Abstract
Hematopoietic differentiation is driven by transcription factors, which orchestrate a finely tuned transcriptional network. At bipotential branching points lineage decisions are made, where key transcription factors initiate cell type-specific gene expression programs. These programs are stabilized by the epigenetic activity of recruited chromatin-modifying cofactors. An example is the association of the transcription factor RUNX1 with protein arginine methyltransferase 6 (PRMT6) at the megakaryocytic/erythroid bifurcation. However, little is known about the specific influence of PRMT6 on this important branching point. Here, we show that PRMT6 inhibits erythroid gene expression during megakaryopoiesis of primary human CD34+ progenitor cells. PRMT6 is recruited to erythroid genes, such as glycophorin A. Consequently, a repressive histone modification pattern with high H3R2me2a and low H3K4me3 is established. Importantly, inhibition of PRMT6 by shRNA or small molecule inhibitors leads to upregulation of erythroid genes and promotes erythropoiesis. Our data reveal that PRMT6 plays a role in the control of erythroid/megakaryocytic differentiation and open up the possibility that manipulation of PRMT6 activity could facilitate enhanced erythropoiesis for therapeutic use.
Introduction
Hematopoietic lineage decisions are driven by transcription factors, which define cell type-specific gene expression and thus instruct lineage specification during terminal differentiation. A subset of transcription factors is important for hematopoietic stem cell emergence and also for later lineage-specific gene expression.1–5 For multilineage differentiation processes, such as hematopoiesis, epigenetic stabilization of gene expression programs is of central importance. In this process of epigenetic gene regulation, transcription factors recruit cofactors with enzymatic activity to target genes. These cofactors are able to change chromatin organization by modification.6,7 The most prominent epigenetic modifications are methylation of DNA on cytidines and a large number of different posttranslational modifications of histones. These interdependent modifications mostly take place at the histone tails and comprise a pattern which can encode distinct functions.6,8,9 The important function of transcriptional regulators in hematopoiesis is highlighted by the observation that alterations of transcriptional regulators can convert one cell type into another.10–15
At the megakaryocytic/erythroid lineage bifurcation transcription factors such as RUNX1, FLI1, KLF1, GATA1 and TAL1 play a decisive role in the establishment of the megakaryocytic or erythroid gene expression program, respectively.16–21 The transcription factor RUNX1 (also known as AML1: acute myeloid leukemia 1) plays a major role in hematopoietic stem cell emergence.22,23 Furthermore, RUNX1 is important for the establishment of the megakaryocytic gene expression program and the repression of erythroid genes.24–28 Depending on the associated cofactors RUNX1 can act as an activator or repressor of gene expression. Importantly, RUNX1 cooperates with central epigenetic complexes such as the trithorax-(MLL)-complex and the polycomb-(PRC)-complex, which trigger the activating trimethylation of lysine 4 on histone 3 (H3K4me3) and the repressive trimethylation of lysine 27 on histone 3 (H3K27me3), respectively.29–32 RUNX1 also interacts with protein arginine methyltransferase 6 (PRMT6).33,34 PRMT6 is a member of the PRMT-family, which consists of enzymes that methylate arginine residues on proteins, including histones.35 PRMT6-mediated asymmetric dimethylation of histone 3 at the arginine at position 2 (H3R2me2a) counteracts the activating H3K4me3, thus PRMT6 acts predominantly as a repressor of gene expression.36–38 It has been demonstrated that PRMT6 has an influence on embryonic stem cell identity.39 In megakaryocytic/erythroid progenitors, PRMT6 is recruited by RUNX1 to target genes and acts as a repressor by setting the H3R2me2a mark. This way RUNX1/PRMT6 contribute to the establishment of bivalent chromatin marking at megakaryocytic differentiation genes, such as CD41, in progenitors.33
Despite the evident role of PRMT6 at megakaryocytic/erythroid branching, its cell type-specific function has not been studied in detail. In light of the notion that small molecule inhibition of epigenetic enzymes could influence in vitro differentiation it is instructive to study the biological processes mediated by PRMT6. We found that decreased PRMT6 activity in primary human CD34+ progenitor cells leads to increased in vitro erythroid differentiation, whereas overexpression of PRMT6 decreases erythroid differentiation. During megakaryocytic differentiation of progenitor cells PRMT6 contributes to the suppression of erythroid genes by establishment of a repressive chromatin environment. Interestingly, PRMT6 inhibition by a small molecule also enhances erythropoiesis. This opens up the possibility of using PRMT6 inhibitors for more effective in vitro differentiation of erythrocytes.
Methods
Cell culture
K562 (ATCC CCL-243) and HEK293T/17 (ATCC CRL-11268) cells were cultured in RPMI-1640 and DMEM medium, respectively. Growth media were supplemented with 10% fetal calf serum, 2 mM glutamine and 1% penicillin/streptomycin. For megakaryocytic differentiation K562 cells were treated with 30 nM 12-o-tetradecanylphorbol-13-acetate (TPA; Sigma, Darmstadt, Germany). The cells were harvested after 3 days and analyzed using flow cytometry.
Samples of granulocyte colony-stimulating factor mobilized peripheral or bone marrow human primary CD34+ cells from healthy donors were used, with approval of the ethics committee (permit #329-10). CD34+ cells were immunomagnetically enriched according to the manufacturer’s instructions (Miltenyi, Bergisch Gladbach, Germany) and expanded under serum-free conditions using Stem Span (SFEMI, Stemcell Technologies, Vancouver, Canada) as described previously.27,28,33 The cells were then subjected to erythroid or megakaryocytic differentiation.33,40 After 6 days the differentiation status was determined by fluorescence activated cell sorting (FACS) and cells were used for mRNA analysis or chromatin immunoprecipitation (ChIP). For overexpression and knockdown experiments expanded cells were transduced with lentiviral vectors. Transduced GFP+ cells were sorted and subsequently subjected to colony-forming unit (CFU) assay in methylcellulose, according to the manufacturer’s instructions (Miltenyi, Bergisch Gladbach, Germany). Colonies were counted 12 days after seeding. For erythroid-megakaryocytic differentiation in liquid culture, isolated bone marrow CD34+ cells were maintained in serum-free expansion medium SFEMII (Stemcell Technologies, Vancouver, Canada) supplemented with 100 ng/mL stem cell factor, 10 ng/mL interleukin-3, 10 ng/mL interleukin-6, 0.5 U/mL erythropoietin and 50 ng/mL thrombopoietin. Differentiation was verified by FACS and mRNA analysis. The PRMT6 inhibitor MS023 was obtained from Biomol (Hamburg, Germany).
