Summary
Some of the most challenging stress conditions that organisms encounter during their lifetime involve the transient accumulation of reactive oxygen and chlorine species. Extremely reactive to amino acid side chains, these oxidants cause widespread protein unfolding and aggregation. It is therefore not surprising that cells draw on a variety of different strategies to counteract the damage and maintain a healthy proteome. Orchestrated largely by direct changes in the thiol oxidation status of key proteins, the response strategies involve all layers of protein protection. Reprogramming of basic biological functions helps decrease nascent protein synthesis and restore redox homeostasis. Mobilization of oxidative stress-activated chaperones and production of stress-resistant non-proteinaceous chaperones prevent irreversible protein aggregation. Finally, redox-controlled increase in proteasome activity removes any irreversibly damaged proteins. Together, these systems pave the way to restore protein homeostasis and enable organisms to survive stress conditions that are inevitable when living an aerobic lifestyle.
Introduction
Proteins are the most diverse and structurally complex macromolecules in the cell. Considered nature’s workhorses, they are involved in almost every aspect of biological function known. The function of proteins is defined by their specific three-dimensional structure, making it a top priority for every cell and organism to ensure that nascent polypeptide chains adopt and native proteins preserve their properly folded conformation even during stressful situations (Balchin et al., 2016). Control over a healthy proteome (i.e., proteostasis) begins with the birth of the polypeptide chain on the ribosome and ends with the coordinated death of the mature protein by degradation. Each step in between is carefully orchestrated and involves a complex and highly dynamic network of proteostasis factors (Hartl et al., 2011; Taipale et al., 2014). Failure of this network to do its job not only renders the entire process of costly protein synthesis futile, but might also lead to the toxic accumulation of misfolded proteins (i.e., aggregation) (Brehme et al., 2014; Kaushik and Cuervo, 2015). At the heart of the proteostasis network is a fleet of molecular chaperones that assist in the folding of client proteins, promote protein assembly and disassembly, mediate protein trafficking and signaling, and protect unfolding proteins against the formation of irreversible protein aggregates during non-stress and stress conditions (Hartl et al., 2011; Taipale et al., 2014). Chaperones are assisted by co-chaperones, which fine-tune the activity of chaperones, and work in concert with catalytic folding factors, proteases and non-proteinaceous chaperones. Over the past few years, it became evident that not all members of this network are equally capable of protecting the proteome under all conditions that organisms might encounter over their lifetime. One particular stressful situation that most aerobically living organisms have to eventually deal with is the accumulation of reactive oxygen (ROS) or chlorine species (RCS). Highly reactive towards all cellular macromolecules, ROS/RCS cause major metabolic, transcriptomic and proteomic changes and greatly challenge the cellular proteastasis network. In this review, we will examine the sources of ROS and RCS, discuss their cellular effects, and summarize the various strategies that organisms developed to maintain a healthy proteome during a life with oxygen.
Cellular Oxidants: Where Do They Come From and What Do They Do?
Aerobic organisms regularly encounter reactive oxygen species (ROS) (Murphy, 2009) or reactive chlorine species (RCS) (Ezraty et al., 2017; Kettle et al., 2014). The physiologically most relevant ROS are superoxide anions (O2−) and peroxide (H2O2). They are constantly produced as byproducts of respiration, during oxidative protein folding in the ER and by members of the NADPH oxidase (Nox) and cytochrome p450 family (Holmstrom and Finkel, 2014; Hrycay and Bandiera, 2015; Tu and Weissman, 2002). Superoxide gets readily dismuted into peroxide by superoxide dismutase, thereby indirectly contributing to the cellular H2O2 reservoir. Peroxide is subsequently detoxified by catalases and peroxidases, reacts with proteins involved in redox-sensitive signal transduction processes (Holmstrom and Finkel, 2014) or converts into hydroxyl radicals (•OH) upon reaction with transition metals, particularly Fe2+ and Cu2+ ions (i.e., Fenton reaction) (Halliwell and Gutteridge, 2007; Jomova and Valko, 2011). Hydroxyl radicals are considered to be one of the most reactive ROS and are responsible for much of the oxidative damage that proteins, lipids and DNA experience once ROS levels increase. Some of these oxidation products, particularly lipid peroxides, such as the electrophile 4-hydroxynonenol (4-HNE), can then further perpetuate the cellular damage (Forman et al., 2008; Gaschler and Stockwell, 2017). Another fate of peroxide is the formation of hypochlorous acid (HOCl), a reactive chlorine species (RCS) that is well known for its microbicidal effects and widely used as the active ingredient of household bleach (Winterbourn et al., 2016). Formation of HOCl, which is catalyzed by enzymes such as myeloperoxidase or lactoperoxidase, occurs primarily as part of the innate immune response in activated neutrophils as well as in milk and other mucosal secretions, and serves as a first line of antibacterial defense (Winterbourn and Kettle, 2013). Not surprisingly, organisms have developed a host of oxidative defense systems to counteract ROS/RCS production and repair the oxidative damage. In addition to ROS detoxifying enzymes and small non-protein antioxidants (e.g., glutathione; vitamins) that quench the oxidants (Marengo et al., 2016), cells use various repair systems (e.g., methionine sulfoxide reductase, thioredoxin, glutaredoxin) to reverse the oxidative modifications and restore redox homeostasis (Lu and Holmgren, 2014).
Oxidative Stress: A Constant Threat When Living an Aerobic Lifestyle
Cells constantly generate oxidants and produce antioxidants (Holmstrom and Finkel, 2014; Miki and Funato, 2012). Despite their attempts to strike a healthy balance, organisms encounter numerous situations in which oxidant levels are no longer in sync with the cell’s detoxification systems, generating a potentially lethal condition, termed oxidative stress. One classical example is during inflammation, when activated macrophages and neutrophils produce large amounts of peroxide and HOCl during the so-called oxidative burst (Winterbourn et al., 2016). Although much of the ROS and RCS are produced within phagosomes in an attempt to kill off invading pathogens, some are also released into the environment where they contribute to the tissue damage frequently observed at sites of chronic inflammations (Holmstrom and Finkel, 2014). Accumulation of ROS has also long been considered to be a major contributor to aging and age-related diseases (Labunskyy and Gladyshev, 2013). Although scientists are still debating why aging organisms accumulate ROS, and whether ROS affect lifespan, the fact that aging tissues show increased oxidative damage remains undisputable (Stadtman, 2001).
Proteins are one of the major cellular targets of ROS and RCS. This is in large part due to the sheer number of oxidation-sensitive amino acid side chains present in proteins (Winterbourn and Kettle, 2013). The most frequently observed oxidative modifications involve cysteine and methionine, which have very high reaction rates with hydroxyl radicals or HOCl (~106–108 M−1s−1) (Winterbourn and Kettle, 2013). Peroxide is generally less reactive (~10 M−1s−1), except for proteins with exquisitely peroxide-sensitive thiols (e.g., peroxiredoxins). In addition to the mostly reversible thiol and methionine oxidation reactions, which are often used to redox-regulate protein activity (Lo Conte and Carroll, 2013), proteins tend to undergo a number of irreversible side chain modifications, including thiol overoxidation, carbonylation and di-tyrosine formation (Heinecke et al., 1993; Miki and Funato, 2012). Newly synthesized polypeptide chains are particularly vulnerable, presumably because side chain modifications directly impact nascent protein folding (Medicherla and Goldberg, 2008). Nonetheless, mature proteins are sensitive as well. However, whereas slow acting oxidants (e.g., peroxide) do not cause substantial protein unfolding, oxidants such as HOCl cause widespread aggregation (Winter et al., 2008). This is likely due to the ability of HOCl to rapidly react with residues during local unfolding events, shifting the equilibrium of proteins towards unfolding and aggregation.
