Abstract
The use of a mouse model to study the breadth of symptoms and disease severity seen in human WNV infection can provide insight into the kinetics of the immune response and the specific pathways responsible for control of WNV infection and viral clearance. Here, we provide protocols for performing WNV infection of mice, as well as complete immunophenotyping analysis of the cellular immune response to infection in both the periphery and the central nervous system in a mouse model of West Nile Virus infection.
Keywords: West Nile Virus, immunophenotyping, clinical disease
UNIT INTRODUCTION
This article provides detailed protocols for using a mouse model of West Nile virus infection to study clinical disease and symptoms, as well as studying the cellular immune responses to infection by extensive immunophenotyping of the tissues. Full details are provided for 1) infection of the mouse by subcutaneous footpad injection, 2) monitoring for clinical disease symptoms, 3) tissue processing of the spleen, brain, and blood, and 4) flow cytometry phenotyping on isolated lymphocytes. Specific flow cytometry markers and protocols are provided for suggested immune phenotyping characterization.
NOTE: All protocols described in this unit have been approved by the author’s Institutional Animal Care and Use Committee (IACUC), conforming to national and local government regulations regarding the care and use of laboratory animals.
STRATEGIC PLANNING
Biosafety
West Nile virus is classified as a Biosafety Level-3 (BSL-3) agent by the CDC (Kauffman, Franke, Wong, & Kramer, 2011). All studies involving WNV in animal models are conducted in IACUC-approved facilities, with all procedures taking place within the containment of biosafety cabinets. Personnel are required to don personal protective equipment including a fluid repellent gown, two pairs of gloves, sleeve protectors, hair bonnet, facial mask as a splash precaution, and booties when conducting infection studies. Individual requirements from the investigator’s governing IACUC and institution must be fulfilled before beginning work with West Nile virus.
Ethical Considerations
As these studies involve infection of mice with West Nile virus, the study animals will experience significant disease symptoms in the forms of weight loss, partial paralysis, and ultimately euthanasia, as well as momentary pain from subcutaneous infection in the footpad of virus inoculum. However, the ultimate goals of the study include identification of immune processes and pathways involved in protection from West Nile virus and clinical disease, which is needed in order to better prevent and treat West Nile virus and other flaviviruses such as Zika virus. Great care should be taken to reduce the suffering of animals through clear criteria for euthanasia prior to experimental endpoint due to clinical disease symptoms. Additionally, careful design of experiments is necessary to determine the smallest number of animals needed to obtain significant results.
Mouse Strain Selection
West Nile virus infection and immunity has been characterized extensively in the C57BL/6 (B6) mouse model. Following subcutaneous infection in the footpad with the WNV-TX neuroinvasive strain, approximately 30% of infected mice succumb to infection within 10–12 days post infection, depending on the inoculum dose (Daffis, Samuel, Keller, Gale, & Diamond, 2007; Daffis, Samuel, Suthar, Gale, & Diamond, 2008; Daffis, Samuel, Suthar, Keller, et al., 2008; Samuel et al., 2006; Suthar et al., 2010). Innate and adaptive immune responses coordinate the protection and viral control of West Nile virus infection and disease (Suthar, Diamond, & Gale, 2013). In innate immunity, RLR and IFN-mediated actions are essential to restrict WNV replication and neuroinvasive CNS disease, while T cells are critical for viral clearance in the CNS (Daffis, Samuel, Suthar, Keller, et al., 2008; Errett, Suthar, McMillan, Diamond, & Gale, 2013; Keller et al., 2006; Samuel et al., 2006; Suthar et al., 2010).
However, given the breadth in symptoms and disease severity seen in human WNV infection, not all are effectively captured in the B6 model, such as gastrointestinal involvement and long-term neurological weakness (Carson et al., 2006; Gottfried, Quinn, & Jones, 2005; Klee et al., 2004; Leis, Stokic, Webb, Slavinski, & Fratkin, 2003; Marciniak, Sorosky, & Hynes, 2004; Sejvar, 2007; H. Wang, Siddharthan, Hall, & Morrey, 2011). To expand the studies of West Nile virus infection and immunity, other strains of mice might be examined, such as the more recently developed Collaborative Cross (CC) strains. The CC model includes eight founder mouse strains: five classical inbred strains (C57BL/6J, A/J, 129S1/SvImJ, NOD/ShiLtJ, and NZO/H1LuJ) and three wild-type-derived strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ) (Churchill et al., 2004; Threadgill & Churchill, 2012; Threadgill, Miller, Churchill, & de Villena, 2011). The founder strains represent the three major Mus musculus subspecies, capturing nearly 90% of common genetic variation in Mus musculus strains (>40 million SNPs, >4 million small insertion/deletions), with this variation randomly distributed across the genome (Roberts, Pardo-Manuel de Villena, Wang, McMillan, & Threadgill, 2007). In recent publications, the use of the CC mouse model has been shown to measure many additional phenotypes of WNV infection in both the periphery and the CNS, to further characterize the adaptive immune response to WNV (Graham et al., 2016; Graham et al., 2015). The choice of the mouse background strain is an important consideration for the individual investigator’s research goals, and a dose titration of the virus used should also be considered for optimum, consistent results.