Chromatin immunoprecipitation
Cell lysates and the ChIP assay were performed according to the X-ChIP protocol from Abcam, with modifications. For immunoprecipitation 3–10 μg of specific antibody were used. ChIP DNA was purified using DNA purification columns ChIP DNA Clean and Concentrator (Zymo Research, Irvine, USA) and analyzed by quantitative polymerase chain reaction (PCR). DNA recovery was calculated as percentage of the input. Error bars represent the standard deviation from at least four determinations. Histone modification ChIP values were corrected for nucleosome density using ChIP values for histone 3 (H3). ChIP-ReChIP was performed as described previously.40 The sequences of primer pairs used for the ChIP-PCR analysis are available upon request. Antibodies used in this study are listed in the Online Supplementary Material.
Gene expression analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized using Omniscript reverse transcriptase (Qiagen). Quantitative reverse transcriptase PCR was performed using SYBR Green PCR Mastermix (Eurogentec, Luettich, Belgium). Relative amounts of mRNA were normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression values. Primer sequences are available upon request.
Knockdown constructs and vector information are given in the Online Supplementary Material. The gene expression array of shPRMT6 K562 cells was analyzed via the limma package of Bioconductor. Differentially expressed genes were filtered to a minimum of two-fold change and Benjamini-Hochberg corrected P-value <0.05 as previously described.40 Data were deposited in the GEO-Expression database, GSE92251. Further functional association of candidate genes was performed with the webtool DAVID using standard settings.41,42
Western blot analysis was performed as described in the Online Supplementary Material.
Statistics
Array data were processed as stated above. ChIP and quantitative reverse transcriptase PCR data were analyzed using PRISM software. The error bars represent the standard deviation from the mean. P values were calculated using the Student t-test from at least four determinations. P values <0.05 were considered statistically significant (*P<0.05; **P<0.01; ***P<0.001).
Results
PRMT6 inhibits erythropoiesis
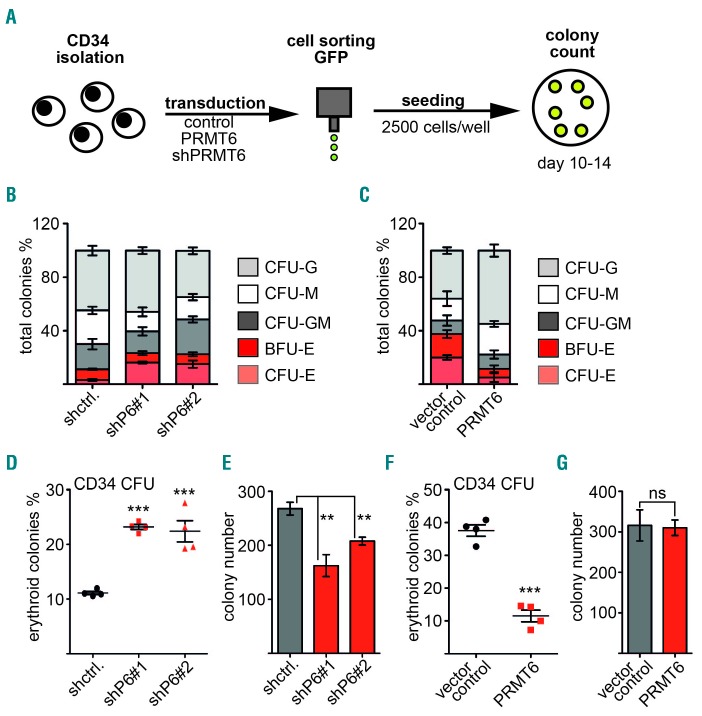
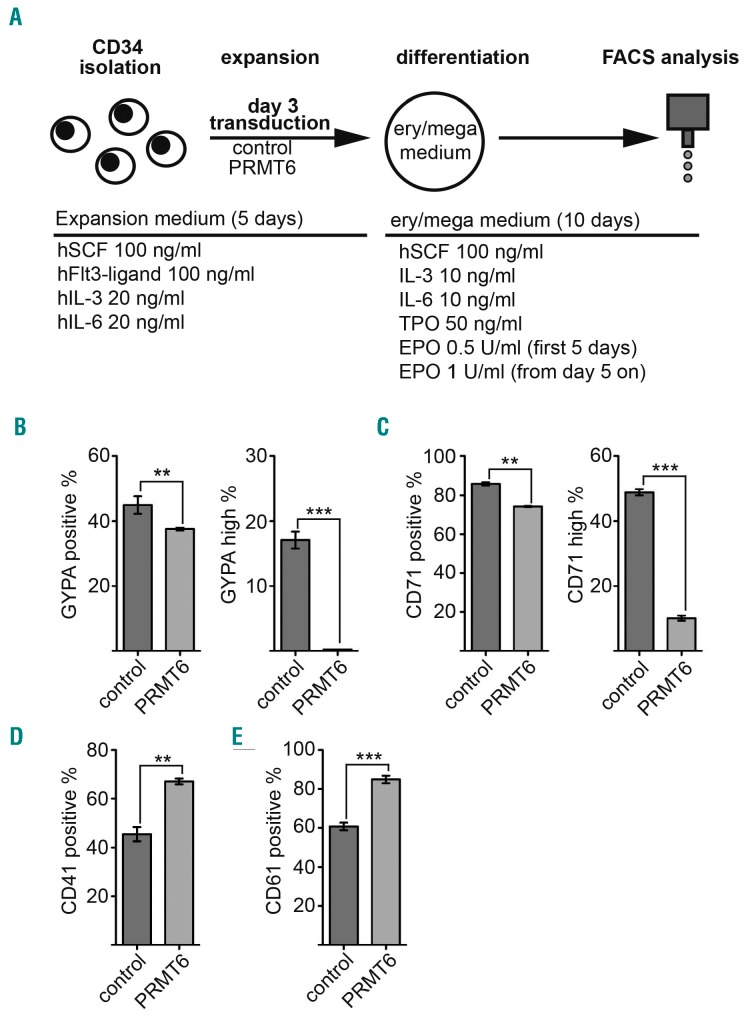
PRMT6 is associated with RUNX1 on megakaryocytic target genes in progenitor cells and present on erythroid genes upon megakaryocytic differentiation.28,33 This shows that PRMT6 plays a role in gene expression control at megakaryocytic/erythroid branching and might, therefore, influence differentiation. To explore this possibility, we performed a CFU assay. Human primary CD34+ cells were transduced with shRNA knockdown vectors and PRMT6 overexpression vectors, respectively (Online Supplementary Figure S1). Transduced cells were sorted and subjected to a CFU assay under conditions which allow myeloid differentiation, including erythropoiesis (Figure 1A). The knockdown of PRMT6 resulted in a decrease of the relative number of monocytic colonies, whereas the overexpression of PRMT6 increased the number of monocytic colonies as well as granulocytic colonies (Figure 1B,C). Interestingly, the knockdown of PRMT6 doubled the number of erythroid colonies (Figure 1D) and resulted in a modest reduction of total colony number (Figure 1E). In contrast, PRMT6 overexpression reduced erythroid colony formation (Figure 1F) and had no influence on colony number (Figure 1G). Under the conditions employed for the CFU assay, megakaryocytic differentiation could not be monitored. However, in a previous study we found that PRMT6 inhibits megakaryocytic genes in progenitor cells but leaves these promoters upon megakaryocytic differentiation.33 Thus, we wondered whether PRMT6 would alter erythroid/megakaryocytic differentiation under conditions that allow for both erythroid and megakaryocytic differentiation. To examine this, we transduced human CD34+ cells with a PRMT6 overexpression vector. Two days after transduction the