Abstract
Background
The 5-year survival rate of esophageal cancer is less than 10% in developing countries, where more than 90% of these cancers are squamous cell carcinomas (ESCC). Endoscopic screening is not feasible on the scale required however non-endoscopic cell-sampling techniques may be feasible. Coupling endoscopic biopsies or non-endoscopic samples with biomarker analysis could reduce the subjectivity associated with both methods and thus improve diagnostic accuracy. The aims of this study were therefore to identify biomarkers for esophageal squamous dysplasia and carcinoma.
Methods
A publically available dataset was used to identify genes with differential expression in ESCC compared with normal esophagus (NE). Each gene was ranked by a support vector machine separation score. Expression profiles were examined, before validation by qPCR and immunohistochemistry.
Results
800 genes were overexpressed in ESCC compared to NE (p<10−5). Of the top 50 genes, 33 were expressed in ESCC epithelium and not in NE epithelium or stroma using the Protein Atlas website. These were taken to qPCR validation and 20 genes were significantly overexpressed in ESCC compared to NE (p<0.05). TNFAIP3 and CHN1 showed differential expression with immunohistochemistry. TNFAIP3 expression increased gradually through NE, mild, moderate and severe dysplasia, and SCC (p<0.0001). CHN1 staining was rarely present in the top third of NE epithelium and extended progressively towards the surface in mild, moderate, and severe dysplasia, and SCC (p<0.0001).
Conclusions
Two novel promising biomarkers for ESCC were identified, TNFAIP3 and CHN1.
Impact
CHN1 and TNFAIP3 may improve diagnostic accuracy of screening methods for ESCC.
Keywords: screening, esophageal squamous cell, cancer, carcinoma, biomarker
Introduction
Cancer of the esophagus is the 6th most common cause of cancer death in the world (1). The squamous-cell carcinoma (ESCC) subtype has been declining in the West but remains the most common subtype in the rest of the world. ESCC accounts for 82% of esophageal cancer cases worldwide, including 90% of cases in the highest risk areas of Iran and China, but only around 30% of cases in the UK and USA (1). Patients with ESCC usually present late, with locally advanced disease or metastases, resulting in very poor survival. The 5-year relative survival rate in the USA is 18% (2), but in most high risk populations, where the medical infrastructure is less well developed, it is <10% (3). Early detection and treatment is associated with improved survival (4, 5). Although lead-time bias must be accounted for, a recent 10-year prospective community assignment study in China has reported a significant reduction in ESCC mortality following a single endoscopic screening followed by appropriate therapy (6). Rapid advances in imaging (7), minimally invasive endoscopic therapies (8–10), and novel chemoradiotherapy regimes (11) provide the opportunity to improve patient outcomes when disease is discovered at an early stage.
In high incidence areas for ESCC screening is an important consideration (12). Endoscopy is the gold standard for diagnosis, and identification of dysplasia can be assisted using Lugol’s iodine staining (12, 13). However, on a population wide scale, endoscopy based methods are not logistically or economically feasible as a method of screening due to their high cost and requirement for expertise (12, 14–16). Non-endoscopic cell-sampling techniques are less invasive and costly, though the sensitivity and specificity of cytological assessment have been disappointing (12, 17–19). Coupling a pan-esophageal non-endoscopic cell-collection device with analysis of biomarkers could improve diagnostic accuracy. Equally, biomarker analysis of endoscopic specimens could reduce both the requirement for histopathological expertise and the risk of sampling bias because of the molecular field defect, thus potentially reducing both the procedure length and the number of samples required. Hence the biomarker assisted analysis could reduce the cost of endoscopic diagnosis to a level where it could be considered for screening high risk populations (15, 20).
There is currently a lack of suitable diagnostic biomarkers for ESCC (15, 21). Recent work has begun to identify candidate genes for differentiating ESCC pre-malignant changes (22–28), however the studies have different designs and rarely examine the same genes, making cross comparisons difficult, and the studies are very limited in quantitative analysis (15).
We hypothesised that protein biomarkers for squamous cell dysplasia and ESCC could be identified that would be suitable for application to clinical human tissue samples. The aims of this study were therefore to identify candidate genes that are upregulated in ESCC and squamous dysplasia compared to normal esophageal epithelium and then to validate the putative targets at both the RNA and protein levels on samples from a cohort of patients with ESCC and healthy controls.
Methods
Microarray analysis
The publically available cDNA microarray data set described in Greenawalt et al. (29) was used to identify gene expression profiles from 65 samples (26 ESCC and 39 normal esophageal epithelium controls, Figure 1). A total of ~9,400 unique cDNA clones were available. Data was available in the form of normalised test:reference hybridisation signal intensity ratios. These were converted to log2 values and three clear outliers were excluded (1 from normal control, 2 from ESCC). A one-dimensional support vector machine (SVM) separation score for each gene was calculated for high expression in ESCC compared to low expression in normal controls with a soft 1-norm margin with weight C=1000. For each gene these scores were divided by the fold change between the geometric average of low and high expression and the geometric average fold change of high expression against control.