Redox Regulation: Controlling Protein Function during Oxidative Stress
With the development of quantitative redox proteomic studies, it is now clear that nearly every physiological process in the cell is either directly or indirectly affected by the cellular redox state (Yang et al., 2016). This level of control is achieved by employing redox-sensitive proteins at crucial checkpoints. Redox-sensitive proteins are typically characterized by the presence of one or more highly conserved (i.e., structurally or functionally important) cysteines, whose thiol groups show high reactivity towards even slow acting oxidants such as peroxide (Roos and Messens, 2011) (Fig. 2). Oxidation of these protein thiols leads to the formation of either intra- or intermolecular disulfide bonds or, in rarer cases, to sulfenic acid, sulfonic acid, or sulfenylamide (Lo Conte and Carroll, 2013). In response, proteins undergo local or global conformational rearrangements, which directly contribute to a loss or gain of function, depending on the protein. Members of the glutaredoxin and thioredoxin family then reverse the thiol modifications and return the proteins and hence the pathways to their original redox and functional states. While some proteins use reversible thiol modification as mechanism to regulate their activity, others undergo reversible oxidation of critical methionine residues (Manta and Gladyshev, 2017). However, since precise quantification of the extent of in vivo methionine oxidation in proteins is still challenging, it is unclear how many proteins and pathways use reversible methionine oxidation as their mode of redox control (Peterfi et al., 2016).
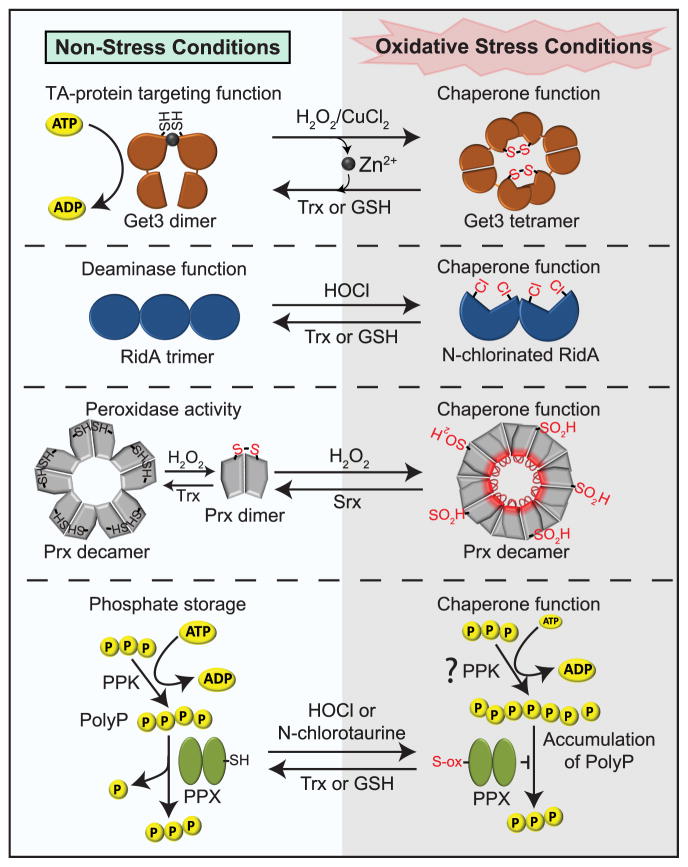
Figure 2. Decreasing Protein Aggregation by Posttranslationally Increasing the Chaperone Pool.
Many of the proteins that serve as chaperones under oxidative stress are dual-function proteins, with distinct enzymatic functions under non-stress conditions. Under oxidative stress conditions, Get3, RidA and peroxiredoxin undergo reversible oxidative modifications that abolish their enzymatic functions and turn them into highly effective ATP-independent chaperones. In addition, bacteria convert ATP into long chains of polyphosphate (polyP), which serve as highly effective chemical chaperones. The transient accumulation of polyP appears to be triggered by a factor that stimulates polyP synthesis during oxidative stress, and mediated by the oxidative inactivation of polyphosphatase PPX, the enzyme that degrades polyP. Please see text for more details.
Metabolic Adaptations to Oxidative Stress
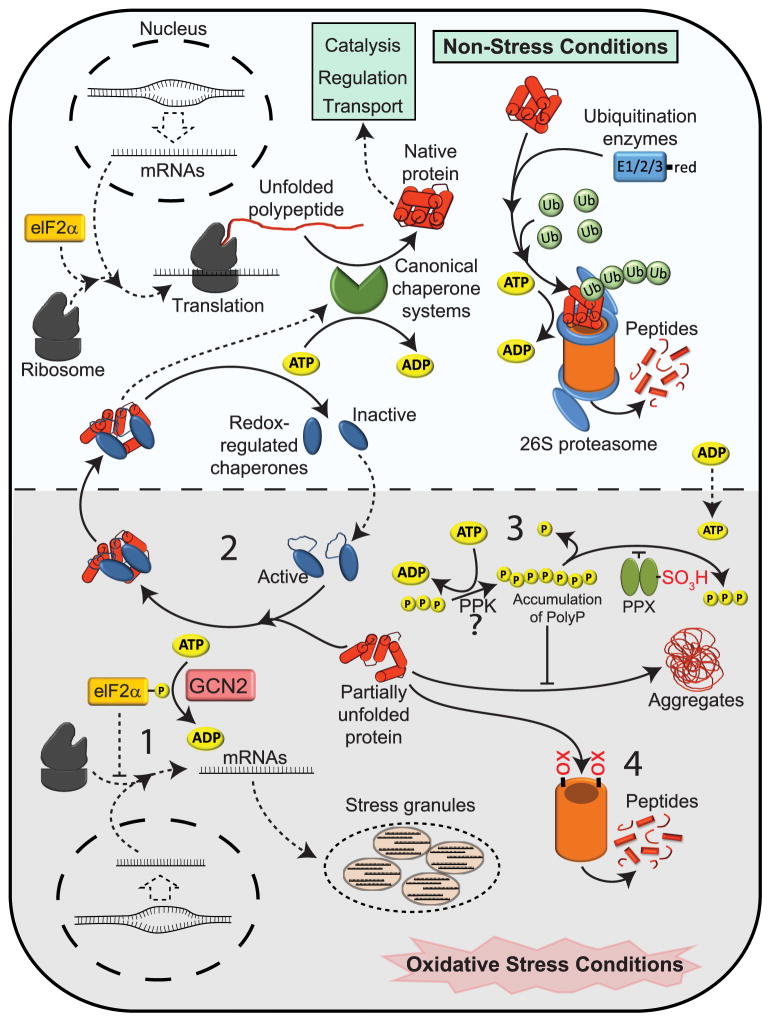
Both newly synthesized as well as mature proteins undergo irreversible oxidative side chain modifications, which can cause protein unfolding and potentially irreversible aggregation. Since damage often cannot be avoided, organisms face the challenge of how to cut their losses. One central metabolic event appears to guide many of the cellular adaptations that are invoked. Organisms, ranging from E. coli to mammalian cells, appear to respond to the accumulation of ROS and/or RCS with an up to 50% decrease in cellular ATP concentrations (Colussi et al., 2000; Kumsta et al., 2011; Winter et al., 2008; Winter et al., 2005). This substantial decrease in the energy levels of the cell has long been attributed to the oxidative inactivation of redox-regulated metabolic enzymes involved in ATP-generating pathways (e.g., GAPDH, F0F1-ATP-synthase) (Hildebrandt et al., 2015; Janero et al., 1994). In fact, oxidative inactivation of key glycolytic enzymes is known to be responsible for the active re-routing of glucose from glycolysis to the pentose phosphate pathway, effectively reducing ATP synthesis at the expense of generating NADPH (Ralser et al., 2007). NADPH is necessary to fuel the thioredoxin and glutaredoxin systems, which are essential for restoring cellular redox homeostasis (Birben et al., 2012). In addition, more recent data from our lab suggest that in some bacteria (and maybe other organisms as well) much of the cellular ATP is actively converted into long chains of polyP, whose protein-stabilizing features contribute to decreased protein aggregation and increased oxidative stress resistance (Gray et al., 2014). The transient depletion of the cell’s ATP content affects most cellular activities and appears to put cells into a quasi-dormant state, helping them to “ride out the storm.” However, it might also affect the activity of ATP-dependent canonical chaperones and proteases, which would explain the need for alternative proteostasis factors that deal with protein homeostasis during oxidative stress.