cells were transferred to growth medium, which contained thrombopoietin and erythropoietin (Figure 2A).43 After 10 days of culture we measured the erythroid differentiation markers GYPA and CD71 by FACS. Furthermore, we determined the megakaryocytic differentiation markers CD41 and CD61. We found that the percentage of GYPA+ cells was about 40% in the control (Figure 2B, left) and 20% of all cells had high GYPA expression in the control (Figure 2B, right). Upon PRMT6 overexpression the number of GYPA+ cells was reduced. Moreover, the GYPAhi population was virtually absent upon PRMT6 overexpression (Figure 2B). Similarly, the number of CD71hi cells (another erythroid marker) was also reduced upon PRMT6 expression (Figure 2C). In contrast, the expression of the megakaryocytic markers, CD41 and CD61, was increased upon PRMT6 expression (Figure 2D,E). Corresponding flow cytometry data are shown in Online Supplementary Figure S2. Taken together, our data indicate that more mature erythroid cells with high GYPA and high CD71 expression are almost absent and the number of cells with megakaryocytic markers is increased upon PRMT6 overexpression. These data indicate that the cells have shifted from an erythroid to a megakaryocytic phenotype upon PRMT6 overexpression.
Figure 1.
PRMT6 inhibits erythroid differentiation. (A) Schematic workflow of the colony-forming unit (CFU) assay. Human CD34+ cells were transduced with PRMT6 knockdown (shPRMT6), PRMT6 expression, or control vector. Transduced GFP+ cells were sorted by FACS and subjected to the CFU assay. Colonies were counted on day 10–14 after seeding. (B, C) CFU assay of CD34+ cells upon PRMT6 knockdown and overexpression. Human CD34+ cells were transduced with PRMT6 knockdown vector (shPRMT6), PRMT6 expression vector, or control vector. Transduced GFP+ cells were sorted by FACS and subjected to a CFU assay. Colonies were counted 10–14 days after seeding. (B) CFU assay upon knockdown of PRMT6 using two different shRNA. Unspecific shRNA was used as a control. (C) CFU assay upon PRMT6 overexpression. Empty vector serves as the control. CFU-G colony-forming unit-granulocyte, CFU-M colony-forming unit-monocyte, CFU-GM colony-forming unit-granulocyte, monocyte, BFU-E burst forming unit-erythroid, CFU-E colony forming unit-erythroid. (D) Knockdown of PRMT6 using two different shRNA (shP6) enhances erythroid differentiation of CD34+ cells in the CFU assay. (E) The total number of colonies in the CFU assay after PRMT6 knockdown is shown. (F) The relative frequency of erythroid colonies (in percent) was decreased upon PRMT6 overexpression compared to the control. (G) The total number of colonies in the CFU assay after PRMT6 overexpression is shown. Error bars show the standard deviation calculated from at least four determinations. The P-values were calculated using the Student t-test. *P<0.05; **P< 0.01; ***P<0.001.
Figure 2.
PRMT6 overexpression alters erythroid/megakaryocytic differentiation (A) Schematic workflow of the differentiation analysis. Human CD34+ cells were isolated and cultured in SFEMI with the indicated supplements (expansion medium). At day 3 the cells were transduced with a PRMT6 expression vector or empty control vector, 2 days later the cells were transferred to erythroid/megakaryocytic differentiation medium (ery/mega medium). Ery/mega medium was SFEMII with the indicated supplements and 2 mM glutamine was also added. The transduced GFP+ cells were analyzed after 10 days in ery/mega medium by FACS. (B) The number of GYPA+ cells (left) and the number of GYPAhi cells was determined (right) with and without PRMT6 overexpression using a GYPA antibody (CD235a). (C) The number of CD71+ (left) and CD71hi (right) cells was determined. (D) The number of CD41+ cells is shown. (E) The number of CD61+ cells is shown. (B–E) Cell numbers are given in percent related to the total number of transduced GFP+ cells. Error bars give the standard deviation from four independent determinations. The P-values were calculated using the Student t-test. **P<0.01; ***P< 0001.
