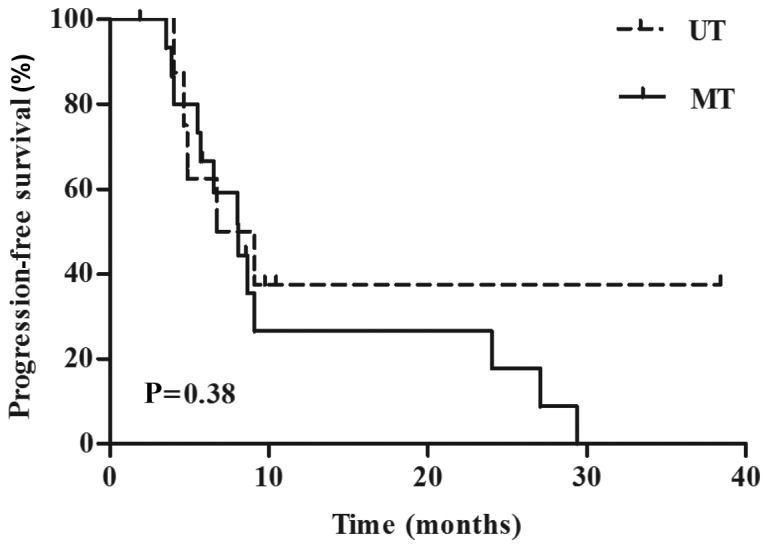
Kaplan-Meier plots of PFS probabilities according to baseline ctDNA BRAFV600E status in cases treated with targeted therapy (n=26). PFS stratified by BRAFV600E in ctDNA were 8.1 months vs. 6.7 months (P=0.38) for the undetectable type and mutant type. The P-value are indicated below the graph. PFS, progression free survival.
Figure 3.
Associations between baseline ctDNA concentrations and response to targeted treatment. Quantitative data were statistically analyzed using Mann-Whitney U tests. The median concentrations of baseline ctDNA for PR, SD and PD groups refered to 0.37, 0.12, 0.24 copies/ml respectively. The P-value are indicated above the graph. PR, partial responses; SD, stable disease; PD, progressive disease.
Discussion
In the present study we used 3D dPCR to detect ctDNA with BRAFV600E in 58 Chinese patients with melanoma and statistically analyzed the correlations between ctDNA level and clinical characteristics, treatment response, and survival. The aim of our study is to know whether ctDNA is a great alternative to biopsy and whether 3D dPCR is a suitable detection method for ctDNA. Furthermore, we want to explore if BRAFV600E ctDNA is a candidated biomarker in patients with melanoma or even a predictive marker after treatment of BRAF inhibitors.
In prior reports, the concordance between BRAFV600E in ctDNA and BRAFV600E in tissue samples was ~70% and the sensitivity was 38–79% (9,11–13). The concordance and sensitivity between ctDNA and tumor tissue in our study were 70.2 and 76%, which were higher than those of prior reports. The main reason for this observation is that our study uniquely used 3D dPCR, which is highly sensitive, quantitative, and real-time. Unlike other detection methods, such as Beads Emulsion Amplification Magnetics (BEAMing), Amplification Refractory Mutation System (ARMS), and Next Generation Sequencing (NGS) (9,11–13), each reaction of 3D dPCR has 20,000 wells and each well is isolated from its neighbors. This ensures the high sensitivity and accuracy of 3D dPCR. In addition, 3D dPCR is simple to perform. In fact we were able to obtain our results within 2 h. The use of 3D dPCR prevents mistakes due to complex operation and calculation errors. Although BRAFV600E in ctDNA was identified by 3D dPCR, which is the most sensitive among all of the detection tools use thus far, 29.8% of the patients had plasmic BRAF statuses that were distinct from those of their tumor tissues. This may have been owing to the mechanisms underlying the transfer of ctDNA into the blood, which are as yet not completely clear. It is also known that the levels of ctDNA vary at different times of the day. In addition, the heterogeneity of the tumors may have led to differences between the ctDNA and the tissue. CtDNA derived from apoptotic or necrotic cell debris of primary tumors, metastatic tumors, or CTCs may better reflect the entire picture of the tumor when compared to using one site to obtain tumor tissue (18). Finally, genetic alterations may appear after different treatments (13). This is important, as ~80% of the patients with melanoma in this study had not been treated with the first-line therapy.