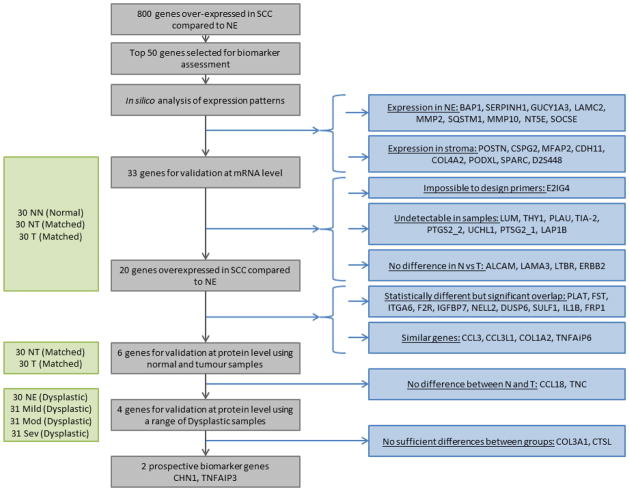
Figure 1.
Overview of discovery-validation pipeline and results at each stage. Samples used at each stage are displayed on the left, with the sample cohort shown in brackets. NN = Normal esophagus from endoscopically normal patients (normal cohort); T = Tumour, NT = Normal Esophagus from ESCC patients (matched cohort); NE = Normal Esophagus, Mild = mild dysplasia, Mod = moderate dysplasia, Sev = severe dysplasia (dysplastic cohort).
In this way, genes were ranked using a support vector machine (SVM) separation score, with a low score reflecting: i) consistent expression in each group of samples, ii) a good separation between normal and ESCC expression levels, and iii) a satisfactory level of expression in ESCC samples.
Protein Atlas Evaluation
The 50 genes with the lowest SVM separation scores were assessed using the expression profiles on the Protein Atlas website (http://www.proteinatlas.org/) to ensure their suitability for paraffin embedded tissues. Genes were excluded based on reported protein expression in the epithelium of normal oesophagus or if they were known to be solely expressed in the stroma.
Human specimens
The putative genes were validated using real-time polymerase chain reaction (RT-PCR) in 30 samples each of: normal esophagus from patients who were endoscopically normal (NN), ESCC from the tumour site of patients (T), and normal esophagus taken from the same patients with ESCC, from as far from the tumour site as possible (NT). The NT and T groups were a ‘matched cohort’, as the corresponding NT and T samples were paired from each patient.
The protein expression of putative biomarkers validated by RT-PCR was confirmed by immunohistochemistry on paraffin-embedded sections from the NT and T samples from the matched cohort, and on 34 paraffin-embedded biopsies of normal esophagus (NE), 31 mild dysplasia (Mild), 31 moderate dysplasia (Mod), and 31 severe dysplasia (Sev) samples from a ‘dysplastic cohort’.
In this study, the NN samples came from patients attending endoscopy at Addenbrooke’s Hospital, Cambridge UK for routine diagnostic procedures with endoscopically-normal esophagus. The NT and T samples came from esophagectomy speciments used in a previous study in Linxian, China (30), and the biopsies of the “dysplastic cohort” came from another previous study in Linxian, China (19). All of the original studies, and the use of collected specimens for future evaluations, were approved by the appropriate IRBs.
RNA extraction and real-time PCR
Total RNA was extracted from frozen samples using an AllPrep DNA/RNA Mini kit (QIAGEN Ltd, Manchester, UK) was then was reverse transcribed using the QuantiTect Reverse Transcription kit (QIAGEN Ltd, Manchester, UK). Quantitative PCR was performed using the LightCycler 480 SYBR Green I Master mix according to manufacturer’s instructions (Roche Diagnostics GmbH, Mannheim, Germany). PCR consisted of 45 cycles of 95°C denaturation (10s), 60°C annealing (10s) and extension (10s). Positive controls were identified for each primer pair. The cycle threshold (Ct) was determined for each sample and the average Ct of the triplicate samples was calculated. The expression of each gene relative to the geometric mean of the triplicate average Ct values for β-actin and 40S ribosomal protein S18 (RPS18) was determined as ΔCt. A melt curve was constructed for each primer.
Immunohistochemistry
Sections of 3.5μm each were stained using a Bond system with the Bond Polymer Refine™ detection kit according to manufacturer’s instructions (Leica Microsystems, Milton Keynes, UK). Origin of the primary antibodies as well as staining conditions are detailed in Table S1. A negative control was performed by omission of the primary antibody.
Each slide was double scored on both extent and intensity. For all genes except CHN1, extent was scored based on percentage of stained epithelium: 0 if absent, 1 for one cell to 33%, 2 for 34–66%, and 3 for ≥67%. For CHN1, extent was scored based on staining from the basal membrane to the epithelial surface: 0 if absent, 1 for any staining in the basal third of the epithelium, 2 for staining in the basal two thirds of the epithelium, and 3 for staining in the superficial third of the epithelium. Intensity was scored as 0 if absent, 1 for weak, 2 for medium, and 3 for strong staining.
Statistical analysis
A one-way ANOVA analysis with a Dunn’s multiple comparisons test was performed to analyse difference in mRNA expression. A Kruskal-Wallis one-way analysis of variance by ranks was performed to analyse difference in IHC scoring between all sample groups. A Wilcoxon matched-pairs signed-rank test was performed to analyse difference in IHC scoring between NT and T samples from the matched cohort. A Mann-Whitney test was used to compare IHC scoring between NN and NT samples. All statistics were performed using Prism (GraphPad Software).