BASIC PROTOCOL 1. WEST NILE VIRUS INFECTION VIA FOOTPAD INJECTION
West Nile Virus is transmitted to mammals subcutaneously through the bite of an infected mosquito. To mimic a mosquito bite, WNV infection is performed by subcutaneous injection in the rear footpad. Previous studies have shown that the footpad route of inoculation best mimics the natural course of infection and immunity in humans, as the virus first encounters dendritic cells and macrophages that initiate the innate immune response (Daffis, Samuel, Suthar, Gale, et al., 2008; Daffis, Samuel, Suthar, Keller, et al., 2008; Daffis, Suthar, Szretter, Gale, & Diamond, 2009). Mice are first anesthetized with a mixture of ketamine/xylazine. Following the foot pinch test, where the hind foot is squeezed and mice are deemed properly anesthetized when they do not have a reflex reaction, mice are then infected with WNV, and monitored until they recover from anesthesia.
Materials
Ketamine (100mg/mL)
Xylazine (20mg/mL)
Phosphate-buffered saline (PBS; Invitrogen)
West Nile Virus (kindly provided by the Gale Lab, University of Washington)
1/2cc insulin syringe (28G, BD Biosciences)
8–10 week old mice (see Strategic Planning)
-
Prepare ketamine/xylazine mouse mix:
0.65 ml ketamine (100 mg/ml)
0.22 ml xylazine (20 mg/ml)
9.13 ml of sterile PBS
-
Prepare appropriate WNV virus dilution from stock aliquot. For footpad injections, limit dosage volume to <50μl.
For example, to deliver a dosage of 100pfu WNV-TX in a 40 μL volume, create a dilution from the stock aliquot that is 2500pfu/mL. -
Anesthetize the mice with 0.02 ml mouse mix per g body weight, which will administer 130 mg/kg ketamine, 8.8 mg/kg xylazine
Monitor the animal during anesthesia for respiration and loss of reflexes. In cages with multiple animals, ensure that mice do not lay on each other as they become anesthetized. -
Once mice are fully anesthetized, deliver infection volume into the rear footpad using insulin syringe.
The needle bevel should face downwards into the skin, with the needle into the skin just past the bevel before injecting. After injection, wait a few seconds before slowly withdrawing the needle to prevent inoculum from leaving the injection site. Return mice to the cage on their backs next to each other to recover from anesthesia, and monitor mice until they are fully awake again (usually within 30 minutes).
BASIC PROTOCOL 2. CLINICAL SCORE AND WEIGHT LOSS
In the B6 mouse model, approximately 30% of WNV-TX infected mice aged 8–10 weeks succumb to infection within 10–12 days, depending on the infectious dose (Daffis, Samuel, Suthar, Gale, et al., 2008; Samuel et al., 2006; Suthar et al., 2010). The kinetics of infection and observed clinical disease must be established for each unique mouse strain and model system, as these vary greatly (Graham et al., 2015). Monitoring weight loss and clinical score over the time course of infection gives immediate insight into the severity of infection, and is also necessary from an ethics standpoint to determine if mice should be removed from the experiment before the anticipated endpoint. In mice, signs of illness include weakened hind limbs, hunched posture, rough or ungroomed fur, decreased activity, labored breathing, weight loss, and decrease in body condition score.
Materials
Infected mouse (from Basic Protocol 1)
Balance and animal weigh boat/container
Data sheet for recording weight, clinical score, and any notes
Data analysis/statistical software (GraphPad Prism)
Hydrogel (ClearH2O)
-
Infect mice as described in Basic Protocol 1.
An age-matched control group (mock infected, with PBS only) should also be included for comparing normal weight gain over the time course of the experiment. -
Record animal weight on day of infection (day 0). Record weights daily or as desired/required by institutional animal regulations.
Mice should be weighed within a consistent time period each day, as they will weigh more in the morning after eating during the night/dark cycle. -
Track progress of infection using a clinical scoring system to monitor mouse health and recovery. For West Nile virus infection, suggested clinical scoring criteria are as follows:
0 Healthy Mouse (baseline)
1 Ruffled fur, lethargy, hunched posture, no paresis, normal gait
2 Altered gait, limited movement in 1 hind limb
3 Lack of movement, paralysis in 1 or both hind limbs
4 Moribund
-
If weight loss greater than 10% or a clinical score of 2 or greater is reached, gel should be provided in the cage, and mice should be monitored daily until they either recover or reach the specific criteria for euthanasia (paralysis, weight loss greater than 20%, etc.).
Euthanasia criteria varies by institution and individual animal protocol. At experimental endpoint, sacrifice mice to perform additional experiments and assays.
Clinical data analysis
Weight loss, clinical score, and survival curves can be created for each cohort in the experiment. Weight curves are generally expressed as percentage of starting weight, so that the d0 weight is 100% and all subsequent dates reference weight loss compared to d0. Clinical score can be graphed on the 0–4 scale, while survival curves are best displayed in a Kaplan-Meier curve(Rich et al., 2010). Additional statistical analysis can reveal differences between different cohorts at a given time point post-infection.