How Cells Deal with Nascent Protein Folding during Oxidative Stress
Down-regulation of global protein synthesis (Fig. 1) is common to different pathophysiological conditions associated with increased levels of cellular ROS, including starvation, exposure to antibiotics (Morano et al., 2012), inflammation (Ravishankar et al., 2015), and aging (Ferraz et al., 2016; Luo et al., 2017). This strategy not only directly reduces protein aggregation by decreasing the concentration of aggregation-prone nascent polypeptides but also leads to global repression of gene expression during potentially error-prone conditions (Morano et al., 2012; Shenton et al., 2006). Global gene repression is mediated by eIF2α kinases, which phosphorylate the translational initiation factor 2 alpha (elF2α). Phosphorylation of elF2α causes a general decrease in cap-dependent mRNA translation. At the same time, translation of cap-independent mRNAs is initiated, leading to the synthesis of proteins involved in adaptation and stress response (Morano et al., 2012; Taniuchi et al., 2016). Although some eIF2α kinases have been shown to be directly activated by peroxide (e.g., yeast GCN2) or arsenite-induced oxidative stress (e.g. heme-regulated eIF2α kinase (HRI), involvement of one or more additional redox-regulated steps in the oxidative elF2α phosphorylation process are likely to contribute to this global reorganization of protein synthesis (Pakos-Zebrucka et al., 2016).
Figure 1. Protein Homeostasis During Oxidative Stress - A Multilayer Approach.
Under non-stress conditions, protein folding occurs both co-and posttranslationally and is supported by ATP-dependent canonical chaperones. Once natively folded, the proteins fulfill their expected functions until degradation signals trigger ubiquitination and subsequent degradation by the ATP-dependent 26S proteasome. Redox-regulated chaperones are inactive but might exert non-stress related enzymatic functions. Under oxidative stress conditions, cellular ATP levels drop, reducing the activity of ATP-dependent processes. To protect cells against the accumulation of irreversible damaged protein aggregates, a number of ROS and/or RCS-mediated changes occur that either directly or indirectly contribute to 1) reduction of new protein synthesis; 2) activation of ATP-independent chaperones; 3) conversion of ATP into polyphosphate, an effective inorganic chaperone; and 4) increase in ATP-independent 20S proteasome activity. Please see text for more details.
When global translation is down, stalled mRNAs, translation initiation factors, and other RNA binding proteins are sequestered into stress granules and P-bodies, whose formation is rapidly induced by oxidative stress (Chen and Liu, 2017) (Fig. 1). Stress granule formation appears to protect mRNA and promote rapid reinitiation of protein translation as soon as stress conditions are over and energy levels are restored (Buchan and Parker, 2009). However, although increased stress granule formation has repeatedly been observed during oxidative stress treatments, the physiological relevance and the extent to which this strategy plays a major role in oxidative stress defense are still under debate (Arimoto-Matsuzaki et al., 2016).
How Cells Deal with Protein Unfolding and Aggregation during Oxidative Stress
Accumulation of ROS and RCS causes substantial protein damage, which can lead to widespread oxidative protein unfolding, aggregation and ultimately cell death. Over the past 15 years, much research has been conducted to investigate how organisms protect their proteome against irreversible damage during oxidative stress. It is now clear that while most known protein unfolding stress conditions are dealt with by upregulating ATP-dependent chaperone systems, such as Hsp60 (Escherichia coli GroEL), Hsp70 (E. coli DnaK), Hsp90 (E. coli HtpG), or Hsp104 (E. coli ClpB) (Hartl et al., 2011), oxidative stress-induced protein unfolding requires special resources. This appears to be due to a combination of i) the nature of stress, which is extremely rapid and affects a wide range of proteins simultaneously, ii) the sensitivity of some canonical chaperones towards oxidative inactivation (Khor et al., 2004), and iii) the observed drop in ATP-levels, which seem to affect at least some of the classical ATP-dependent foldases (Winter et al., 2005). As a consequence, cells evoke a number of strategies to specifically deal with oxidative protein damage, which appear to all serve one common goal: to rapidly increase the pool of ATP-independent chaperone holdases using existing resources. In the following section, we will discuss some of these tactics, focusing on 1) the specific redox-mediated activation of proteins that serve as potent chaperone once activated; 2) the transformation of well-known and abundant enzymes into binding platforms for unfolding proteins; 3) the conversion of ATP-dependent folding chaperones into ATP-independent holding chaperones; and 4) the synthesis of inorganic polyphosphate, which serves as redox resistant protein-stabilizing scaffold. Each of these systems contributes to the prevention of toxic protein aggregation during oxidative stress, and to the cell’s recovery once non-stress conditions are restored.
Redox Regulation Par Excellence: The Bacterial Chaperone Hsp33
More than 15 years ago, studies in E. coli uncovered a cytosolic 33 kDa heat shock protein, Hsp33, which gained potent ATP-independent molecular chaperone activity specifically during oxidative stress (Jakob et al., 1999). The ROS sensor turned out to consist of a highly conserved, zinc-coordinating four-cysteine motif located in the far C-terminus of Hsp33 (Jakob et al., 2000). Under non-stress conditions, when the cysteines are reduced and zinc is coordinated, Hsp33 is compactly folded and displays no discernable chaperone activity. During oxidative stress conditions that trigger protein unfolding in vitro and/or vivo (e.g., HOCl, bile salts, peroxide at elevated temperatures), Hsp33 rapidly forms two intramolecular disulfide bonds, releases the bound zinc and undergoes widespread conformational rearrangements that cause activation of its chaperone function (Ilbert et al., 2007; Reichmann et al., 2012; Winter et al., 2008). The finding that Hsp33, and several other stress-activated chaperones, gain chaperone activity by converting from a fully folded to a partially disordered conformation, coined the term “conditionally disordered chaperones” (Bardwell and Jakob, 2012). This mechanism allows for a very rapid and extremely stress-specific posttranslational response. According to mutational and structural studies, the critical functional switch in Hsp33 is mediated by the folding status of a central ~50 aa linker region, which is compactly folded in inactive Hsp33, but rapidly unfolds upon oxidation of the nearby cysteine-motif. Once unfolded, the hydrophilic linker region appears to employ long-range electrostatic interactions to capture and bind unfolding client proteins (Groitl et al., 2016; Reichmann et al., 2012; Rimon et al., 2017). Hydrophobic interactions between unfolding proteins and a compactly folded N terminal platform of Hsp33 appear to further stabilize the complex (Groitl et al., 2016). Inactivation of oxidized Hsp33 and client release are also carefully orchestrated and not only require reducing conditions but also the presence of folding chaperones and ATP, which support refolding of the bound client proteins (Hoffmann et al., 2004). This mechanism likely protects against premature client release under conditions that are reducing but not yet conducive for protein folding.
The Dual Function Protein Get3: Hsp33 Counterpart in Yeast
Recently, the targeting factor Get3 was identified as a eukaryotic counterpart of Hsp33 in yeast (Fig. 2). Although structurally completely unrelated, Get3 contains two pairs of cysteines, coordinates zinc, is chaperone-inactive under reducing conditions, and turns into an effective ATP-independent molecular chaperone upon oxidant-induced formation of two disulfide bonds (Voth et al., 2014). Moreover, like Hsp33, disulfide bond formation causes loss of zinc binding, major conformational rearrangements consistent with partial unfolding, and the formation of chaperone-active higher oligomeric species, which bind a wide range of different client proteins with no obvious client specificity (Voth et al., 2014). Finally, yeast cells lacking get3 are highly sensitive to copper stress, a condition well known for its potentially toxic ROS production (Powis et al., 2013; Voth et al., 2014).