Results
Identification of putative targets
The SVM separation score analysis (examples shown in figure S1) yielded 800 genes which were overexpressed in ESCC compared to normal esophagus (p < 10−5, adjusted for multiple comparison). Expected expression profiles were evaluated using the Protein Atlas website for the 50 most significant genes: 9 genes were found to be expressed in normal esophagus (BAP1, SERPINH1, GUCY1A3, LAMC2, MMP2, SQSTM1, MMP10, NT5E and SOCSE) and 8 were expressed only in the stroma and would therefore not be suitable for a biopsy or cytology screening test (POSTN, CSPG2, MFAP2, CDH11, COL4A2, PODXL, SPARC and D2S448), (figure 1). These 17 genes were excluded from the study. A total of 33 genes were taken to mRNA validation.
mRNA validation
Altered expression in ESCC compared to normal esophagus was confirmed at the mRNA level by real-time qPCR in 30 histopathologically verified tissues each from normal esophagus (NN), normal esophagus from ESCC patients (NT), and ESCC (T). The matched cohort of NT and T samples allowed analysis for the specificity of biomarkers compared to histologically confirmed ESCC within and between cancer patients.
A suitable primer pair could not be designed for E2Ig4. Validation of this target gene was therefore not taken any further. Eight genes (LUM, THY1, PLAU, TIA-2, PTGS2_2, UCHL1, PTSG2_1, LAP1B) were not detected in either normal (NN, NT) or ESCC (T) samples. Four genes (ALCAM, LAMA3, LTBR, ERBB2) had no statistical difference in expression between groups (figure 1 and 2A). These genes were excluded from the study.
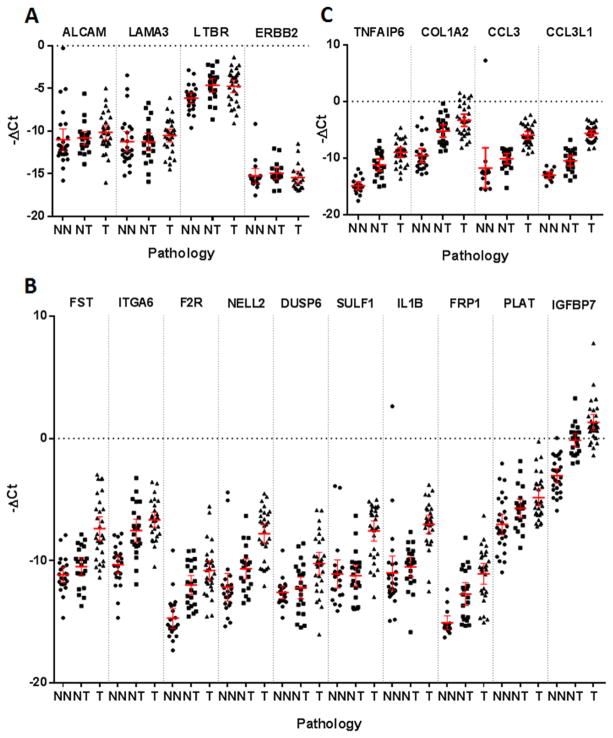
Figure 2.
−ΔCt qPCR data relative to housekeepers β-actin and RPS18 from normal esophagus (NN), normal esophagus from ESCC patients (NT) and ESCC (T) samples. The mean and 95% confidence intervals are displayed. A: Genes with no statistical difference in expression between sample groups. B: Genes which showed significant overlap in expression between sample groups. C: Genes with less clear separation compared to their analogous gene counterparts.
The expression of 20 genes was significantly higher in ESCC compared to normal samples (range p = 0.0002 to p < 0.0001) but despite the significant difference in expression, 10 of these genes (FST, ITGA6, F2R, NELL2, DUSP6, SULF1, IL1B, FRP1, PLAT, IGFBP7) displayed marked overlap in expression between sample groups. It was therefore unlikely that the difference in mRNA expression would translate to a clear difference in protein expression and were therefore not taken forward to validation at the protein level (figure 1 and 2B). It is very interesting to note that 11 out of these 20 genes also displayed a significant over expression in NT compared to NN (Figure 1, Figure 2 and Table S2) suggesting a strong field defect around ESCC which could be advantageous for a non-endoscopic cytology sample method.
The remaining 10 genes contained groups of analogous genes: tumour necrosis factor alpha-induced protein 3 and 6 (TNFAIP3, TNFAIP6), collagens type III alpha 1 and type I alpha 2 (COL3A1, COL1A2), and C-C motif chemokine ligands 18 and 3 and 3-like 1 (CCL18, CCL3, CCL3L1). Only TNFAIP3, COL3A1 and CCL18 were taken to protein validation, based on better separation in expression between sample groups compared to TNFAIP6, COL1A2, and CCL3 and CCL3L1 respectively; the remainder were excluded from the study (figure 1 and 2C).