PRMT6 represses erythroid genes
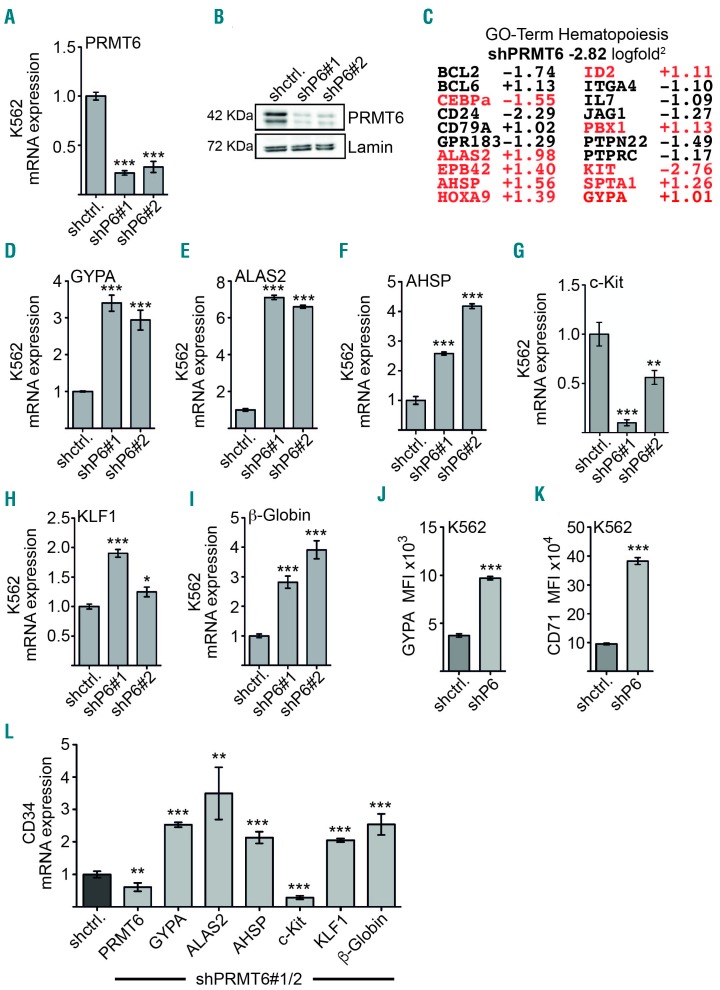
To investigate how PRMT6 influences erythroid differentiation, we analyzed gene expression downstream of PRMT6 upon knockdown of PRMT6. For this we used K562 erythroleukemia cells, which express GYPA and low levels of the erythroid master regulator KLF1. Gene expression analysis was studied by array analysis 5 days after transduction of shPRMT6 expression vectors (Figure 3A,B, Online Supplementary Figure S3). PRMT6 knockdown resulted in changed expression of more than 1,000 genes (Online Supplementary Figure S3). About half of the genes were upregulated and the other half downregulated upon PRMT6 knockdown (Online Supplementary Figure S3). Gene ontology analysis (GO-terms) using DAVID41,42 revealed that PRMT6 influences genes with distinct functions. The most significant GO-categories were “response to wounding” and “negative regulation of cell growth” (Online Supplementary Figure S3). The GO-category hematopoiesis was also enriched (Figure 3C). Ten of the 20 genes involved in hematopoiesis have a known function in erythropoiesis. Erythroid-specific genes were mostly upregulated. This includes ALAS2 (delta-aminolevulinate synthase 2), which plays a role in heme biosynthesis, AHSP (alpha hemoglobin stabilizing protein) and the erythroid differentiation marker GYPA (glycophorin A) (Figure 3C). CEBPα and c-Kit were downregulated upon PRMT6 knockdown (Figure 3C).
Figure 3.
Gene expression analysis upon PRMT6 knockdown. (A, B) K562 cells were transduced with two different shRNA constructs against PRMT6 (shP6) and the knockdown was evaluated by quatitative reverse transcriptase q-RT-PCR and western blot. (C) Gene expression array analysis was performed with shPRMT6 K562 cells 5 days after transduction. Hematopoiesis-associated genes are shown. The numbers give the changes upon PRMT6 knockdown as logfold2. PRMT6 expression was reduced −2.82 logfold2 compared to the control, expressing a non-targeting shRNA. Genes marked in red have a described role in erythropoiesis. (D–I) A subset of genes from the array analysis was reanalyzed by quantitative real-time PCR 7 days after PRMT6 transduction. Error bars represent the standard deviation from at least four determinations and two independent knockdowns. (J,K) PRMT6 knockdown in K562 cells led to an increase of the GYPA (CD235a) and CD71 cell surface expression measured by FACS. The median fluorescence intensity (MFI) of GYPA-APC and CD71-APC staining in sh-control (shctrl.) and shPRMT6 (shP6) cells is shown. (L) The expression of the genes was measured by qRT-PCR in CD34+ cells upon knockdown of PRMT6. Knockdown cells were sorted and maintained in ery/mega medium for 5 days. Gene expression was determined by quantitative reverse transcriptase PCR. The knockdown values (shPRMT6#1/2) represent the combined data from two different knockdown constructs. Error bars display the standard deviation calculated from at least four determinations. The P-values were calculated using the Student t-test. *P<0.05; **P<0.01; ***P<0.001.
To further examine the influence of PRMT6 on erythroid gene expression we measured the expression of erythroid genes by quantitative reverse transcriptase PCR 7 days after transduction of K562 cells (Online Supplementary Figure S3). PRMT6 knockdown resulted in a marked increase of the erythroid markers GYPA, ALAS2 and AHSP (Figure 3D–F). Similar to the array data c-Kit expression was decreased upon PRMT6 knockdown (Figure 3G). The erythroid genes KLF1 and β-globin were significantly increased at this time point after knockdown (Figure 3H,I). Like KLF1, the erythroid transcription factors TAL1 and GATA1 were also influenced by the level of PRMT6 in K562 cells (Online Supplementary Figure S4). The effect of PRMT6 knockdown on GYPA expression was also detectable at the level of the cell surface (Figure 3J, Online Supplementary Figure S5). Furthermore, expression of the early erythroid surface marker CD71 was increased (Figure 3K, Online Supplementary Figure S5). The K562 cells displayed a reddish color upon PRMT6 knockdown, indicating increased heme production (Online Supplementary Figure S5). Moreover, the knockdown of PRMT6 in human CD34+ cells influenced the expression of GYPA, ALAS2, AHSP, c-Kit, KLF1 and β-globin in the same direction as in K562 cells (Figure 3L). These data indicate that PRMT6 has a repressive influence on the expression of some erythroid genes, which is released upon PRMT6 knockdown.