In contrast to Hsp33, however, Get3 has a well-defined function under reducing conditions, where it serves as the central ATP-dependent member of machinery that shuttles newly synthesized tail-anchored (TA) proteins to the endoplasmic reticulum (ER) (Mariappan et al., 2011; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). Get3 accepts TA proteins from soluble members of the Get complex and releases them into the ER membrane using ATP binding and hydrolysis to regulate the cycle. When reduced and active as a targeting factor, Get3 exists as a functional dimer that uses one of the cysteine pairs of each subunit to stabilize the dimer-dimer interface via zinc coordination (Stefer et al., 2011). The binding site for TA proteins has been identified as a hydrophobic groove that spans both Get3 subunits (Mateja et al., 2015). When oxidized and active as a chaperone, the ATPase activity of Get3 is nearly abolished, the TA binding site is buried, and new hydrophobic surfaces are exposed that likely serve as the binding sites for unfolding intermediates (Voth et al., 2014). Mutation in the TA protein binding site abrogated the targeting activity (Mateja et al., 2009) without affecting the redox-regulated chaperone function or the ability of this mutant variant to complement the stress-sensitive phenotype of a get3 deletion mutant (Voth et al., 2014).
These results suggest that Get3 serves as a dual function protein, targeting TA proteins during non-stress conditions and preventing protein aggregation under ATP-depleted oxidative stress conditions (Voth and Jakob, 2017). Much work, however, remains to be done. Little is known about the precise activation mechanism of Get3 in vitro or in vivo, the extent to which a similar mode of regulation applies to Trc40 (the Get3 homologue in higher eukaryotic cells) (Stefanovic and Hegde, 2007), the structural rearrangements that confer chaperone function, or the physiological conditions that support activation of its chaperone function. However, the tools seem to be on hand and will hopefully shed light into these questions in the near future.
Other Dual Function Proteins that Serve as Oxidative Stress-Activated Chaperones
Recently, another type of reversible redox activation mechanism was uncovered that converts “regular proteins” into potent ATP-independent chaperones during HOCl stress. One of these proteins is the highly conserved bacterial enzyme reactive intermediate deaminase A (RidA). Under non-stress conditions, RidA serves as a deaminase and accelerates the release of ammonia from reactive enamine/imine intermediates, including the IlvA enzyme (Lambrecht et al., 2012) (Fig. 2). Upon treatment with RCS, however, RidA undergoes reversible N-chlorination of positively charged amino acids. These modifications inactivate the enzyme function while turning RidA into an effective chaperone that prevents aggregation of misfolded substrates, including its native substrate IlvA (Muller et al., 2014). The thiol-independent activation is strictly dependent on treatment with distinct RCS (e.g., hypochloric acid or monochloramine), and is fully reversible by dithiotheitol (DTT), ascorbic acid, or other physiological antioxidants, including thioredoxin and glutathione (Muller et al., 2014). Similar to most other stress-activated chaperones, RCS-dependent activation of RidA leads to conformational changes associated with an increase in surface hydrophobicity and the formation of higher oligomeric structures. Deletion of the ridA gene increases bacterial oxidative stress sensitivity, demonstrating the physiological relevance of RidA’s chaperone function (Lindemann et al., 2013).
N-chlorination-mediated activation of chaperone function is not restricted to bacterial proteins, but has recently also been observed for eukaryotic α-macroglobulin. HOCl-induced N-chlorination triggered the dissociation of α-macroglobulin into dimers with increased hydrophobic surfaces, converting it into a general, ATP-independent chaperone (Wyatt et al., 2014). Chaperone-active α-macroglobulin lacks any distinct client specificity as it prevents aggregation of a wide range of misfolded client proteins forming either amorphous or amyloid-like aggregates (Wyatt et al., 2014). One of the most central questions that remain to be investigated concerns the mechanism by which one or more side chain chlorinations turns proteins into chaperones. Given the fully reversible nature of this modification, it would not be surprising to find many more proteins that use a similar mechanism to protect organisms against protein aggregation during oxidative stress.
The third group of dual function proteins that give up their “regular function” to switch into molecular chaperones during oxidative stress are eukaryotic peroxiredoxins (PRXs) (Fig. 2). During non-stress or mild oxidative stress conditions, PRXs use their highly conserved catalytic cysteine to react with peroxide or peroxynitrite derivatives, thereby either detoxifying the oxidants to harmless water or directly oxidizing specific target proteins in redox-regulated signaling pathways (Poole et al., 2011; Rhee et al., 2012). Upon severe oxidative stress, however, some PRXs (e.g., yeast Tsa1, human Prx1, and Prx2) lose their peroxidase function and turn into ATP-independent chaperones (Jang et al., 2004; Moon et al., 2005). ROS-mediated activation of the chaperone function is triggered by the overoxidation of the active site cysteine to sulfinic acid and is accompanied by changes in the oligomeric state of the protein. Enzymes specifically dedicated to reduce these overoxidized PRXs (i.e., sulfiredoxin) guarantee the reversibility of this process (Rhee et al., 2007). The physiological relevance of this reversible activation during oxidative stress became evident in recent studies in yeast, where overoxidized of yeast Prx (Tsa1) was found to associate with aggregated proteins, recruit Hsp70/Hsp104 to the aggregates, and promote clearance of the aggregates upon re-reduction by sulfiredoxin (Hanzen et al., 2016). However, it also raises the question how organisms, such as C. elegans, which do not encode sulfiredoxins prevent peroxiredoxin’s overoxidation (Thamsen et al., 2011) and/or compensate for the lack of their chaperone activity during oxidative protein unfolding. However, since activation of PRX’s chaperone function is not restricted to oxidative stress (Saccoccia et al., 2012; Teixeira et al., 2015), it is possible that these organisms use a different route to activate the chaperone during oxidative stress.
Oxidative Stress - Turning a Foldase into an ATP-Independent Holdase
Members of the Hsp70 family play some of the most crucial roles in cellular proteostasis, assisting in protein folding, trafficking, (dis) assembly and (dis)aggregation, and many excellent reviews have been published discussing function and mechanism of this ATP-dependent protein-folding chaperone (Balchin et al., 2016). We will therefore focus primarily on recent studies, which revealed that members of this family contribute to increased oxidative stress resistance by converting into ATP-independent chaperone holdases (Wang and Sevier, 2016). One redox sensitive member appears to be the endoplasmic reticulum (ER)-resident Hsp70-homologue BiP (Grp94). Triggered by the expression of an overactive variant of Ero1, the primary producer of peroxide in the ER, BiP was found to undergo reversible oxidative modification of its single conserved cysteine (Wang et al., 2014). Modification of this cysteine, which is located in BiP’s ATP binding pocket, negatively affects its ATPase activity and appears to lock BiP in its high-affinity binding state. Under these conditions, BiP effectively prevents protein aggregation while avoiding potentially futile cycles of binding and release, which would otherwise waste valuable ATP. Reduction of BiP is, at least in part, mediated by the co-chaperone Sil1, which appears to serve a dual function as well; as BiP’s nucleotide exchange factor and as reducing agent converts BiP to its regular ATP-dependent chaperone-function upon return to non-stress conditions (Siegenthaler et al., 2017). These results were preceded by earlier reports in oxidatively stressed mammalian cells, where Grp78 was found to undergo disulfide bond formation that increases its chaperone functions (Wei et al., 2012). However, more detailed studies are needed to define the precise functional consequences of this oxidative modification.