BASIC PROTOCOL 3. TISSUE HARVEST AND PROCESSING
To measure immune phenotypes in both the periphery and the CNS, lymphocytes are isolated from tissues of interest. Here we outline protocols for preparing single-cell suspensions from both spleen and brain that may be used for downstream flow cytometry analysis. The dLN (popliteal) could be assayed in a similar manner to the spleen. This protocol relies on mechanical homogenization of the CNS rather than the use of collagenase or other digestive enzymes to avoid cleaving any cell surface markers that would be needed in flow cytometry assays.
Materials
Dissection instruments (scissors, forceps, etc.)
70% ethanol
Absorbent disposable pads
200 μL pipette and filter tips
2mL Eppendorf tubes
15mL conical tubes
30mL syringe
18G1½ needles
Complete RPMI media (see recipe below)
PBS
6 well plates
cell strainers (70μm)
frosted slides
hypertonic Percoll (see recipe below)
0.5% FACS buffer (see recipe below)
70mm filter top FACS tubes
Harvest
-
1
Euthanize mice with CO2 for 5 minutes.
-
2
Mouse prep: Cover work surface with absorbent disposable pad. Lay mouse on back; spray thoroughly with 70% ethanol. Using forceps, lift up the skin just above the urethral opening and use scissors to cut along the midline up to the xyphoid. Use scissors to cut up the sternum and and expose the chest cavity.
Lift up gently with forceps while cutting to avoid nicking any internal organs. -
3
Blood collection: Sever hepatic artery using scissors. Using a 200uL pipette with filter tip, collect pooled blood and transfer to labeled eppendorf tube. Be careful of clots plugging the pipette tip. Set aside at room temperature for later processing.
We prefer this method for blood collection as it is efficient and leaves the heart intact for perfusion in upcoming steps, which is necessary to remove circulating blood from the brain before collection. -
4
Spleen removal: Cut skin further down the right side (the mouse’s left side) just below the rib cage for better visualization. Gently lift spleen using tweezers, severing connections and any fat attached using scissors. Place spleen in prepared 15mL conical vial containing 5mL of sterile PBS and place on ice until ready for further processing.
-
5
Perfusion: Cut the rib cage so you can visualize the heart, being careful not to nick the heart or arteries. Prepare a 30mL syringe with an 18-G, 1½-inch needle filled with sterile PBS. Insert needle into the left ventricle of the heart. Do not pierce all the way through the heart. Slowly depress the plunger, allowing approximately 10mL of PBS to perfuse through the mouse. This may be difficult to start depending on the level of clotting that has already occurred.
-
6
Brain removal: While holding the mouse by the head, remove the head by cutting at the base of the skull. Peel skin from skull, pulling towards the front of head (do not need to remove completely). To cut the skull, insert scissors at the base of the skull and cut around the sides towards the face being careful to cut the brain as little as possible. Lift up skull flap. If it does not lift easily, cut further towards the front of the skull. Place tweezers under the brain, removing the brain to a prepared 15mL conical vial containing RPMI complete media. Place on ice until ready for further processing.
Brain processing
-
7
Invert tube with brain 2–3 times before pouring contents of tube into one of the wells on a 6-well plate.
-
8
Dissociate brain using frosted microscope slides, crushing the tissue between the slides by holding them in parallel.
Repeat until all tissues have been reduced to a small enough size to be pipetted using a 5mL pipette. -
9
Using a 5mL pipette, mix up and down gently a couple times in the 6-well plate before returning to the 15mL tube.
-
10
Wash well of 6-well plate with 600μL RPMI complete and add to 15mL tube. Return tube to ice while processing additional samples.
-
11
Once all samples have been disassociated and returned to the conical tubes, add 2.4mL hypertonic Percoll (see recipe) to sample, bringing the final volume to 8mL and resulting in a 30% percoll solution. Vortex sample to ensure the solution is fully mixed.
-
12
Centrifuge brain samples over the Percoll gradient at 350xg for 30 minutes at 4°C.
-
13
Aspirate supernatant to remove as much debris as possible without disturbing the cell pellet.
Additional neuronal tissue may stick to the sides of the tube, in which case the cell suspension can be transferred to a new tube to avoid re-introducing this material. -
14
Resuspend cells in 2mL of 0.5% FACS buffer.
-
15
Pass cells through 70μm filter top FACS tubes using a 1mL pipette.
Pipette slowly as any remaining neuronal tissue can quickly clog the filter. Depending on the clarity of the sample, this may require pipetting up and down across free space in the filter. -
16
Dilute 10uL of cells in 90μL trypan blue to count using a hemacytometer
-
17
Centrifuge cells at 500xg for 5 minutes at 4°C. Decant and remove excess supernatant using a 1mL pipette, being careful to not disturb the cell pellet. Resuspend cells at 10 million cells per mL in RPMI complete media. Place cells on ice until ready for use.
Spleen processing
-
18
Prepare 6-well plates with 70μm strainers in each well.