GYPA is a direct target of PRMT6
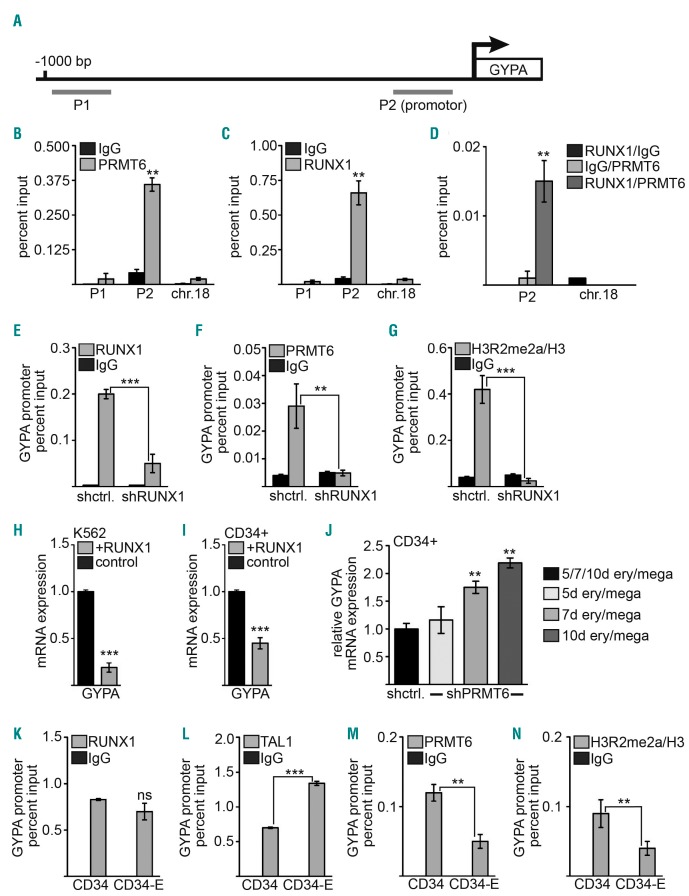
For further analysis of PRMT6 function we focused on this protein’s influence on the glycophorin A gene (GYPA). GYPA is a membrane protein and the main marker of erythroid differentiation. Expression of GYPA is directly controlled by the transcription factors TAL1 and GATA1.44 Our data show that PRMT6 inhibits erythroid differentiation and the expression of the erythroid differentiation gene GYPA. Examination of published ChIP-Seq data revealed that the promoter of GYPA also harbors functional RUNX1 binding sites in addition to TAL1 and GATA1 sites (Online Supplementary Figure S6). These transcription factors are known to be associated with PRMT6.28 By ChIP we detected PRMT6 and RUNX1 at the proximal promoter region of GYPA in K562 cells (Figure 4A–C, Online Supplementary Figure S6). As RUNX1 is able to recruit PRMT6 to target genes,33 we examined whether RUNX1 and PRMT6 co-occupy the GYPA promoter. Using ChIP-ReChIP we did in fact detect RUNX1 and PRMT6 together at this promoter, as indicated by the enrichment of GYPA promoter DNA in the RUNX1/PRMT6 ChIP-ReChIP, but not at a control locus (Figure 4D). The notion that RUNX1 is important for PRMT6 recruitment was further supported by a ChIP-assay after RUNX1 knockdown. RUNX1 knockdown led to decreased RUNX1 binding at the GYPA promoter (Figure 4E). PRMT6 occupancy of the GYPA promoter was decreased (Figure 4F) and H3R2me2a was diminished, an effect mediated by PRMT6 (Figure 4G). When we overexpressed RUNX1, the GYPA levels were reduced in K562 and CD34+ cells (Figure 4H,I). Furthermore, RUNX1 repressed the GYPA promoter in a reporter gene assay (Online Supplementary Figure S6). Interestingly, the knockdown of PRMT6 in CD34+ cells under differentiation conditions, which on its own induced some GYPA expression, led to increased GYPA expression (Figure 4J). When we induced CD34+ cells towards the eythroid lineage, we found that RUNX1 binding remained unchanged on the promoter (Figure 4K), TAL1 binding increased (Figure 4L) and PRMT6 binding decreased (Figure 4M). Concomitantly, the PRMT6-mediated H3R2me2a histone mark was decreased (Figure 4N). Taken together, our data show that RUNX1 contributes to the binding of PRMT6 to the GYPA promoter and support the notion that PRMT6 is a repressor of GYPA expression.
Figure 4.
GYPA is a direct target of hematopoietic transcription factors and PRMT6. (A) Scheme of the GYPA promoter showing the position of the ChIP-primers. (B) ChIP with K562 cells indicates binding of PRMT6 to the promoter region of GYPA (P2) but not to an upstream region (P1) or an unrelated control region (chr.18). (C) RUNX1 binds to the promotor region (P2) of GYPA but not to an upstream region (P1) or an unrelated control region on chromosome 18 (chr.18). (D) Quantitative ChIP-ReChIP of RUNX1 and PRMT6 with the given antibody combinations shows co-occupancy of RUNX1 with PRMT6 at the GYPA promoter (left) but not at a control region (chr.18) in K562 cells. (E, F) ChIP assay after RUNX1 knockdown shows reduced RUNX1 and PRMT6 binding to the GYPA promoter. (G) H3R2me2a modification at the GYPA promoter is decreased upon RUNX1 knockdown. (H, I) RUNX1 overexpression decreased GYPA mRNA expression in K562 and CD34+ cells measured by quantitative reverse transcriptase PCR. (J) Knockdown of PRMT6 in CD34+ cells cultured in ery/mega medium results in increased GYPA mRNA expression with time. The GYPA expression level of the corresponding time point was set as one. (K–N) Changes at the GYPA promoter upon erythroid differentiation of CD34+ cells. (K) RUNX1 binding to the GYPA promoter remains unchanged upon erythroid differentiation. (L) TAL1 binding to the GYPA promoter is increased upon erythroid differentiation. (M) PRMT6 binding is reduced upon erythroid differentiation. (N) Upon erythroid differentiation the repressive H3R2me2a modification at the GYPA promoter is decreased. Note that in K–N the values for IgG are small, so that the bar for the IgG control is not visible. Quantitative ChIP-PCR values are shown as percentage input. Values gathered for histone modification H3R2me2a were normalized with a ChIP against unmodified histone H3. Error bars show the standard deviation from at least four independent evaluations. The P-values were calculated using the Student t-test. *P<0.05; **P<0.01; ***P<0.001.