Redox sensitivity appears not to be restricted to the ER-resident Hsp70 homologues but also affects cytosolic members, including bacterial DnaK (Winter et al., 2005) and yeast Ssa1 (Wang et al., 2012). In contrast to BiP, however, glutathionylation of the single cysteine in DnaK’s ATP binding site or disulfide bond formation in Ssa1 appears to down-regulate their chaperone function, causing release from their respective heat shock factors and hence triggering activation of the heat shock response (Wang et al., 2012; Zhang et al., 2016). Therefore, while not directly beneficial, redox regulation of cytosolic Hsp70 homologues ultimately also seems to protect the proteome during oxidative stress.
Polyphosphate: A Powerful Inorganic Chaperone
Protein-based chaperones are not the only mechanism by which organisms protect themselves against proteotoxic stress conditions. We recently discovered that polyP, a universally conserved polyanion composed of up to 1000 phosphoanhydride bond-linked phosphate monomers (Kornberg et al., 1999), functions as an effective protein-stabilizing scaffold both in vitro and in vivo (Gray et al., 2014). PolyP is found in all pro- and eukaryotic organisms and is reported to be involved in many seemingly unrelated processes, ranging from general stress protection and stimulation in biofilm and persister formation in bacteria to blood clotting and mTOR signaling in mammals (Rao et al., 2009),(Morrissey, 2012). The mechanism(s) by which this simply-structured polymer affects these diverse processes, however, has remained enigmatic for many years (Lehman, 2008).
Our observation that E. coli (and other Gram-negative bacteria) actively convert up to 40% of their cellular ATP pool into polyP in response to physiological oxidants, such as HOCl, instigated our studies into the role(s) of polyP in oxidative stress resistance (Gray et al., 2014). To our surprise, we found that polyP prevented aggregation of oxidatively-damaged proteins or otherwise unfolding proteins both in vitro and in vivo (Gray et al., 2014) (Fig. 2). The fact that low micromolar concentrations of polyP protected purified proteins against aggregation in vitro argued against an indirect effect and suggested that direct interactions between polyP and unfolding proteins maintains their solubility. Indeed, biophysical studies revealed the intriguing results that polyP stabilizes unfolding proteins in a soluble β-sheet-rich conformation (Gray et al., 2014). These results suggested that polyP serves as a scaffold that potentially folds in register with and hence stabilizes β-sheets. This result likely also explains the previously observed stimulatory effect of polyP on processes such as biofilm formation and blood clotting, in which proteins need to be stabilized in a cross β-sheet formation in order to form amyloid-like fibrils or fibrin clots, respectively (DePas and Chapman, 2012). Like stress-activated chaperones, polyP binding to soluble unfolding proteins maintains them in a folding competent conformation. Client release is triggered by polyP hydrolysis, and effective refolding is supported by canonical chaperones.
There are still many open questions regarding polyP’s mechanism of action, its regulation, and its role in eukaryotic stress defenses and other processes. Much progress has been made in bacteria, such as E. coli, where the enzymes synthesizing polyP from ATP (polyP kinase, PPK) or converting them back to either ATP (PPK) or Pi (polyP phosphatase, PPX) have long been known (Kornberg et al., 1999). It appears that oxidant-induced accumulation of polyP is triggered by at least one redox-mediated event, i.e., the oxidative inactivation of polyphosphatase PPX (Fig. 2). This oxidation transiently prevents polyP hydrolysis and promotes its accumulation during oxidative stress. In addition, however, oxidative stress likely also involves a stimulatory effect on polyP synthesis, the nature of which is still unknown. The situation is much more challenging in eukaryotic organisms, where, apart from the vacuolar protein Vtc4 in yeast (Hothorn et al., 2009), no polyP-synthesizing enzyme has been identified so far. Extensive homology searches and attempts to purify the activity have thus far been unsuccessful (Kornberg et al., 1999). However, with the availability of high-throughput RNAi screens, it is hopefully just a matter of time until the responsible enzymes are finally discovered in eukaryotes, and the many functions of this “forgotten polymer” and its roles in stress responses can finally be revealed.
The Last Resort: Degradation of Oxidatively-Damaged Proteins
Another pillar in protein homeostasis is the proteolytic destruction of proteins that either have limited lifetimes or are damaged beyond repair. Eukaryotic cells rely on proteasome-dependent protein degradation, which plays a major role in both ubiquitin-dependent and ubiquitin-independent protein turnover (Collins and Goldberg, 2017), as well as on autophagy (self-eating); the latter involves a lysosome-mediated degradation of damaged and potentially toxic cytosolic protein aggregates and organelles (Dikic, 2017). Given the recent, well-deserved attention that autophagy has enjoyed and the many excellent reviews that have been written about the role and regulation of autophagy during oxidative stress (Kiffin et al., 2006; Lee et al., 2012; Pajares et al., 2015), we focus our attention on the proteasome and discuss how cells adjust this system to preferentially degrade oxidatively-damaged proteins (Fig. 3).
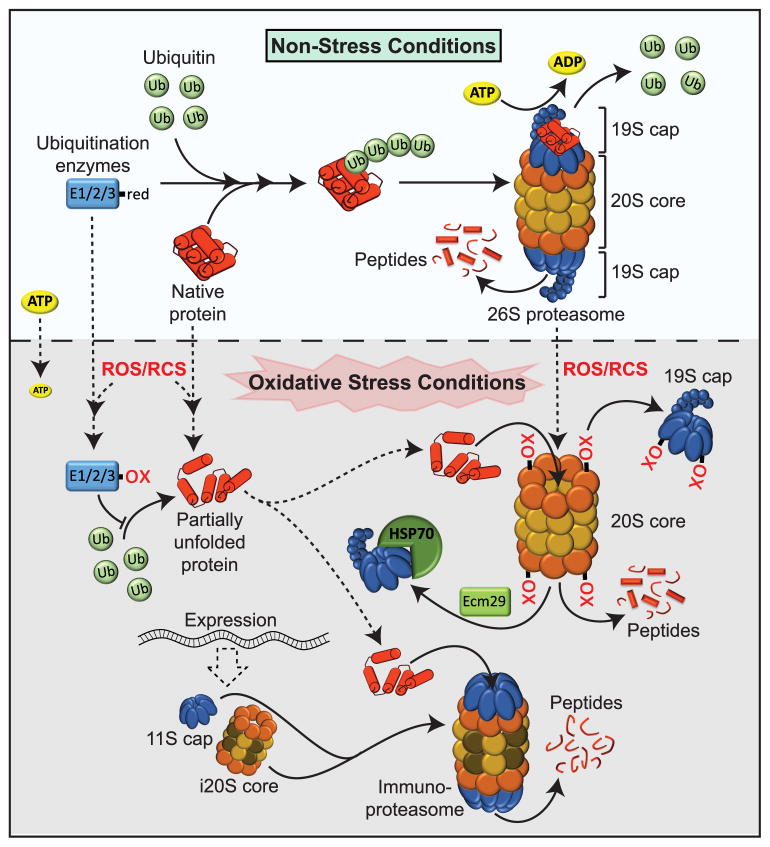
Figure 3. Protein Degradation During Oxidative Stress.
Under non-stress conditions, the majority of proteins targeted for destruction are ubiquitinated and degraded by the 26S proteasome, a hollow barrel-shaped 20S core particle that is capped by two 19S (blue) ATP-dependent regulatory particles. Two outer α-rings (orange) tightly control the gate opening and two inner β-rings (yellow) harbor the proteolytic activity. Proteins of the 19S cap recognize the ubiquitinated client proteins, remove the ubiquitin moiety and mediate the opening of the α-ring gate structure. They furthermore provide the energy to unfold and feed the polypeptide chain into the interior of the barrel-like 20S structure for proteolysis. Under oxidative stress conditions (gray shaded area), partially unfolded proteins directly interact with the α-rings of the 20S proteasome through hydrophobic interactions, eliminating the need for ATP as well as 19S cap or ubiquitination enzymes, both of which contain redox sensitive cysteines. Thiol oxidation of proteins in the α-ring promote further opening of the 20S gate structure, and support disassembly of the 19S caps, which are sequestered by Ecm29 and Hsp70. Chronic oxidative stress induces the expression of an alternative regulatory 11S cap and three alternative β—subunits (brown), which assemble into the 20S immunoproteasome (i20S). Proteolytoc digest by the i20s core generates distinct peptides. The 11S caps binds to both existing 20S and newly assembled i20S proteasomes, functions ATP-independently and increases the protein turnover of the proteasome.