-
19
Invert tube several times so that the spleen is in motion before pouring contents of tube into cell strainer.
-
20
Using a sterile plunger, mash spleen through the cell strainer until no pieces remain. Discard strainer and plunger into bleach.
Several types of mechanical disruption can be used to create single-cell suspensions, including the use of frosted slides as in the brain processing above. We find that 70μm filters remove the greatest amount of fascia and connective tissue while maintaining high cell numbers. -
21
Use a 5mL pipette to transfer the cell suspension back to the 15mL tube, gently pipetting up and down before transferring, and place on ice until all samples are processed.
-
22
Centrifuge cell suspension at 500xg for 5 minutes at 4°C.
-
23
Decant supernatant, then briefly vortex cell pellet.
-
24
Add 3mL ACK lysis buffer to the cell suspension, mix well and incubate at room temperature for 3 minutes.
-
25
Add approximately 10mL PBS to stop the reaction, capping and then inverting tubes to ensure complete mixing.
-
26
Centrifuge cell suspension at 500xg for 5 minutes at 4°C.
-
27
Decant and remove excess supernatant using a 1mL pipette, being careful to not disturb the cell pellet.
-
28
Resuspend cell pellet in 1mL PBS, pipetting up and down to mix. Remove any additional clumps of cell debris and connective tissue that do not go into solution upon mixing by using the 1mL pipette. Add 9mL PBS to bring the final volume to 10mL.
Optionally, perform a second round of isolation by passing the cell suspension over 70μm strainers to 6-well plates. This may be helpful to remove any smaller pieces of tissue and debris that have aggregated together following ACK lysis. -
29
Dilute 10μL cell suspension in 90μL trypan blue and count cells using a hemacytometer.
-
30
Centrifuge cells at 500xg for 5 minutes at 4°C. Decant and remove excess supernatant using a 1mL pipette, being careful to not disturb the cell pellet. Resuspend cells at 10 million cells per mL in RPMI complete. Place cells on ice until ready for use in downstream flow cytometry assays (cell viability may decrease after 6+ hours).
Blood Processing
-
31
After allowing blood to clot at room temperature, centrifuge blood in table top centrifuge at 1000xg for 10 minutes at room temperature.
-
32
Using either a 20μL or 200μL pipette (depending on the amount of blood collected), remove the serum from on top of the clotted blood. Pipette up slowly as to not disturb the clotted blood. The serum should be fairly clear.
-
33
Place the recovered serum into a new labeled Eppendorf tube and place in the −80°C freezer until ready for further use in WNV-specific ELISAs or other assays.
BASIC PROTOCOL 4. Flow cytometry phenotyping for identification of immune subsets, chemokines, and cytokines
The use of multicolor flow cytometry panels has allowed us to characterize the immune response to West Nile virus infection in both tissues and the CNS. Here we outline protocols for an extensive analysis of T cell subsets, activation markers, chemokine receptors, and cytokine production. For intercellular cytokine staining (ICS), the CD8 immumodominant NS4b peptide, as well as heat-inactivated WNV (HI-WNV) are used to re-stimulate cells ex vivo. The use of HI-WNV allows us to examine the breadth of the T cell response more fully than looking at only the T cell cytokine response to an immunodominant peptide, which is especially helpful in mouse strains beyond the C57BL/6 background.
Materials
Single-cell suspensions (see Basic Protocol 3)
Cytokine stimulations
0.5% FACS buffer (see Reagents)
Intracellular Fixation/Permeabilization Buffer set (Ebioscience, 88-8824-00)
Brefeldin A Solution (1000x) (Ebioscience, 00-4506-51)
Live/dead fixable Aqua stain (Ebioscience, L34957)
Antibodies
ICS setup
-
1
For each sample and organ, there will be four different stimulations:
DMSO (negative control)
NS4b peptide (FHCRC Shared Resources)
HI-WNV
aCD3/CD28 (positive control, polyclonal stimulus)
Prepare 100μL of cells (10×106cells/ml) + 100μL stimulations per well in a 96-well U-bottom plate.
-
2
Prep 100μL stims per sample:
-
Negative control: 10μg/mL DMSO + 1:500 1000X BFA + RPMI complete to bring volume to 100μL/sample
DMSO is used as a negative control because the peptide is prepared in DMSO. If no peptide stimulation is used, the negative control would be media. NS4b peptide: 10 μg/mL NS4b peptide + 1:500 1000X BFA + RPMI complete to bring volume to 100μL/sample
Positive control: 1μg/well CD3 + 500ng/well CD28 + 1:500 1000X BFA + RPMI complete to bring volume to 100μL/sample
Heat inactivated WNV (HI-WNV): MOI=5 + RPMI complete to bring volume to 100μL/sample
-
-
3
Place the negative control, NS4b peptide, and positive control stimulations on the same plate. Once both the cells and stims have been plated, place in an incubator at 37°C and 5% CO2 for 5 hours. Plate the HI-WNV stimulated samples on a separate plate and place in an incubator at 37°C and 5% CO2 overnight.
-
4
After 5 hours remove the plate containing the negative control, NS4b peptide, and positive control from the incubator, wrap in foil, and place in a 4°C refrigerator overnight until ready for staining.