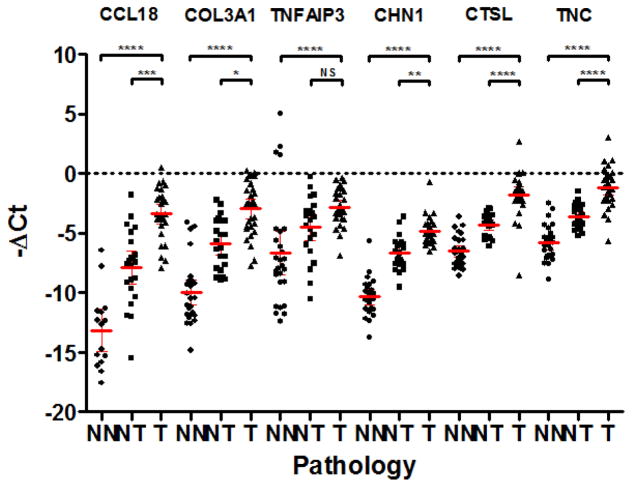
Therefore, 6 genes (CCL18, COL3A1, TNFAIP3, CHN1, CTSL, TNC) which were all overexpressed in ESCC compared to normal samples (p < 0.0001) were selected to be taken to protein validation (figure 1 and 3). It is interesting to note that for 5 of these genes (CCL18 p = 0.0233, COL3A1 p = 0.0008, CHN1 p = 0.0004, CTSL p < 0.0001, TNC p < 0.0001), their expression in normal esophagus from normal patients was lower than in normal esophagus from cancer patients. Again, this suggests some degree of field defect in pathologically normal epithelium adjacent to the cancer.
Figure 3.
−ΔCt qPCR data from normal esophagus (NN), normal esophagus from ESCC patients (NT) and ESCC (T) samples relative to housekeepers β-actin and RPS18. The mean and 95% confidence intervals are displayed. Adjusted p values are as follows: **** p<0.0001, *** p=0.0003, ** p=0.0025, * p=0.01, NS p>0.05.
Protein validation
Increased expression in ESCC compared to normal esophagus was validated at the protein level by immunohistochemistry in histopathologically verified sections from the ‘matched’ and ‘dysplastic’ cohorts of samples, containing normal esophagus from ESCC patients (NT), ESCC (T), normal esophagus (NE), and mild, moderate and severe dysplasia. CCL18 was not statistically overexpressed in ESCC compared to matched normal (figure 1 and figure S2). While TNC was statistical upregulated in ESCC compared to normal esophagus (p<0.0002), in nearly 38% of cases TNC was not expressed and its expression was limited to small foci of tumour cells in 45% of samples (figure 1 and figure S2). Therefore both CCL18 and TNC were excluded from further IHC (figure 1 and figure S2).
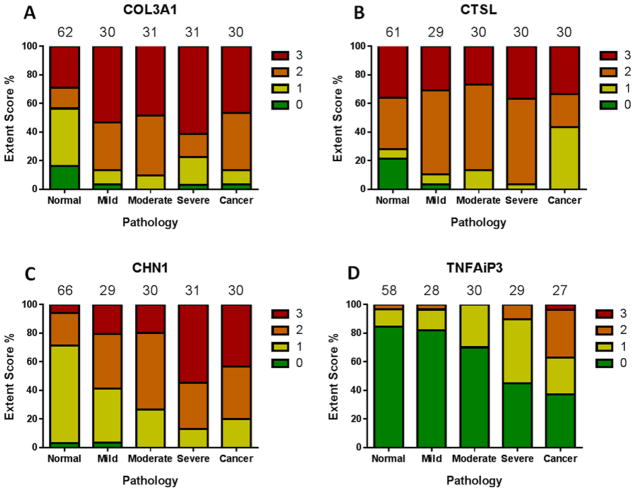
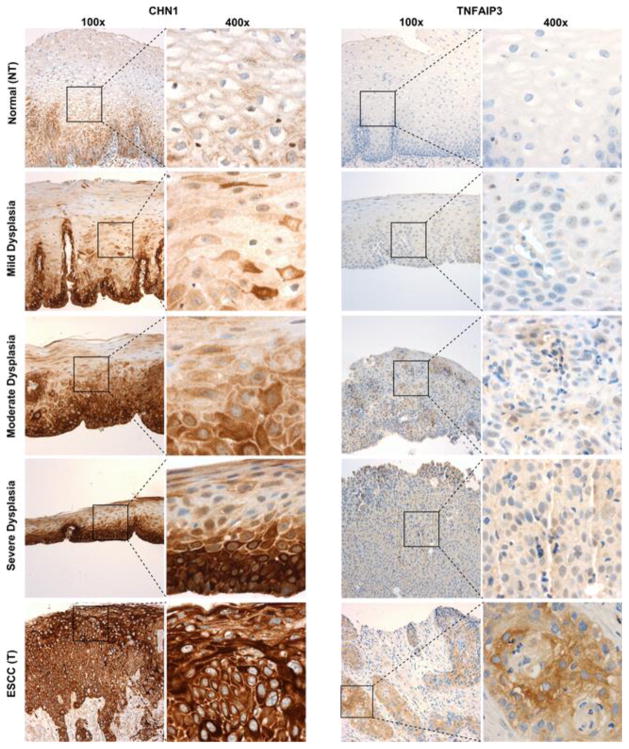
COL3A1 (figure 4A) showed a significant difference (p = 0.0001) in staining across the normal, dysplastic and cancer groups, however a Dunn’s multiple comparisons test showed this was only true for the normal samples copared to any of the other groups, with no change in staining with progression through severity of dysplasia. COL3A1 was expressed mainly in the stromal compartment with very limited epithelial staining (figure S3). COL3A1 was therefore decided to be unsuitable for use as a diagnostic biomarker. CTSL (figure 4B) showed no significant difference (p = 0.3586) in staining across the normal, dysplastic and cancer groups. CTSL was expressed through out the tissue in both stroma and epithelium in normal and cancer samples (figure S4). In contrast, staining of CHN1 (figures 4C and 5) showed a progressive extent of staining towards the superficial layers of the esophageal epithelium, with staining in the superficial third of the epithelium (i.e. a score of 3) in 25% of normal esophagus, 34% of mild dysplasia, 65% of moderate dysplasia, 84% of severe dysplasia, and 97% of ESCC samples (p < 0.0001). TNFAIP3 (figure 4D and 5) also demonstrated stronger staining with increasing dysplasia (p < 0.0001), with staining scores of ≥1 (i.e. at least 1 cell staining positively) in 16% of normal esophagus, 18% of mild dysplasia, 30% of moderate dysplasia, 55% of severe dysplasia, and 63% of ESCC samples.