Differentiation-associated epigenetic changes
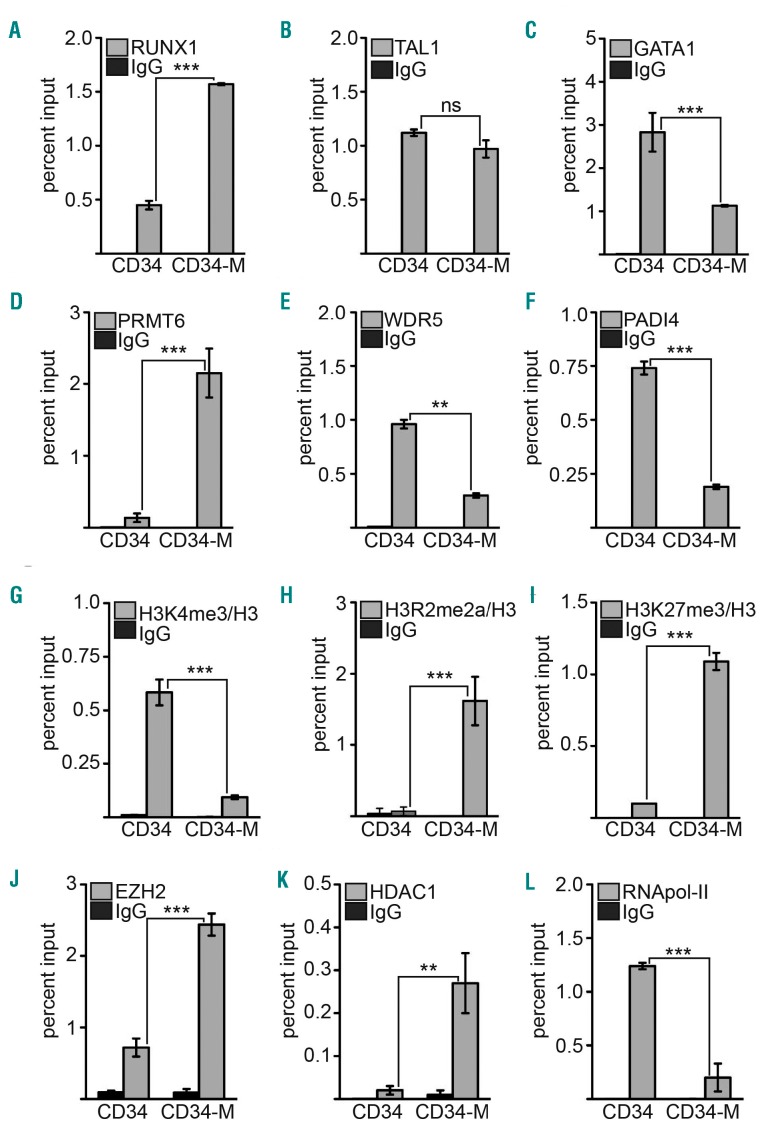
The erythroid gene GYPA is upregulated upon erythroid differentiation and downregulated during megakaryocytic differentiation of human CD34+ cells (Online Supplementary Figure S7). Furthermore, PRMT6 expression is increased during erythroid and megakaryocytic differentiation, whereas RUNX1 is only increased during megakaryocytic differentiation (Online Supplementary Figure S7). Our data show that PRMT6 is associated with repression of GYPA. Thus, we wondered whether PRMT6 is connected to the downregulation of GYPA expression during megakaryocytic differentiation of human primary progenitor cells (Online Supplementary Figures S7 and S8). We found that RUNX1 binding to the GYPA promoter increased during megakaryocytic differentiation of CD34+ cells (Figure 5A), whereas TAL1 binding remained unchanged and GATA1 binding decreased (Figure 5B,C). In line with a repressor function of PRMT6 we found that PRMT6 binding to the GYPA promoter increases upon megakaryocytic differentiation of hCD34+ cells (Figure 5D). It was suggested that PRMT6-mediated H3R2me2a negatively influences WDR5 binding and that the protein arginine deaminase PADI4 can counteract PRMT6 activity.34,40 Accordingly, WDR5 and PADI4 binding decreases at the GYPA promoter (Figure 5E,F). As a consequence, the activating H3K4me3 modification decreases and the repressive H3R2me2a and H3K27me3 methylation marks increase (Figure 5G–I). Concomitant to the increase of the repressive histone modification H3K27me3, the binding of EZH2, which mediates this modification, increases (Figure 5J). In line with the notion that a repressive chromatin environment is established, binding of the repressive histone deacetylase 1 (HDAC1) is increased upon megakaryocytic differentiation (Figure 5K) and occupancy of RNA-polymerase II is decreased (Figure 5L).
Figure 5.
Occupancy of the GYPA promoter upon megakaryocytic differentiation of human CD34+ cells. Binding of transcription factors and histone modifications at the GYPA promoter were determined before and after megakaryocytic differentiation (CD34/CD34-M) by ChIP. (A) RUNX1 binding was increased upon megakaryocytic differentiation. (B) TAL1 binding remained similar after megakaryocytic differentiation. (C) GATA1 binding was decreased upon megakaryocytic differentiation. (D) PRMT6 binding was increased after megakaryocytic differentiation. (E) WDR5 binding was decreased upon megakaryocytic differentiation. (F) PADI4 binding was decreased after megakaryocytic differentiation. (G) H3K4me3 was decreased after megakaryocytic differentiation. (H) H3R2me2a was increased upon megakaryocytic differentiation. (I) H3K27me3 was increased upon megakaryocytic differentiation. (J) EZH2 binding increased after megakaryocytic differentiation. (K) HDAC1 binding increased upon megakaryocytic differentiation. (L) Binding of RNA-polymerase II to the GYPA promoter decreased upon megakaryocytic differentiation. Quantitative PCR values of ChIP experiments are shown as percentage input. Values gathered for histone H3 modifications were normalized with ChIP against unmodified histone H3. Error bars depict the standard deviation from at least four evaluations. The P-values were calculated using the Student t-test. *P<0.05; **P<0.01; ***P<0.001.
Similar changes can also be observed during megakaryocytic differentiation of K562 cells (Online Supplementary Figure S9). In summary, our data demonstrate that PRMT6 and associated repressors contribute to the repression of GYPA expression during megakaryocytic differentiation.