Recruiting the 20S Proteasome to Degrade Oxidatively-Damaged Proteins
During non-stress conditions, most proteins that are targeted for destruction are ubiquitinated and degraded by the 26S proteasome (Collins and Goldberg, 2017; Dikic, 2017; Yu and Matouschek, 2017). This multiprotein complex consists of a 20S core particle and two 19S ATP-dependent regulatory particles that cap the ends of the hollow 20S cylinder (Fig. 3). The 20S core forms a barrel-like structure consisting of two outer α-rings and two inner β-rings each composed of seven different α or β subunits, respectively. While the α-rings mediate client and regulator binding and, in the absence of regulator proteins, form tightly controlled gates that prevent unwanted degradation of proteins, subunits of the β-rings harbor three different proteolytic activities (trypsin-like, chymotrypsin-like, caspase-like) necessary for protein degradation (for excellent recent reviews, see (Collins and Goldberg, 2017; Yu and Matouschek, 2017). Proteins of the 19S cap serve primarily regulatory purposes: (1) they recognize the ubiquitinated client proteins and remove the ubiquitin moiety, (2) they mediate the opening of the α-ring gate structure, and (3) they provide the energy in the form of ATP to unfold and subsequently feed the unraveled polypeptide chain into the interior of the barrel-like 20S structure for degradation (Fig. 3).
Recent studies have shown that the presence of peroxide abolishes 26S-mediated degradation in vitro, whereas 20S-mediated degradation is largely unaffected even at very high (up to 5 mM) peroxide levels (Reinheckel et al., 2000). These results fit well with the finding that oxidatively-damaged proteins are by and large not ubiquitinated in vivo (Kastle et al., 2012) and instead use the 20S core particle for degradation. Oxidized, partially unfolded proteins appear to directly interact with the α-rings of the 20S proteasome through hydrophobic interactions, causing the opening of the 20S gate structure (Fig. 3). This mechanism, which eliminates the need for regulators such as the 19S caps, ubiquitin, or ATP (Jung et al., 2014), makes excellent physiological sense for several reasons: (1) proteins with active site thiols, such as those involved in (poly)ubiquitination reactions, are intrinsically prone to oxidative inactivation (Jung et al., 2014), (2) oxidatively-damaged proteins are already partially unfolded, hence should no longer require 19S caps and ATP for gate opening and unfolding, and (3) protein degradation is uncoupled from ATP levels and is therefore not affected by the ATP-depleted conditions of oxidative stress (Pajares et al., 2015). In addition, ROS trigger several other changes that increase 20S accumulation and remove the need for 19S. Several 20S proteasomal subunits themselves undergo reversible thiol modifications that positively affect ATP-independent degradation by the 20S proteasome. For example, S-glutathionylation of cysteines on the α5 subunit enhances 20S proteosomal activity by further supporting ring opening (Silva et al., 2012), whereas oxidation events near the 20S-19S interaction sites probably directly contribute to the disassembly of the 26S proteasome during oxidative stress (Livnat-Levanon et al., 2014). Treatment with reducing agents was sufficient to reconstitute the functional 26S complex without the involvement of any other chaperones. Other factors that might contribute to the 26S to 20S switch under oxidative stress conditions include Ecm29 in yeast, which together with Hsp70 was found to support disassembly of the 26S proteasome during acute oxidative stress (Aiken et al., 2011), small molecule inhibitors, such as IU1, which prevent deubiquitination by Ubp6, a protein crucial for 26S activity, and Hsp70-mediated sequestration of the 19S cap for extended periods of time during oxidative stress (Aiken et al., 2011) (Fig. 3).
Immunoproteasome i20S: An Alternative Proteasome Induced by Oxidative Stress
In addition to the regular 20S proteasome, it appears that oxidized proteins are also selectively degraded by the related immunoproteasome (i20S), which significantly increases in abundance during oxidative stress (Fig. 3)(Johnston-Carey et al., 2015). The major difference between the 20S and i20S proteasome is the incorporation into the i20S of three alternative β subunits whose expression levels are markedly increased during inflammatory processes and oxidative stress (Johnston-Carey et al., 2015). The three subunits that replace the corresponding subunits in 20S to generate i20S confer additional chymotrypsin activity at the expense of 20S caspase activity (Ferrington and Gregerson, 2012). This leads to a general increase in proteolytic activity and to the generation of distinct peptides that are optimized for binding to the major histocompatibility complex I (Ferrington and Gregerson, 2012; Johnston-Carey et al., 2015). In addition, the same signals that induce the β subunit expression of i20S also trigger expression of an alternative regulator (i.e., 11S), which replace the 19S caps and bind to both existing 20S and newly assembled i20S proteasomes. As expected during oxidative stress, they function ATP-independently and increase the turnover of oxidized, partially unfolded proteins by stimulating i20S proteasome activity.
Concluding Remarks and Future Directions
Over the last three decades, much progress has been made to increase our understanding of the complex and intricate interplay between members of the proteostasis network that is essential in adapting proteostasis to sudden and long-term changes in the environment. These studies have made clear that some stress conditions, including oxidative stress, are more challenging to deal with than others. We have discussed some of the major strategies that pro- and eukaryotic organisms employ to protect their proteome and survive oxidative stress conditions. However, despite the many fascinating breakthroughs that have been made over the past several years, many questions remain unanswered. For instance, we do not know how many more proteins, disguised as enzymes, transcriptional regulators, or structural proteins under non-stress conditions, turn into effective chaperones or proteases under oxidative stress. Since thiol-independent activation mechanisms are being reported more frequently, additional global methods are needed to discover chaperones and members of the degradation machinery that utilize thiol-independent regulation mechanisms during oxidative stress. Equally intriguing is the question of whether these thiol-independent mechanisms can also govern distinct signaling pathways. In either case, we need more systematic approaches to not only identify redox-sensitive proteins but also to investigate their functions under both reducing and oxidizing conditions. It is becoming increasingly obvious that loss/gain of activity upon oxidation/reduction is no longer sufficient to declare that a protein is redox-regulated. While certainly not incorrect, we have to be aware that any loss in one activity might lead to the gain of another activity. Although the mechanism by which such distinct functionalities might have evolved is still unknown, proteins have certainly had ample time to develop multiple activities. It is therefore also tempting to speculate that we might be able to design novel enzymatic functions by using existing chaperones or vice versa. Another aspect that is worth pondering is the fact that many strategies that bacteria have evolved in order to survive oxidative stress are bacteria-specific. Given the immensely important role that oxidative stress plays in host defense and innate immunity, these proteins might be suitable targets for novel antimicrobial therapies. Since bacteria seem to only depend on these protective systems during infection, it is likely that targeting these proteins might reduce the development of resistance while increasing the host’s own ability to fend off infections. Obviously, a lot more needs to be done to reveal all the strategies that organisms use to keep their proteome healthy during oxidative stress. But as we all know, it’s the journey that makes science so exciting, not the destination.
Reichmann et al review the various cellular strategies that organisms employ to protect their proteome during oxidative stress. The authors discuss how cells use oxidants to reprogram basic biological functions, activate specific chaperone systems, and increase proteolytic functions in order to survive stress conditions that are inevitable when living an aerobic lifestyle.