Figure 4.
IHC staining extent scoring for putative genes across matched and dysplastic cohorts of samples. The number of samples in each pathology group is noted above each column. The ‘normal’ group is comprised of NT samples from the matched cohort and NE samples from the dysplastic cohort.
Figure 5.
Representative images of immunohistochemistry staining for normal esophagus from ESCC patients (NT), mild, moderate, and severe dysplastic, and ESCC samples at 100x and 400x magnification.
Discussion
We have demonstrated that microarray analysis can be applied to biomarker discovery in the context of squamous-cell carcinoma of the esophagus. We identified CHN1 andTNFAIP3 as candidate biomarkers for ESCC using commercially available cDNA microarray data. We demonstrated that these were overexpressed in ESCC compared to normal esophageal epithelium at the mRNA and protein levels.
Initial validation was chosen to be at the mRNA level as reverse transcription quantitive polymerase chain reaction (RT-qPCR) is notably cheaper and less labour intensive than immunohistochemistry (IHC), allowing parallel throughput of multiple prospective biomarkers. RNA species are relatively unstable, however, and rarely survive the paraffin embedding process, so samples must be snap frozen. Proteins are much more stable and are readily tested in paraffin embedded samples, so they make for more clinically suitable biomarkers, particularly in remote rural high incidence areas which is why the secondary validation was performed by IHC.
Out of the 50 markers selected for validation, only two were validated at the protein level. This may appear like a low validation rate especially given that 20 out of 33 genes validated at the mRNA level. This is a example of the difficulties faced when trying to identify biomarkers for a particular cancer. While significant differences can be seen in mRNA expression, they do not necessarily translate to levels of protein expression. While the expression level or staining extent of TNFAIP3 or CHN1 respectively increased along the progression from normal esophagus to SCC, neither marker is perfect at defining the dysplastic or cancer states. Combining both markers might however offer a specific and sensitive test for esophageal dysplasia and early squamous cell cancer of the esophagus.
There were some limitations to the microarray experiments conducted, however these did not detract from the results obtained. The microarray experiments were not designed specifically to identify markers distinguishing between ESCC and normal esophagus, but rigorous statistical measures were employed to reduce the effect of this shortfall which also reduced the number of putative genes. It is interesting to note that only 20 of the 33 targets were validated by qPCR. Eight of the excluded genes were due to undetectable expression levels in biopsy samples. This is most likely due to the amplification of the RNA extracted from samples in the microarray protocol (29) which could account for the observed difference in base expression values. In the microarray data available to us there was no data on dysplasic tissue. This could have allowed us to identify more appropriate biomarkers for identifying high-grade squamous dysplasia, which is the primary screening target for ESCC.
The expression level of 13 out of the 20 increased gradualy between normal oesophagus from normal patients, normal oesophagus from cancer patients and cancer samples (figures 2 and 3). The pathology of all samples was confirmed by an expert pathologist, it is therefore unlikely that dysplasia or cancer was present in the normal samples from cancer patients. This intermediate level of SCC biomarkers suggests that a field defect exists in SCC patients. This field defect could be utilised diagnostically. Even in the event of biopsy or cytological collection that misses the area of cancer and/or dysplasia, an abnormal biomarker could still be detected and patients could be recalled for further investivation.