-
5
Following overnight incubation at 37°C and 5% CO2, add BFA to the HI-WNV stimulation samples:
1:200 1000X BFA + RPMI complete to bring volume per sample to 50uL
Add 50μL BFA solution to each well. Place plate back in the incubator for 4 hours at 37°C and 5% CO2.
Flow cytometry staining
The flow cytometry staining protocol below can be performed on cells following ICS stimulation, or directly ex vivo.
-
6
All of the staining should be done in the dark. Centrifuge stimulated cells in 96-well U-bottom plates at 500xg for 3 minutes at 4°C.
-
7
Decant by flicking out the supernatant into a bleach bucket within the biosafety hood and gently tap the excess liquid once on a paper towel.
-
8
Add 200μL PBS per well to wash the excess RPMI complete off the cells.
This step is important to remove any remaining protein that may interfere with the live/dead stain. -
9
Centrifuge cells at 500xg for 3 minutes at 4°C.
-
10
Prepare live/dead stain. Dilute resuspended Aqua live/dead stain 1:1000 in PBS. Prepare enough for 100uL per well.
-
11
Decant cells and resuspend in 100uL of the prepared live dead stain. Gently pipette up and down, trying to avoid making bubbles. Incubate on ice for 30 minutes with foil covering the plate.
-
12
Prepare Fc block. Dilute CD16/32 1:500 in FACS buffer, making enough for 50μL per sample.
-
13
After 30 minutes, add 100μl of PBS to each well and centrifuge at 500xg for 3 minutes at 4°C.
-
14
Decant and resuspend cells in 50μL Fc block by pipetting gently up and down. Place on ice for 10 minutes covered in foil.
-
15
During the incubation, prepare the extracellular antibodies. Add the correct dilution of each antibody in FACS buffer, making enough for 50μL per well.
-
16
Add 150μL FACS buffer to bring the final volume to 200μL. Centrifuge the cells at 500xg for 3 minutes at 4°C.
-
17
Decant and resuspend cells by gently pipetting up and down in 50μL of prepared antibodies. Incubate on ice for 15 minutes.
-
18
Wash cells 1X in FACS buffer, and centrifuge the cells at 500xg for 3 minutes at 4°C.
-
19
Resuspend cells in 200μl of Ebioscience Fixation/Permeabilization solution for at least 30 minutes, or overnight.
Cells can remain in fix/perm overnight (~18 hours) without increases in cell death, making this a good stopping point if needed during the staining process. -
20
Prepare 1x Perm Buffer (from 10X Perm Buffer in Ebioscience kit). Prepare at least 200μl per sample. Prepare the intracellular antibodies, adding the correct dilution of each antibody in 1X Perm buffer, making enough for 50μL per well.
-
21
Centrifuge samples in 200μl fixation/permeabilizaiton solution at 900xg for 2 minutes at 4°C. Decant and resuspend cells in 50μl of prepared intraceullar antibodies. Incubate on ice for 30 minutes.
-
22
Add 150μl 1X Perm Buffer to each well, and centrifuge samples at 900xg for 2 minutes at 4°C.
-
23
Decant, resuspend cells in 200μl of PBS for flow cytometry acquisition and analysis.
REAGENTS AND SOLUTIONS
ACK Lysis Buffer
1 L Deionized water
8.29 g NH4Cl
1 g KHCO3
37.2 mg Na2-EDTA
**Adjust pH of solution if needed(using a strong acid or base) to 7.2–7.4, sterilize using 0.2 μm filter and store at 4°C
10% RPMI complete media
50 mL FBS (heat inactivated)
5 mL L-Glutamine (200mM, 100X)
5 mL pen-strep (10,000 units/ml each)
5 mL Sodium Pyruvate (100mM)
5 mL HEPES (1M)
430 mL RPMI
0.5% FACS buffer
2.5 mL FBS (heat inactivated)
497.5 mL 1X PBS
1:10 Percoll
9 parts Percoll (GE Life Sciences, Catalog #17-0891-01)
1 part 10X PBS
Pre-stains
Live/dead-Aqua (Invitrogen, Catalog #L34957)
CD16/32 (Ebioscience, Catalog #E0359-1630)
ICS stims
DMSO
1000X Brefeldin A (Ebioscience, Catalog #00-4506-51)
NS4b peptide (Fred Hutchinson Cancer Research Center Immune Monitoring Core)
Heat-inactivated WNV (heat inactivated at 57°C for 1 hr)
CD3e (Ebioscience, Catalog #16-0031-85)
CD28 (Ebioscience, Catalog #16-0281-85)
COMMENTARY
Background Information
WNV is an ssRNA virus that cycles between mosquitos and birds, with humans and other mammals serving as incidental hosts. Approximately 20% of infected individuals experience a limited febrile illness, with 1% developing a more severe neuroinvasive disease characterized by encephalitis and meningitis (Diamond, Mehlhop, Oliphant, & Samuel, 2009). However, there is limited knowledge available regarding the immune response to WNV in humans due to the high prevalence of subclinical infection that hinders identification of WNV-infected individuals and follow-up clinical and immune response evaluations. Thus, most knowledge of anti-WNV immunity must come from the study of mouse models of WNV infection.