Our results are in agreement with those of previous studies that have shown that BRAFV600E mutant ctDNA correlates with tumor burden (19). Patients with lower or undetectable amounts of BRAFV600E mutant ctDNA tend to be those with less disease burden as measured by LDH, RECIST sum of diameters, and ECOG performance (12). LDH is the only blood-based biomarker incorporated into the staging system, as elevated levels of LDH are associated with significantly decreased survival (19). Both BRAFV600E in ctDNA and LDH in the blood are good prognostic markers and are thought to be useful in the follow-up of patients with melanoma (20). LDH is neither sensitive nor specific, and other studies have shown that ctDNA is more consistent and informative than LDH (21,22). In addition, ctDNA is significantly more accurate for tracking disease status than traditional serum markers, such as carcinoembryonic antigen (CEA) and cancer antigen 15-3 (CA15-3), which are used to diagnose colorectal and breast cancer, respectively (23,24). In the future, we will monitor the dynamic status of ctDNA and compare the results thus obtained with those of LDH in a large scale study. Here, 18 patients that had surgery before in our study collected their blood samples during the period between the metastatic lesions determined and the first-line therapy. Of note, DFS stratified by BRAFV600E in ctDNA was 26.4 months vs. 9.1 months (P=0.013) for UT and MT, respectively. This indicates that patients with longer DFS before recurrences tended to have lower levels of BRAFV600E mutant ctDNA. Some experts in the breast cancer field have also found that level of ctDNA are reduced or even disappeared after surgery, although some patients have remaining ctDNA (25). This highlights a new direction for ctDNA research. Specifically, it will be interesting to determine whether ctDNA detected after surgery identifies patients at risk for recurrence. This may then guide adjuvant therapy decisions for individual patients.
Another objective of our study was to determine whether BRAFV600E in ctDNA can be used as criteria to select patients for BRAF-targeted therapy. Some previous reports have shown that low baseline ctDNA is a good predictor of response to treatment, longer PFS, and even OS in targeted therapy (12,13,20). Although there was no statistical significance between BRAFV600E in ctDNA and clinical benefit, patients with wild-type BRAF ctDNA have longer PFS and OS than mutant type which supports the findings of previous reports. Three factors may explain the reason of no statistical significance. First, the major factors influenced the effect of BRAF inhibitor is unknown. A decrease of BRAFV600E in ctDNA indicated response to therapy (13,26). Also the increased concentration of mutant copies observed following disease progression (26) and the state of a secondary resistance to the treatment may associated with effect of therapy (13). The effect of BRAF inhibitors would be affected by many factors such as a secondary resistance, so baseline ctDNA of BRAFV600E maybe not the best predictive marker to target therapy, but the variation of this mutation or other gene associated with resistance. Second, it will take time for the PFS and OS to mature. Since only 75% of the patients have reached their endpoints, mortality only accounts for 43.1% of the outcomes. Finally, our study was carried out on a small scale. In fact, there was only one patient who did not have a response after targeted therapy. Therefore, it remains to determine the realistic relationship between clinical benefit and BRAFV600E in ctDNA.
Due to the limitation of our study, we hope a large scale research in the future which includes multiple plasma samples during the treatment and progression. CtDNA from different periods can reflect different conditions of disease and predict clinical benefits of treatment. In addition, experts can make comparison between ctDNA and traditional biomarkers.
In conclusion, our data confirm that 3D dPCR is suitable for ctDNA detection and that BRAFV600E ctDNA is a non-invasive prognostic marker in patients with melanoma.
Acknowledgements
The present study was supported by grants from National Natural Science Foundation of China (grant no. 81672696) and Beijing Municipal Natural Science Foundation (grant no. 7152033).