Pharmacological inhibition of PRMT6 increases erythroid gene expression
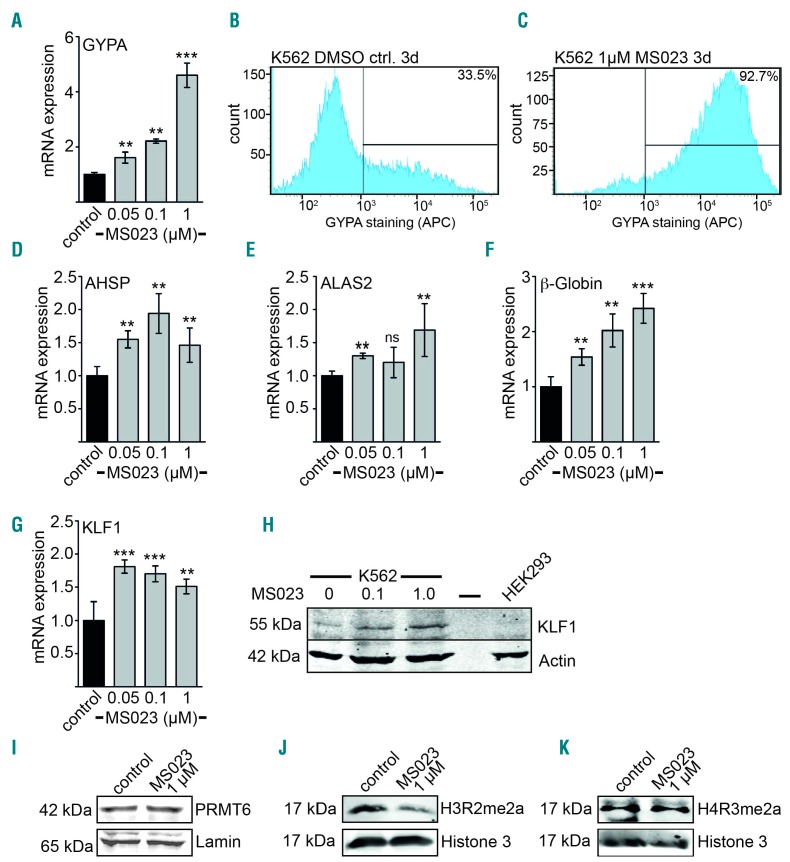
We have shown that binding of the RUNX1-associated repressor PRMT6 is upregulated during megakaryopoiesis and decreased during erythropoiesis at the GYPA locus. Furthermore, knockdown of PRMT6 increases erythropoiesis. Thus, inhibition of PRMT6 enzymatic function might lead to a shift in differentiation. Recently, small molecule inhibitors of PRMT6, which decrease the repressive H3R2me2a methylation in cells, were introduced.45,46 Accordingly, treatment of K562 cells with the PRMT6 inhibitor MS023 for 3 days increased GYPA expression at the mRNA level already at a concentration of 0.05 μM and reached its plateau at 1 μM (Figure 6A). Induction of the erythroid surface marker GYPA was also detected by flow cytometry (Figure 6B,C). Furthermore, other erythroid genes such as AHSP, ALAS2 and β-globin were upregulated 3 days after treatment with inhibitor (Figure 6D–F), resembling the effect of knockdown of PRMT6. Furthermore, KLF1 mRNA and protein levels were increased upon inhibitor treatment (6G, H). Inhibitor treatment had no influence on the amount of PRMT6 protein (Figure 6I), but reduced H3R2 asymmetric methylation as expected (Figure 6J). H4R3me2a, which is mediated by PRMT other than PRMT6, remained unchanged by inhibitor treatment (Figure 6K).
Figure 6.
Inhibition of PRMT6 increases erythroid gene expression. (A) GYPA expression increases at the mRNA level after treatment of K562 cells with the indicated concentrations of the PRMT6 inhibitor MS023 for 3 days. The control was treated with solvent only (DMSO). Expression was measured by quantitative reverse transcriptase PCR. (B,C) GYPA expression at the cell surface upon treatment of K562 cells with PRMT6 inhibitor for 3 days was determined by flow cytometry using an anti-CD235a-APC antibody. GYPA positivity is given in percent according to the indicated gating. (D–F) Expression of the erythroid genes AHSP, ALAS2 and β-globin increased upon treatment of K562 cells with the indicated concentrations of PRMT6 inhibitor for 3 days. (G,H) Expression of KLF1 increased upon inhibitor treatment on the mRNA and protein level. Expression was measured by quantitative reverse transcriptase PCR and western blot analysis. (I) Western blot analysis of PRMT6 protein expression upon inhibitor treatment of K562 cells for 3 days. (J) Western blot analysis of histone 3 methylation (H3R2me2a) upon inhibitor treatment of K562 cells for 3 days. (K) Western blot analysis of histone 4 methylation (H4R3me2a) upon inhibitor treatment of K562 cells for 3 days. Error bars indicate the standard deviation from four independent determinations. The P values were calculated using the Student t-test. *P<0 .05; **P<0.01; ***P<0.001.
The increased expression of GYPA, AHSP and ALAS2 upon inhibitor treatment was inhibited by PRMT6 overexpression, but not in the case of β-globin and KLF1 (Online Supplementary Figure S10). The induction towards erythroid differentiation by PRMT6 inhibition was also seen in an increase of the reddish color of the cell pellet upon inhibitor treatment of K562 cells, indicating increased heme production (Online Supplementary Figure S5).
Inhibition of PRMT6 increases erythroid differentiation of CD34+ cells
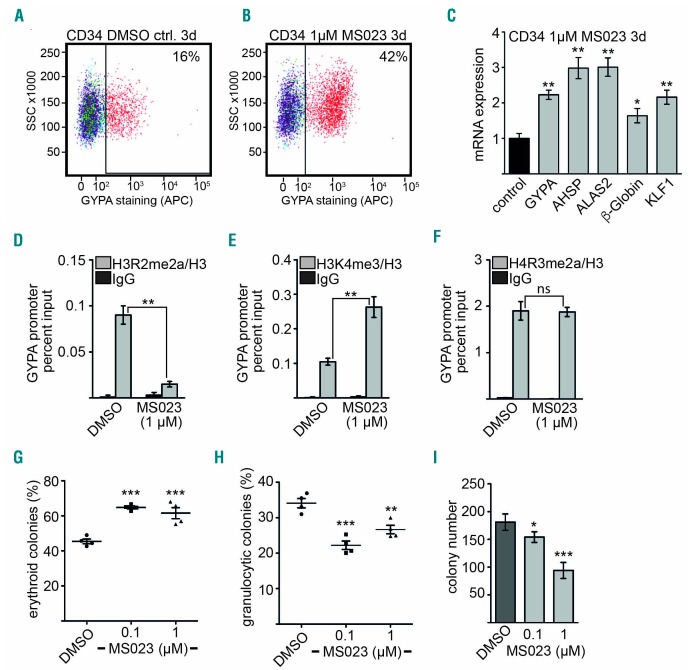
Our data indicate that PRMT6 inhibition might enhance erythroid differentiation. To investigate this notion directly, we treated primary human CD34+ cells with PRMT6 inhibitor in liquid culture under conditions which allow erythroid or megakaryocytic differentiation. This treatment shifted differentiation towards erythropoiesis, as indicated by the higher levels of GYPA and CD71 surface markers (Figure 7A,B and Online Supplementary Figure S11). Expression of other PRMT6-associated erythroid genes was also increased upon PRMT6 inhibition at the mRNA level (Figure 7C). Treatment of CD34+ cells with PRMT6 inhibitor led to decreased H3R2me2a and increased H3K4me3. H4R3me2a remained unchanged at the GYPA promoter (Figure 7D–F). Similarly, at the established PRMT6 target KLF1,28 H3R2me2a was reduced upon inhibitor treatment (Online Supplementary Figure S12). Subsequently, we analyzed human CD34+ cell differentiation upon PRMT6 inhibition in a CFU assay to examine differentiation independently of cell surface markers. We detected an increase of erythroid colonies upon PRMT6 inhibition (Figure 7G) and a decrease of granulocytic colonies (Figure 7H). These alterations were accompanied by an almost 50% decrease of total colonies at high inhibitor concentration (Figure 7I). Taken together, these data indicate that inhibition of PRMT6 increases erythropoiesis during differentiation of progenitor cells.