WNV likely traffics from the periphery to the CNS via axonal spread or by a hematogenous route across the blood–brain barrier (Samuel, Wang, Siddharthan, Morrey, & Diamond, 2007). The generation of immune responses within the CNS are critical to clear the virus; however, at the same time, they must be regulated such that damage to non-renewing populations of neurons is limited. Postinfection, CD8+ T cells migrate to the brain, and their presence correlates with viral clearance (T. Wang et al., 2003). Regulatory T cells (Tregs) can negatively regulate T cell cytokine function in response to virus infection in the brain, but are also necessary to generate a pool of resident memory T cells with which to protect the host from reinfection (Graham, Da Costa, & Lund, 2014).
In order to develop a more complete understanding of both the clinical disease course and immune response to WNV infection, studies in mouse models of infection can further probe immune correlates of protection, particularly in strains of mice that model a wider range of phenotypes that recapitulates human symptoms and disease outcomes (Graham et al., 2015; Kumar et al., 2014). Through extensive flow cytometry analysis, as well as other immunological assays, researchers can further elucidate these correlates of protection from neuroinvasive viral infections to inform therapeutic targets and vaccine strategies in the human population.
Critical Parameters
Control groups
The use of proper control groups is crucial for the clinical measurements of WNV disease. Weight loss during WNV infection is measured as an output of clinical disease, in relation to each mouse’s initial body weight, but a control group of uninfected mice should also be weighed to establish normal weight gain over the time course of the study. This weight gain will vary between strains of mice, so a control group will be needed for each strain in the study.
Optimum reagents
Fresh media preparations (<2 weeks old) should be used in all studies involving ICS staining to ensure maximal cell viability during the longer incubations. For flow cytometry staining, the fixation/permeabilization solution and permeabilization buffer should be prepared fresh at the time of each experiment. We have found that our tetramer staining is optimized with fresh reagent every 6 months.
Troubleshooting
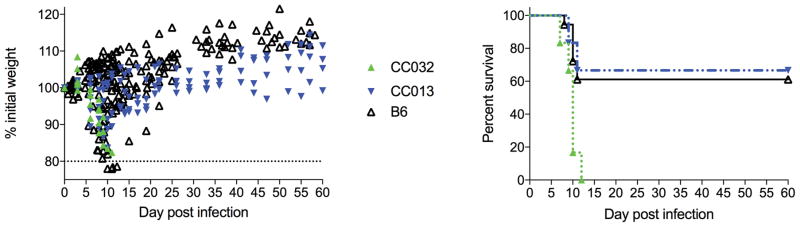
While we have provided a timeline of the typical course of West Nile virus infection in the B6 model, clinical disease will vary by strain (Figure 1). When beginning new studies, a kinetics experiment to examine the response over a time course is advised to establish time points for maximum response in different tissues. To maximize cell viability throughout the course of the experiments, all tissues and cell suspensions should be kept on ice unless otherwise noted. The use of multi-color flow cytometry panels inherently will lead to increased challenges in compensating the flow data. We suggest initially performing FMO (fluorescence minus one) to establish the background in each channel. Additionally, single stain controls should be performed for each experiment, to be used to create the compensation matrix for analysis. Running single stain controls in each experiment also allows the investigator to monitor laser voltages over time, which can help address problems related to the flow cytometer itself.
Figure 1.
Cohorts of mice were infected with 100 pfu WNV-TX as described in Basic Protocol 1. Collaborative cross strains CC032 and CC013 were studied, as well as B6. Percent of initial weight (A) and survival (B) was monitored out to d60 post infection.
Understanding Results
Clinical disease course
In a C57BL/6 mouse model, mice infected by subcutaneous footpad injected with 100 pfu WNV-TX generally lose body weight between days 8–14 post-infection, and may also see an increase in clinical score. Surviving mice begin to recover weight loss 10–14 days after infection, typically returning to starting weight by day 14–18 after infection. This disease course will vary by mouse strain (Figure 1).
Example flow cytometry staining
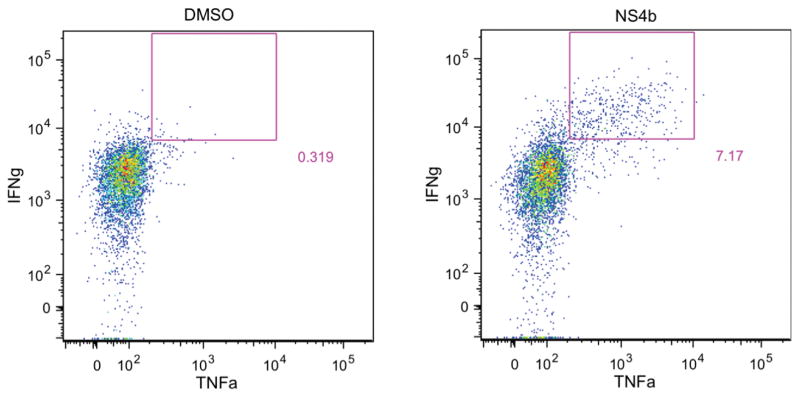
Lymphocyte isolation established in these protocols will provide sufficient cells for multiple flow cytometry panels per sample, including lymphocytes from the brain. This is especially important for intracellular cytokine staining, where negative control samples are necessary to provide the background level of non-specific staining inherently present in the assay. For the data shown in Figure 2, C57BL/6 mice were infected s.c. in the footpad with 100pfu WNV-TX, and brain was harvested 12 days post-infection. Single-cell suspensions were prepared and restimulated prior to staining for flow cytometry analysis. In this example, lymphocytes are gated on CD8+ cells, and the gate denotes IFNg+ TNFa+ cells after either DMSO (left) or peptide (right) stimulation.