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Chi Z, Li S, Sheng X, Si L, Cui C, Han M, Guo J. Clinical presentation, histology, and prognoses of malignant melanoma in ethnic Chinese: A study of 522 consecutive cases. BMC cancer. 2011;11:85. doi: 10.1186/1471-2407-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network, corp-author. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Jr, Kaempgen E, et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 6.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coit DG, Thompson JA, Algazi A, Andtbacka R, Bichakjian CK, Carson WE, III, Daniels GA, DiMaio D, Fields RC, Fleming MD, et al. NCCN guidelines insights: Melanoma, version 3.2016. J Natl Compr Canc Netw. 2016;14:945–958. doi: 10.6004/jnccn.2016.0051. [DOI] [PubMed] [Google Scholar]
- 8.Si L, Kong Y, Xu X, Flaherty KT, Sheng X, Cui C, Chi Z, Li S, Mao L, Guo J. Prevalence of BRAF V600E mutation in Chinese melanoma patients: Large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur J Cancer. 2012;48:94–100. doi: 10.1016/j.ejca.2011.06.056. [DOI] [PubMed] [Google Scholar]
- 9.Molina-Vila MA, de-Las-Casas CM, Bertran-Alamillo J, Jordana-Ariza N, González-Cao M, Rosell R. cfDNA analysis from blood in melanoma. Ann Transl Med. 2015;3:309. doi: 10.3978/j.issn.2305-5839.2015.11.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, De Mattos-Arruda L, Muñoz-Couselo E, Manzano JL, Cortes J, Berros JP, Drozdowskyj A, Sanmamed M, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res. 2015;25:486–495. doi: 10.1097/CMR.0000000000000187. [DOI] [PubMed] [Google Scholar]
- 12.Santiago-Walker A, Gagnon R, Mazumdar J, Casey M, Long GV, Schadendorf D, Flaherty K, Kefford R, Hauschild A, Hwu P, et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res. 2016;22:567–574. doi: 10.1158/1078-0432.CCR-15-0321. [DOI] [PubMed] [Google Scholar]
- 13.Gray ES, Rizos H, Reid AL, Boyd SC, Pereira MR, Lo J, Tembe V, Freeman J, Lee JH, Scolyer RA, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6:42008–42018. doi: 10.18632/oncotarget.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid AL, Freeman JB, Millward M, Ziman M, Gray ES. Detection of BRAF-V600E and V600K in melanoma circulating tumour cells by droplet digital PCR. Clin Biochem. 2015;48:999–1002. doi: 10.1016/j.clinbiochem.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Sakaizawa K, Goto Y, Kiniwa Y, Uchiyama A, Harada K, Shimada S, Saida T, Ferrone S, Takata M, Uhara H, Okuyama R. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br J Cancer. 2012;106:939–946. doi: 10.1038/bjc.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidess-Sigal E, Liu HE, Triboulet MM, Che J, Ramani VC, Visser BC, Poultsides GA, Longacre TA, Marziali A, Vysotskaia V, et al. Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: Comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget. 2016;7:85349–85364. doi: 10.18632/oncotarget.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jennings LJ, George D, Czech J, Yu M, Joseph L. Detection and quantification of BCR-ABL1 fusion transcripts by droplet digital PCR. J Mol Diagn. 2014;16:174–179. doi: 10.1016/j.jmoldx.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knol AC, Vallée A, Herbreteau G, Nguyen JM, Varey E, Gaultier A, Théoleyre S, Saint-Jean M, Peuvrel L, Brocard A, et al. Clinical significance of BRAF mutation status in circulating tumor DNA of metastatic melanoma patients at baseline. Exp Dermatol. 2016;25:783–788. doi: 10.1111/exd.13065. [DOI] [PubMed] [Google Scholar]
- 21.Tsao SC, Weiss J, Hudson C, Christophi C, Cebon J, Behren A, Dobrovic A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci Rep. 2015;5:11198. doi: 10.1038/srep11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang GA, Tadepalli JS, Shao Y, Zhang Y, Weiss S, Robinson E, Spittle C, Furtado M, Shelton DN, Karlin-Neumann G, et al. Sensitivity of plasma BRAFmutant and NRASmutant cell-free DNA assays to detect metastatic melanoma in patients with low RECIST scores and non-RECIST disease progression. Mol Oncol. 2016;10:157–165. doi: 10.1016/j.molonc.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 25.Beaver JA, Jelovac D, Balukrishna S, Cochran R, Croessmann S, Zabransky DJ, Wong HY, Toro PV, Cidado J, Blair BG, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20:2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanmamed MF, Fernández-Landázuri S, Rodríguez C, Zárate R, Lozano MD, Zubiri L, Perez-Gracia JL, Martín-Algarra S, González A. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem. 2015;61:297–304. doi: 10.1373/clinchem.2014.230235. [DOI] [PubMed] [Google Scholar]