The biological reason for alterations in the expression of CHN1 and TNFAIP3 remain unclear. Chimerin 1 (CHN1) is expressed in neurones and is predominantly found in the cerebral cortex. It codes for a Rho GTPase-activating protein which is important for dendritic morphology (31) and axon guidance (32). Missense mutations in CHN1 have been associated with variants of Duane’s retraction syndrome (33, 34) and cranial nerve abnormalities (35). The possible role of CHN1 in aiding cellular remodelling in dysplastic and cancer cells would need to be investigated to fully comprehend the full impact of the striking increase in expression of the gene along the progression to cancer. Tumour necrosis factor, alpha-induced protein 3 (TNFAIP3) codes for a ubiquitin editing enzyme which inhibits NFκB and TNF-mediated apoptosis. It is associated with many autoimmune conditions (36–39) and has been noted to have tumour suppressor functions in lymphomas and colorectal cancer (40–43). However, TNFAIP3 also has oncogenic properties, with implication in tamoxifen resistance in breast cancer and developing resistance to apoptosis to promote cancer cell survival (44). It would interesting to understand the role of TNFAIP3 in squamous cell cancer and its link with possible resistance to chemotherapy.
In summary, the biomarker discovery/validation pipeline successfully identified markers for esophageal squamous dysplasia and squamous cell carcinoma of the esophagus. A clinical study applied to endoscopic biopsies or non-endoscopic cell collaction device of the biomarkers CHN1 and TNFAIP3 is now warranted. If used in high risk regions through out the world, such an approach has the potential to identify most patients with moderate or severe dysplasia who would benefit most from endoscopic treatments to prevent the development of invasive squamous cell cancers.
Supplementary Material
Representative examples of SVM separation score data selected from the 100 genes with the highest SVM scores using a random number generator. The x-axis shows the sample number; normal samples are on the left of each graph, tumour samples on the right separated by a red bar. The horizontal lines represent the means of the expression scores for the normal and tumour samples respectively.
IHC staining extent scoring for normal esophagus from ESCC patients (NT) and ESCC (T) matched samples.
Representative images of immunohistochemistry staining for normal esophagus from ESCC patients (NT) and ESCC samples (T) at 100x magnification.
Antibodies used and the conditions for immunohistochemistry.
Summary of known the functions and roles in cancer of the 20 genes overexpressed in tumour compared to normal tissue from normal patients. The p value indicates the level of significance for the difference in expression between normal from normal patients (NN) and normal from normal tissue from patients with esophageal cancer.
Acknowledgments
Financial support:
The Addenbrooke’s Hospital Human Research Tissue Bank, supported by the NIHR Cambridge Biomedical Research Centre, supported this study. This study was funded by the NIHR and the Evelyn Trust. This study was also supported in part by the intramural research program of the National Cancer Institute. RCF has programmatic funding from the Medical Research Council and infrastructure support from the Biomedical Research Centre and the Experimental Medicine Centre.
Footnotes
Conflict of Interest:
Rebecca C Fitzferald and Pierre Lao-Sirieix are named inventors on patents pertaining to a non-endoscopic screening device, the Cytosponge, and related assays. The technology was licensed to Covidien GI Solutions (now Medtronic). RCF and PLS have not received any financial benefits to date. Since December 2013, PLS is now employed partly by Covidien GI Solutions. All other authors have no conflicts of interest to declare.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Aghcheli K, Marjani HA, Nasrollahzadeh D, Islami F, Shakeri R, Sotoudeh M, et al. Prognostic factors for esophageal squamous cell carcinoma--a population-based study in Golestan Province, Iran, a high incidence area. PLoS One. 2011;6:e22152. doi: 10.1371/journal.pone.0022152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen LQ, Hu CY, Ghadirian P, Duranceau A. Early detection of esophageal squamous cell carcinoma and its effects on therapy: an overview. Dis Esophagus. 1999;12:161–7. doi: 10.1046/j.1442-2050.1999.00039.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang GQ, Jiao GG, Chang FB, Fang WH, Song JX, Lu N, et al. Long-term results of operation for 420 patients with early squamous cell esophageal carcinoma discovered by screening. Ann Thorac Surg. 2004;77:1740–4. doi: 10.1016/j.athoracsur.2003.10.098. [DOI] [PubMed] [Google Scholar]