Figure 2.
Intracellular cytokine staining of brain cells 12d after WNV infection. C57BL/6 mice were infected s.c. in the footpad with 100pfu WNV-TX, and brain was harvested 12 days post-infection as previously described. Single cell suspensions were prepared and restimulated prior to staining for flow cytometry analysis. Cells are gated on CD8+ cells, and gate denotes IFNg+ TNFa+ cells after either DMSO (left) or peptide (right) stimulation.
Time Considerations
Upon arrival to the investigator’s colony (if ordered from an outside institution), mice should be allowed to acclimate for one week in their new environment before infection. Time must be allotted for weight measurements and clinical scoring over the time course of infection, and this should be done at around the same time each day to avoid variations due to the normal fluctuation of mice eating during the dark hours.
For the harvest and processing of tissues, the number of mice which can be included in an experiment will be dependent on the number of personnel available to work on the study. It is especially important that the brain samples be processed within a few hours of harvest, so ideally the harvest itself would not take more than 2–3 hours.
When processing samples from both the periphery and the CNS, brain samples should be processed first due to the more delicate nature of the tissue. Additionally, since the Percoll centrifugation step is 30 minutes, spleen or other tissues can be homogenized and further processed in this time frame. Once the cells are in the incubator starting the clock on the 5 hour incubation with BFA, researchers can then proceed with other flow cytometry stainings directly ex vivo.
Table 1.
Suggested Flow Cytometry Staining Panels
| Treg panel | T cell ICS | CD8 T cell panel | |
|---|---|---|---|
| BV605 | CD4 (Biolegend, Cat.#100547) | CD4 (Biolegend, Cat.#100547) | CD4 (Biolegend, Cat.#100547) |
| APC Cy7 | CD29 (Ebioscience, Cat. #47-0291-82) | CD25 (Ebioscience, Cat. #47-0251-82) | CD44 Ebioscience, Cat. #47-0441-82 |
| PerCP eFluor710 (PerCP-Cy5.5) | CXCR3 (Ebioscience, Cat. #45-1831-82) | * IFNg (Ebioscience, Cat. #45-7311-82) | CXCR3 (Ebioscience, Cat. #45-1831-82) |
| FITC | CD44 (Biolegend, Cat. #103006) | * IL-17 (Biolegend, Cat. #506908) | * Ki67 (Ebioscience, Cat. #11-5698-82) |
| PE | CCR5 (BD, Cat. #555306) | CD162/PSGL-1 (BD, Cat. #555306) | CCR5 (Biolegend, Cat. #107006) |
| BUV395 | * CD3 (BD, Cat. #740268) | * CD3 (BD, Cat. #740268) | |
| Ax700 | * Foxp3 (Ebioscience, Cat. #56-5773-82) | CD62L (Ebioscience, Cat. #56-0621-82) | CD62L (Ebioscience, Cat. #56-0621-82) |
| PECy7 | GITR (Ebioscience, Cat. #25-5874-82) | * Tbet (Ebioscience, Cat. #25-5825-82) | KLRG-1 (Ebioscience, Cat. #25-5893-82) |
| PECy5 | ICOS (Ebioscience, Cat. #15-9942-82) | CD44 (Ebioscience, Cat. #15-0441-83) | CD127 (Ebioscience, Cat. #15-1271-83) |
| APC | * CTLA-4 | * TNFa (Biolegend, Cat. #506308) | NS4b class I tetramer (FHCRC shared resources) |
| BV421 (Pac Blue) | CD73 (Biolegend, Cat. #127212) | Ly6C (Biolegend, Cat. #128014) | CCR7 (Ebioscience, Cat. #48-1971-82) |
| BV650 | CD25 (Biolegend, Cat. #102037) | CD8 (Biolegend, Cat. #100741) | CD8 (Biolegend, Cat. #100741) |
| AmCyan | Live/dead (Invitrogen, Cat.#L34957) | Live/dead (Invitrogen, Cat.#L34957) | Live/dead (Invitrogen, Cat.#L34957) |
denotes intracellular stain
Acknowledgments
Funding for this study was provided by National Institutes of Health grants U19AI100625 to JML.