Acknowledgments
This work was supported by grants from the Binational Science Foundation (2015056), Israel Science Foundation (1765/13 and 2629/16), and Human Frontier Science Program (CDA00064/2014) to D.R., and from the National Institutes of Health (GM122506) to U.J., and by a fellowship from the German Research Foundation to W.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiken CT, Kaake RM, Wang X, Huang L. Oxidative stress-mediated regulation of proteasome complexes. Molecular & cellular proteomics: MCP. 2011;10:R110006924. doi: 10.1074/mcp.M110.006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto-Matsuzaki K, Saito H, Takekawa M. TIA1 oxidation inhibits stress granule assembly and sensitizes cells to stress-induced apoptosis. Nat Commun. 2016;7:10252. doi: 10.1038/ncomms10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37:517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehme M, Voisine C, Rolland T, Wachi S, Soper JH, Zhu Y, Orton K, Villella A, Garza D, Vidal M, et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014;9:1135–1150. doi: 10.1016/j.celrep.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu B. Relationships between Stress Granules, Oxidative Stress, and Neurodegenerative Diseases. Oxidative medicine and cellular longevity. 2017;2017:1809592. doi: 10.1155/2017/1809592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Goldberg AL. The Logic of the 26S Proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi C, Albertini MC, Coppola S, Rovidati S, Galli F, Ghibelli L. H2O2-induced block of glycolysis as an active ADP-ribosylation reaction protecting cells from apoptosis. FASEB J. 2000;14:2266–2276. doi: 10.1096/fj.00-0074com. [DOI] [PubMed] [Google Scholar]
- DePas WH, Chapman MR. Microbial manipulation of the amyloid fold. Res Microbiol. 2012;163:592–606. doi: 10.1016/j.resmic.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. Proteasomal and Autophagic Degradation Systems. Annu Rev Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- Ezraty B, Gennaris A, Barras F, Collet JF. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol. 2017;15:385–396. doi: 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- Ferraz RC, Camara H, De-Souza EA, Pinto S, Pinca AP, Silva RC, Sato VN, Castilho BA, Mori MA. IMPACT is a GCN2 inhibitor that limits lifespan in Caenorhabditis elegans. BMC Biol. 2016;14:87. doi: 10.1186/s12915-016-0301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrington DA, Gregerson DS. Immunoproteasomes: structure, function, and antigen presentation. Prog Mol Biol Transl Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman HJ, Fukuto JM, Miller T, Zhang H, Rinna A, Levy S. The chemistry of cell signaling by reactive oxygen and nitrogen species and 4-hydroxynonenal. Arch Biochem Biophys. 2008;477:183–195. doi: 10.1016/j.abb.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschler MM, Stockwell BR. Lipid peroxidation in cell death. Biochem Biophys Res Commun. 2017;482:419–425. doi: 10.1016/j.bbrc.2016.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JC, et al. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53:689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groitl B, Horowitz S, Makepeace KA, Petrotchenko EV, Borchers CH, Reichmann D, Bardwell JC, Jakob U. Protein unfolding as a switch from self-recognition to high-affinity client binding. Nat Commun. 2016;7:10357. doi: 10.1038/ncomms10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 4. Oxford; New York: Oxford University Press; 2007. [Google Scholar]
- Hanzen S, Vielfort K, Yang J, Roger F, Andersson V, Zamarbide-Fores S, Andersson R, Malm L, Palais G, Biteau B, et al. Lifespan Control by Redox-Dependent Recruitment of Chaperones to Misfolded Proteins. Cell. 2016;166:140–151. doi: 10.1016/j.cell.2016.05.006. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Heinecke JW, Li W, Daehnke HL, 3rd, Goldstein JA. Dityrosine, a specific marker of oxidation, is synthesized by the myeloperoxidase-hydrogen peroxide system of human neutrophils and macrophages. J Biol Chem. 1993;268:4069–4077. [PubMed] [Google Scholar]
- Hildebrandt T, Knuesting J, Berndt C, Morgan B, Scheibe R. Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol Chem. 2015;396:523–537. doi: 10.1515/hsz-2014-0295. [DOI] [PubMed] [Google Scholar]
- Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. EMBO J. 2004;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, et al. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science. 2009;324:513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- Hrycay EG, Bandiera SM. Involvement of Cytochrome P450 in Reactive Oxygen Species Formation and Cancer. Adv Pharmacol. 2015;74:35–84. doi: 10.1016/bs.apha.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nature structural & molecular biology. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Eser M, Bardwell JC. Redox switch of hsp33 has a novel zinc-binding motif. J Biol Chem. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- Janero DR, Hreniuk D, Sharif HM. Hydroperoxide-induced oxidative stress impairs heart muscle cell carbohydrate metabolism. Am J Physiol. 1994;266:C179–188. doi: 10.1152/ajpcell.1994.266.1.C179. [DOI] [PubMed] [Google Scholar]
- Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Johnston-Carey HK, Pomatto LC, Davies KJ. The Immunoproteasome in oxidative stress, aging, and disease. Crit Rev Biochem Mol Biol. 2015;51:268–281. doi: 10.3109/10409238.2016.1172554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomova K, Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011;283:65–87. doi: 10.1016/j.tox.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Jung T, Hohn A, Grune T. The proteasome and the degradation of oxidized proteins: Part II - protein oxidation and proteasomal degradation. Redox Biol. 2014;2:99–104. doi: 10.1016/j.redox.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastle M, Reeg S, Rogowska-Wrzesinska A, Grune T. Chaperones, but not oxidized proteins, are ubiquitinated after oxidative stress. Free Radic Biol Med. 2012;53:1468–1477. doi: 10.1016/j.freeradbiomed.2012.05.039. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21:1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- Kettle AJ, Albrett AM, Chapman AL, Dickerhof N, Forbes LV, Khalilova I, Turner R. Measuring chlorine bleach in biology and medicine. Biochim Biophys Acta. 2014;1840:781–793. doi: 10.1016/j.bbagen.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Khor HK, Fisher MT, Schoneich C. Potential role of methionine sulfoxide in the inactivation of the chaperone GroEL by hypochlorous acid (HOCl) and peroxynitrite (ONOO-) J Biol Chem. 2004;279:19486–19493. doi: 10.1074/jbc.M310045200. [DOI] [PubMed] [Google Scholar]
- Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Kumsta C, Thamsen M, Jakob U. Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxid Redox Signal. 2011;14:1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labunskyy VM, Gladyshev VN. Role of reactive oxygen species-mediated signaling in aging. Antioxid Redox Signal. 2013;19:1362–1372. doi: 10.1089/ars.2012.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht JA, Flynn JM, Downs DM. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J Biol Chem. 2012;287:3454–3461. doi: 10.1074/jbc.M111.304477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: crosstalk and redox signalling. Biochem J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman IR. Historical perspective: Arthur Kornberg, a giant of 20th century biochemistry. Trends in Biochemical Sciences. 2008;33:291–296. doi: 10.1016/j.tibs.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Lindemann C, Lupilova N, Muller A, Warscheid B, Meyer HE, Kuhlmann K, Eisenacher M, Leichert LI. Redox proteomics uncovers peroxynitrite-sensitive proteins that help Escherichia coli to overcome nitrosative stress. J Biol Chem. 2013;288:19698–19714. doi: 10.1074/jbc.M113.457556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnat-Levanon N, Kevei E, Kleifeld O, Krutauz D, Segref A, Rinaldi T, Erpapazoglou Z, Cohen M, Reis N, Hoppe T, et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014;7:1371–1380. doi: 10.1016/j.celrep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- Lo Conte M, Carroll KS. The redox biochemistry of protein sulfenylation and sulfinylation. J Biol Chem. 2013;288:26480–26488. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- Luo H, Chiang HH, Louw M, Susanto A, Chen D. Nutrient Sensing and the Oxidative Stress Response. Trends Endocrinol Metab. 2017;28:449–460. doi: 10.1016/j.tem.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manta B, Gladyshev VN. Regulated methionine oxidation by monooxygenases. Free Radic Biol Med. 2017;109:141–155. doi: 10.1016/j.freeradbiomed.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marengo B, Nitti M, Furfaro AL, Colla R, Ciucis CD, Marinari UM, Pronzato MA, Traverso N, Domenicotti C. Redox Homeostasis and Cellular Antioxidant Systems: Crucial Players in Cancer Growth and Therapy. Oxidative medicine and cellular longevity. 2016;2016:6235641. doi: 10.1155/2016/6235641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Mateja A, Dobosz M, Bove E, Hegde RS, Keenan RJ. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 2015;347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Funato Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J Biochem. 2012;151:255–261. doi: 10.1093/jb/mvs006. [DOI] [PubMed] [Google Scholar]