- 6.Wei WQ, Chen ZF, He YT, Feng H, Hou J, Lin DM, et al. Long-Term Follow-Up of a Community Assignment, One-Time Endoscopic Screening Study of Esophageal Cancer in China. J Clin Oncol. 2015;33:1951–7. doi: 10.1200/JCO.2014.58.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddymasu SC, Sharma P. Advances in Endoscopic Imaging of the Esophagus. Gastroenterology Clinics of North America. 2008;37:763. doi: 10.1016/j.gtc.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Inoue H, Minami H, Kaga M, Sato Y, Kudo SE. Endoscopic mucosal resection and endoscopic submucosal dissection for esophageal dysplasia and carcinoma. Gastrointest Endosc Clin N Am. 2010;20:25–34. v–vi. doi: 10.1016/j.giec.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Bergman JJ, Zhang YM, He S, Weusten B, Xue L, Fleischer DE, et al. Outcomes from a prospective trial of endoscopic radiofrequency ablation of early squamous cell neoplasia of the esophagus. Gastrointest Endosc. 2011;74:1181–90. doi: 10.1016/j.gie.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haidry RJ, Butt MA, Dunn J, Banks M, Gupta A, Smart H, et al. Radiofrequency ablation for early oesophageal squamous neoplasia: outcomes form United Kingdom registry. World J Gastroenterol. 2013;19:6011–9. doi: 10.3748/wjg.v19.i36.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass GA, Furlong H, O’Sullivan KE, Hennessy TP, Walsh TN. Chemoradiotherapy, with adjuvant surgery for local control, confers a durable survival advantage in adenocarcinoma and squamous cell carcinoma of the oesophagus. Eur J Cancer. 2014;50:1065–75. doi: 10.1016/j.ejca.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 12.Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol. 2012;9:278–87. doi: 10.1038/nrclinonc.2012.35. [DOI] [PubMed] [Google Scholar]
- 13.Dawsey SM, Fleischer DE, Wang GQ, Zhou B, Kidwell JA, Lu N, et al. Mucosal iodine staining improves endoscopic visualization of squamous dysplasia and squamous cell carcinoma of the esophagus in Linxian, China. Cancer. 1998;83:220–31. [PubMed] [Google Scholar]
- 14.Gerson LB, Groeneveld PW, Triadafilopoulos G. Cost-Effectiveness Model of Endoscopic Screening and Surveillance in Patients With Gastroesophageal Reflux Disease. Clinical Gastroenterology and Hepatology. 2004;2:868–79. doi: 10.1016/s1542-3565(04)00394-5. [DOI] [PubMed] [Google Scholar]
- 15.Taylor PR, Abnet CC, Dawsey SM. Squamous dysplasia--the precursor lesion for esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2013;22:540–52. doi: 10.1158/1055-9965.EPI-12-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roshandel G, Khoshnia M, Sotoudeh M, Merat S, Etemadi A, Nickmanesh A, et al. Endoscopic screening for precancerous lesions of the esophagus in a high risk area in Northern Iran. Arch Iran Med. 2014;17:246–52. [PMC free article] [PubMed] [Google Scholar]
- 17.Boller D, Spieler P, Schoenegg R, Neuweiler J, Kradolfer D, Studer R, et al. Lugol chromoendoscopy combined with brush cytology in patients at risk for esophageal squamous cell carcinoma. Surg Endosc. 2009;23:2748–54. doi: 10.1007/s00464-009-0489-0. [DOI] [PubMed] [Google Scholar]
- 18.Roth MJ, Liu SF, Dawsey SM, Zhou B, Copeland C, Wang GQ, et al. Cytologic detection of esophageal squamous cell carcinoma and precursor lesions using balloon and sponge samplers in asymptomatic adults in Linxian, China. Cancer. 1997;80:2047–59. doi: 10.1002/(sici)1097-0142(19971201)80:11<2047::aid-cncr3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 19.Pan QJ, Roth MJ, Guo HQ, Kochman ML, Wang GQ, Henry M, et al. Cytologic detection of esophageal squamous cell carcinoma and its precursor lesions using balloon samplers and liquid-based cytology in asymptomatic adults in Llinxian, China. Acta Cytol. 2008;52:14–23. doi: 10.1159/000325430. [DOI] [PubMed] [Google Scholar]
- 20.di Pietro M, Boerwinkel DF, Shariff MK, Liu X, Telakis E, Lao-Sirieix P, et al. The combination of autofluorescence endoscopy and molecular biomarkers is a novel diagnostic tool for dysplasia in Barrett’s oesophagus. Gut. 2015;64:49–56. doi: 10.1136/gutjnl-2013-305975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–52. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 22.Takikita M, Hu N, Shou JZ, Giffen C, Wang QH, Wang CY, et al. Fascin and CK4 as Biomarkers for Esophageal Squamous Cell Carcinoma. Anticancer Research. 2011;31:945–52. [PMC free article] [PubMed] [Google Scholar]
- 23.Xue LY, Hu N, Song YM, Zou SM, Shou JZ, Qian LX, et al. Tissue microarray analysis reveals a tight correlation between protein expression pattern and progression of esophageal squamous cell carcinoma. BMC Cancer. 2006;6:296. doi: 10.1186/1471-2407-6-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashyap MK, Marimuthu A, Kishore CJ, Peri S, Keerthikumar S, Prasad TS, et al. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol Ther. 2009;8:36–46. doi: 10.4161/cbt.8.1.7090. [DOI] [PubMed] [Google Scholar]
- 25.Zhou JH, Zhang B, Kernstine KH, Zhong L. Autoantibodies against MMP-7 as a novel diagnostic biomarker in esophageal squamous cell carcinoma. World Journal of Gastroenterology. 2011;17:1373–78. doi: 10.3748/wjg.v17.i10.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su H, Hu N, Yang HH, Wang C, Takikita M, Wang QH, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17:2955–66. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang WC, Wu TT, Chandan VS, Lohse CM, Zhang L. Ki-67 and ProExC are useful immunohistochemical markers in esophageal squamous intraepithelial neoplasia. Hum Pathol. 2011;42:1430–7. doi: 10.1016/j.humpath.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Takala H, Kauppila JH, Soini Y, Selander KS, Vuopala KS, Lehenkari PP, et al. Toll-Like Receptor 9 Is a Novel Biomarker for Esophageal Squamous Cell Dysplasia and Squamous Cell Carcinoma Progression. Journal of Innate Immunity. 2011;3:631–38. doi: 10.1159/000329115. [DOI] [PubMed] [Google Scholar]