LITERATURE CITED
- Carson PJ, Konewko P, Wold KS, Mariani P, Goli S, Bergloff P, Crosby RD. Long-term clinical and neuropsychological outcomes of West Nile virus infection. Clin Infect Dis. 2006;43(6):723–730. doi: 10.1086/506939. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J … Complex Trait, C. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36(11):1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Keller BC, Gale M, Jr, Diamond MS. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 2007;3(7):e106. doi: 10.1371/journal.ppat.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Suthar MS, Gale M, Jr, Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008;82(21):10349–10358. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Suthar MS, Keller BC, Gale M, Jr, Diamond MS. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J Virol. 2008;82(17):8465–8475. doi: 10.1128/JVI.00918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Suthar MS, Szretter KJ, Gale M, Jr, Diamond MS. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 2009;5(10):e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Mehlhop E, Oliphant T, Samuel MA. The host immunologic response to West Nile encephalitis virus. Front Biosci (Landmark Ed) 2009;14:3024–3034. doi: 10.2741/3432. [DOI] [PubMed] [Google Scholar]
- Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol. 2013;87(21):11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried K, Quinn R, Jones T. Clinical description and follow-up investigation of human West Nile virus cases. South Med J. 2005;98(6):603–606. doi: 10.1097/01.SMJ.0000155633.43244.AC. [DOI] [PubMed] [Google Scholar]
- Graham JB, Da Costa A, Lund JM. Regulatory T cells shape the resident memory T cell response to virus infection in the tissues. J Immunol. 2014;192(2):683–690. doi: 10.4049/jimmunol.1202153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JB, Swarts JL, Wilkins C, Thomas S, Green R, Sekine A, … Lund JM. A Mouse Model of Chronic West Nile Virus Disease. PLoS Pathog. 2016;12(11):e1005996. doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JB, Thomas S, Swarts J, McMillan AA, Ferris MT, Suthar MS, … Lund JM. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio. 2015;6(3):e00493–00415. doi: 10.1128/mBio.00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EB, Franke MA, Wong SJ, Kramer LD. Detection of West Nile virus. Methods Mol Biol. 2011;665:383–413. doi: 10.1007/978-1-60761-817-1_21. [DOI] [PubMed] [Google Scholar]
- Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M., Jr Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol. 2006;80(19):9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee AL, Maidin B, Edwin B, Poshni I, Mostashari F, Fine A, … Nash D. Long-term prognosis for clinical West Nile virus infection. Emerg Infect Dis. 2004;10(8):1405–1411. doi: 10.3201/eid1008.030879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Roe K, Nerurkar PV, Orillo B, Thompson KS, Verma S, Nerurkar VR. Reduced immune cell infiltration and increased pro-inflammatory mediators in the brain of Type 2 diabetic mouse model infected with West Nile virus. J Neuroinflammation. 2014;11:80. doi: 10.1186/1742-2094-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis AA, Stokic DS, Webb RM, Slavinski SA, Fratkin J. Clinical spectrum of muscle weakness in human West Nile virus infection. Muscle Nerve. 2003;28(3):302–308. doi: 10.1002/mus.10440. [DOI] [PubMed] [Google Scholar]
- Marciniak C, Sorosky S, Hynes C. Acute flaccid paralysis associated with West Nile virus: motor and functional improvement in 4 patients. Arch Phys Med Rehabil. 2004;85(12):1933–1938. doi: 10.1016/j.apmr.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Rich JT, Neely JG, Paniello RC, Voelker CC, Nussenbaum B, Wang EW. A practical guide to understanding Kaplan-Meier curves. Otolaryngol Head Neck Surg. 2010;143(3):331–336. doi: 10.1016/j.otohns.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007;18(6–7):473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Wang H, Siddharthan V, Morrey JD, Diamond MS. Axonal transport mediates West Nile virus entry into the central nervous system and induces acute flaccid paralysis. Proc Natl Acad Sci U S A. 2007;104(43):17140–17145. doi: 10.1073/pnas.0705837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, … Diamond MS. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol. 2006;80(14):7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejvar JJ. The long-term outcomes of human West Nile virus infection. Clin Infect Dis. 2007;44(12):1617–1624. doi: 10.1086/518281. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Diamond MS, Gale M., Jr West Nile virus infection and immunity. Nat Rev Microbiol. 2013;11(2):115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, … Gale M., Jr IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6(2):e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. Ten years of the collaborative cross. G3 (Bethesda) 2012;2(2):153–156. doi: 10.1534/g3.111.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Miller DR, Churchill GA, de Villena FP. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR J. 2011;52(1):24–31. doi: 10.1093/ilar.52.1.24. [DOI] [PubMed] [Google Scholar]
- Wang H, Siddharthan V, Hall JO, Morrey JD. Autonomic nervous dysfunction in hamsters infected with West Nile virus. PLoS One. 2011;6(5):e19575. doi: 10.1371/journal.pone.0019575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Scully E, Yin Z, Kim JH, Wang S, Yan J, … Fikrig E. IFN-gamma-producing gamma delta T cells help control murine West Nile virus infection. J Immunol. 2003;171(5):2524–2531. doi: 10.4049/jimmunol.171.5.2524. [DOI] [PubMed] [Google Scholar]