- Moon JC, Hah YS, Kim WY, Jung BG, Jang HH, Lee JR, Kim SY, Lee YM, Jeon MG, Kim CW, et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JH. Polyphosphate: a link between platelets, coagulation and inflammation. Int J Hematol. 2012;95:346–352. doi: 10.1007/s12185-012-1054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Langklotz S, Lupilova N, Kuhlmann K, Bandow JE, Leichert LI. Activation of RidA chaperone function by N-chlorination. Nat Commun. 2014;5:5804. doi: 10.1038/ncomms6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M, Jimenez-Moreno N, Dias IH, Debelec B, Vucetic M, Fladmark KE, Basaga H, Ribaric S, Milisav I, Cuadrado A. Redox control of protein degradation. Redox Biol. 2015;6:409–420. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfi Z, Tarrago L, Gladyshev VN. Practical guide for dynamic monitoring of protein oxidation using genetically encoded ratiometric fluorescent biosensors of methionine sulfoxide. Methods. 2016;109:149–157. doi: 10.1016/j.ymeth.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Hall A, Nelson KJ. Overview of peroxiredoxins in oxidant defense and redox regulation. Curr Protoc Toxicol. 2011;Chapter 7(Unit7):9. doi: 10.1002/0471140856.tx0709s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis K, Schrul B, Tienson H, Gostimskaya I, Breker M, High S, Schuldiner M, Jakob U, Schwappach B. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. J Cell Sci. 2013;126:473–483. doi: 10.1242/jcs.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H, et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NN, Gomez-Garcia MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annual Review of Biochemistry. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- Ravishankar B, Liu H, Shinde R, Chaudhary K, Xiao W, Bradley J, Koritzinsky M, Madaio MP, McGaha TL. The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc Natl Acad Sci U S A. 2015;112:10774–10779. doi: 10.1073/pnas.1504276112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheckel T, Ullrich O, Sitte N, Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch Biochem Biophys. 2000;377:65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl. 2007:S3–8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Woo HA, Kil IS, Bae SH. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimon O, Suss O, Goldenberg M, Fassler R, Yogev O, Amartely H, Propper G, Friedler A, Reichmann D. A Role of Metastable Regions and Their Connectivity in the Inactivation of a Redox-Regulated Chaperone and Its Inter-Chaperone Crosstalk. Antioxid Redox Signal. 2017;27:1252–1267. doi: 10.1089/ars.2016.6900. [DOI] [PubMed] [Google Scholar]
- Roos G, Messens J. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med. 2011;51:314–326. doi: 10.1016/j.freeradbiomed.2011.04.031. [DOI] [PubMed] [Google Scholar]
- Saccoccia F, Di Micco P, Boumis G, Brunori M, Koutris I, Miele AE, Morea V, Sriratana P, Williams DL, Bellelli A, et al. Moonlighting by different stressors: crystal structure of the chaperone species of a 2-Cys peroxiredoxin. Structure. 2012;20:429–439. doi: 10.1016/j.str.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- Siegenthaler KD, Pareja KA, Wang J, Sevier CS. An unexpected role for the yeast nucleotide exchange factor Sil1 as a reductant acting on the molecular chaperone BiP. Elife. 2017;6 doi: 10.7554/eLife.24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GM, Netto LE, Simoes V, Santos LF, Gozzo FC, Demasi MA, Oliveira CL, Bicev RN, Klitzke CF, Sogayar MC, et al. Redox control of 20S proteasome gating. Antioxid Redox Signal. 2012;16:1183–1194. doi: 10.1089/ars.2011.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Stefer S, Reitz S, Wang F, Wild K, Pang YY, Schwarz D, Bomke J, Hein C, Lohr F, Bernhard F, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333:758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, Larsen B, Choi H, Berger B, Gingras AC, Lindquist S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi S, Miyake M, Tsugawa K, Oyadomari M, Oyadomari S. Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci Rep. 2016;6:32886. doi: 10.1038/srep32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F, Castro H, Cruz T, Tse E, Koldewey P, Southworth DR, Tomas AM, Jakob U. Mitochondrial peroxiredoxin functions as crucial chaperone reservoir in Leishmania infantum. Proc Natl Acad Sci U S A. 2015;112:E616–624. doi: 10.1073/pnas.1419682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamsen M, Kumsta C, Li F, Jakob U. Is overoxidation of peroxiredoxin physiologically significant? Antioxid Redox Signal. 2011;14:725–730. doi: 10.1089/ars.2010.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- Voth W, Jakob U. Stress-Activated Chaperones: A First Line of Defense. Trends Biochem Sci. 2017 doi: 10.1016/j.tibs.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth W, Schick M, Gates S, Li S, Vilardi F, Gostimskaya I, Southworth DR, Schwappach B, Jakob U. The protein targeting factor Get3 functions as ATP-independent chaperone under oxidative stress conditions. Mol Cell. 2014;56:116–127. doi: 10.1016/j.molcel.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pareja KA, Kaiser CA, Sevier CS. Redox signaling via the molecular chaperone BiP protects cells against endoplasmic reticulum-derived oxidative stress. eLife. 2014;3:e03496. doi: 10.7554/eLife.03496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sevier CS. Formation and Reversibility of BiP Protein Cysteine Oxidation Facilitate Cell Survival during and post Oxidative Stress. The Journal of biological chemistry. 2016;291:7541–7557. doi: 10.1074/jbc.M115.694810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gibney PA, West JD, Morano KA. The yeast Hsp70 Ssa1 is a sensor for activation of the heat shock response by thiol-reactive compounds. Mol Biol Cell. 2012;23:3290–3298. doi: 10.1091/mbc.E12-06-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei PC, Hsieh YH, Su MI, Jiang X, Hsu PH, Lo WT, Weng JY, Jeng YM, Wang JM, Chen PL, et al. Loss of the oxidative stress sensor NPGPx compromises GRP78 chaperone activity and induces systemic disease. Mol Cell. 2012;48:747–759. doi: 10.1016/j.molcel.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Kettle AJ. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal. 2013;18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Kettle AJ, Hampton MB. Reactive Oxygen Species and Neutrophil Function. Annu Rev Biochem. 2016;85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- Wyatt AR, Kumita JR, Mifsud RW, Gooden CA, Wilson MR, Dobson CM. Hypochlorite-induced structural modifications enhance the chaperone activity of human alpha2-macroglobulin. Proc Natl Acad Sci U S A. 2014;111:E2081–2090. doi: 10.1073/pnas.1403379111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Carroll KS, Liebler DC. The Expanding Landscape of the Thiol Redox Proteome. Molecular & cellular proteomics: MCP. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Matouschek A. Recognition of Client Proteins by the Proteasome. Annu Rev Biophys. 2017;46:149–173. doi: 10.1146/annurev-biophys-070816-033719. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang J, Wu S, Gong W, Chen C, Perrett S. Glutathionylation of the Bacterial Hsp70 Chaperone DnaK Provides a Link between Oxidative Stress and the Heat Shock Response. The Journal of biological chemistry. 2016;291:6967–6981. doi: 10.1074/jbc.M115.673608. [DOI] [PMC free article] [PubMed] [Google Scholar]