- 29.Greenawalt DM, Duong C, Smyth GK, Ciavarella ML, Thompson NJ, Tiang T, et al. Gene expression profiling of esophageal cancer: Comparative analysis of Barrett’s esophagus, adenocarcinoma, and squamous cell carcinoma. International Journal of Cancer. 2007;120:1914–21. doi: 10.1002/ijc.22501. [DOI] [PubMed] [Google Scholar]
- 30.Koshiol J, Kreimer AR. Lessons from Australia: Human Papillomavirus Is Not a Major Risk Factor for Esophageal Squamous Cell Carcinoma. Cancer Epidemiology Biomarkers & Prevention. 2010;19:1889–92. doi: 10.1158/1055-9965.EPI-10-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buttery P, Beg AA, Chih B, Broder A, Mason CA, Scheiffele P. The diacylglycerol-binding protein alpha1-chimaerin regulates dendritic morphology. Proc Natl Acad Sci U S A. 2006;103:1924–9. doi: 10.1073/pnas.0510655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark C, Austen O, Poparic I, Guthrie S. alpha2-Chimaerin regulates a key axon guidance transition during development of the oculomotor projection. J Neurosci. 2013;33:16540–51. doi: 10.1523/JNEUROSCI.1869-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyake N, Demer JL, Shaaban S, Andrews C, Chan WM, Christiansen SP, et al. Expansion of the CHN1 strabismus phenotype. Invest Ophthalmol Vis Sci. 2011;52:6321–8. doi: 10.1167/iovs.11-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyake N, Chilton J, Psatha M, Cheng L, Andrews C, Chan WM, et al. Human CHN1 mutations hyperactivate alpha2-chimaerin and cause Duane’s retraction syndrome. Science. 2008;321:839–43. doi: 10.1126/science.1156121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang JM, Seong MW, Kim JH, Park SS. Absence of CHN1 in two patients with a bilateral absence of cranial nerves IV and VI. Graefes Arch Clin Exp Ophthalmol. 2014 doi: 10.1007/s00417-014-2828-7. [DOI] [PubMed] [Google Scholar]
- 36.Lee YH, Song GG. Associations between TNFAIP3 gene polymorphisms and systemic lupus erythematosus: a meta-analysis. Genet Test Mol Biomarkers. 2012;16:1105–10. doi: 10.1089/gtmb.2012.0096. [DOI] [PubMed] [Google Scholar]
- 37.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Associations between TNFAIP3 gene polymorphisms and rheumatoid arthritis: a meta-analysis. Inflamm Res. 2012;61:635–41. doi: 10.1007/s00011-012-0455-5. [DOI] [PubMed] [Google Scholar]
- 38.Nititham J, Taylor KE, Gupta R, Chen H, Ahn R, Liu J, et al. Meta-analysis of the TNFAIP3 region in psoriasis reveals a risk haplotype that is distinct from other autoimmune diseases. Genes Immun. 2014 doi: 10.1038/gene.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song RH, Yu ZY, Wang Q, Muhali FS, Jiang WJ, Xiao L, et al. Polymorphisms of the TNFAIP3 region and Graves’ disease. Autoimmunity. 2014;47:459–65. doi: 10.3109/08916934.2014.914504. [DOI] [PubMed] [Google Scholar]
- 40.Honma K, Tsuzuki S, Nakagawa M, Tagawa H, Nakamura S, Morishima Y, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:2467–75. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- 41.Novak U, Rinaldi A, Kwee I, Nandula SV, Rancoita PM, Compagno M, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113:4918–21. doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–9. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ungerback J, Belenki D, Jawad ul-Hassan A, Fredrikson M, Fransen K, Elander N, et al. Genetic variation and alterations of genes involved in NFkappaB/TNFAIP3- and NLRP3-inflammasome signaling affect susceptibility and outcome of colorectal cancer. Carcinogenesis. 2012;33:2126–34. doi: 10.1093/carcin/bgs256. [DOI] [PubMed] [Google Scholar]
- 44.Vendrell JA, Ghayad S, Ben-Larbi S, Dumontet C, Mechti N, Cohen PA. A20/TNFAIP3, a new estrogen-regulated gene that confers tamoxifen resistance in breast cancer cells. Oncogene. 2007;26:4656–67. doi: 10.1038/sj.onc.1210269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative examples of SVM separation score data selected from the 100 genes with the highest SVM scores using a random number generator. The x-axis shows the sample number; normal samples are on the left of each graph, tumour samples on the right separated by a red bar. The horizontal lines represent the means of the expression scores for the normal and tumour samples respectively.
IHC staining extent scoring for normal esophagus from ESCC patients (NT) and ESCC (T) matched samples.
Representative images of immunohistochemistry staining for normal esophagus from ESCC patients (NT) and ESCC samples (T) at 100x magnification.
Antibodies used and the conditions for immunohistochemistry.
Summary of known the functions and roles in cancer of the 20 genes overexpressed in tumour compared to normal tissue from normal patients. The p value indicates the level of significance for the difference in expression between normal from normal patients (NN) and normal from normal tissue from patients with esophageal cancer.