Figure 7.
Inhibition of PRMT6 increases erythroid differentiation of CD34+ cells. (A,B) GYPA expression increases upon treatment of primary human CD34+ cells with PRMT6 inhibitor as measured by flow cytometry using an anti-CD235a-APC antibody. (C) Expression of the erythroid genes AHSP, ALAS2, β-globin and KLF1 increases upon treatment of hCD34+ cells with the indicated concentration of PRMT6 inhibitor for 3 days. Expression was measured by quantitative reverse transcriptase PCR. (D–F) ChIP assay upon PRMT6 inhibitor treatment of CD34+ cells for 3 days. H3R2me2a was decreased upon inhibitor treatment and H3K4me3 was increased upon inhibitor treatment. H4R3me2a remained unchanged upon inhibitor treatment. (G) Treatment of human CD34+ cells with PRMT6 inhibitor MS023 enhances erythroid differentiation at the given inhibitor concentrations in a CFU assay. Error bars give the standard deviation from four independent inhibitor treatments. (H) Treatment of human CD34+ cells with PRMT6 inhibitor MS023 reduced granulocytic differentiation at the given inhibitor concentrations in a CFU assay. Error bars give the standard deviation from four independent inhibitor treatments. (I) The total number of colonies in the CFU assay with human CD34+ cells upon treatment with PRMT6 inhibitor is shown. Error bars indicate the standard deviation from four independent determinations. The P values were calculated using the Student t-test. *P<0.05; **P<0.01; ***P<0.001.
Discussion
The interplay between transcription factors and their epigenetic cofactors is decisive for the establishment and maintenance of a cell type-specific gene expression program. In this process, the chromatin environment at cell type-specific genes is adjusted according to cell fate decisions taken at key lineage fate bifurcations. Consequently, alterations in DNA and histone modification patterns activate one gene expression program at the expense of the other.
In this study, we made some significant novel observations regarding gene expression control during megakaryopoietic/erythroid lineage differentiation. Our data demonstrate that PRMT6 inhibits erythroid gene expression during lineage differentiation. Under conditions that allow erythroid or megakaryocytic differentiation, the knockdown of PRMT6 enhances erythropoiesis, whereas PRMT6 overexpression inhibits erythropoiesis in CFU assays. Furthermore, we showed that PRMT6 mediates the repressive H3R2me2a modification at erythroid genes such as GYPA and KLF1. We detected low levels of PRMT6 present on the GYPA promoter in progenitor cells, which increase upon megakaryopoiesis. Concomitantly, H3R2me2a is increased and this goes hand in hand with the establishment of a repressive histone modification pattern with reduced H3K4me3 at the promoter upon megakaryopoiesis. An analysis of PRMT6 function at the megakaryocytic/erythroid branching with a hematopoietic knockout mouse model would be very attractive. Our data also indicate that the transcription factor RUNX1 contributes to PRMT6 recruitment to GYPA promoter as knockdown of RUNX1 reduces PRMT6 occupancy. Interestingly, PRMT6 is present on the promoter of megakaryocytic differentiation genes such as CD41 in progenitor cells. In this case, loss of PRMT6 leads to upregulation of CD41 in stem cell expansion medium.33 Moreover, upon megakaryopoiesis RUNX1 activates these megakaryocytic genes33 and in the same cells RUNX1 is present together with PRMT6 in a repressive complex on erythroid genes.28 In combination, our data indicate that there are two distinct facets of the RUNX1/PRMT6 complex, one associated with megakaryocytic genes in progenitors33 and the other with erythroid genes upon megakaryopoiesis.27,28 Furthermore, we detected genes, that are upregulated or downregulated upon PRMT6 knockdown. This hints towards repressive and activating roles of PRMT6 depending on the gene, as recently proposed.47 How the formation of distinct RUNX1 complexes is regulated is not known; however, different promoter contexts and the modification status of RUNX1 could have a regulatory influence.48–50 Furthermore, different isoforms of RUNX1 could convey altered protein:protein interactions of RUNX1 on distinct promoters. It is also conceivable that RUNX1 itself is methylated by PRMT6 as it was described that PRMT1 is able to perform histone and non-histone methylation in conjunction with RUNX1.48 Recently, it has been shown that the expression of RUNX1 isoform differs between megakaryocytic cells and erythoid cells.51 Our observations hint towards an essential function of PRMT6 in the shutdown of the erythroid gene expression program during megakaryocytic differentiation. The notion that a RUNX1/PRMT6 complex mediates this repression is also supported by our observation that RUNX1 knockdown or PRMT6 knockdown similarly lead to increased GYPA and KLF1 expression (this study and28). Given that PRMT6 cannot bind DNA directly, its recruitment is dependent on a physical interaction with transcription factors such as RUNX1, as well as possibly with other transcription factors present at regulatory sites. Our recent observation that PRMT6 can also be associated with the important transcription factor, TAL1,40 supports the notion that PRMT6 can be present in distinct gene regulatory complexes, depending on the promoter and the cell type. Recently, PRMT6 has been found to interact with members of the polycomb complex (PRC) and to contribute to PRC-mediated repression.52 It currently remains unclear whether PRMT6 is stably associated with a larger transcriptional complex. It was, however, shown that PRMT6 regulates cell proliferation and senescence.53–56 Our data show that there is also a decrease in colony formation upon knockdown of PRMT6 in human CD34+ cells or when the cells are treated with the PRMT6 inhibitor. However, upon inhibitor treatment no major increase in cell death was observed. For a definite statement on the effect of MS023 on cell growth a detailed analysis of apoptosis, senescence and cell cycle distribution would be essential. The anti-proliferative effect of the loss of PRMT6 merits further investigation in relevant mouse leukemia models. Knockdown of PRMT6 enhances erythropoiesis and pharmacological inhibition of PRMT6 also supports erythropoiesis of primary human CD34+ cells. Significant efforts are being made worldwide to develop efficient in vitro protocols for the production of therapeutic cells from hematopoietic or embryonic stem cells.57 Epigenetic compounds, which target epigenetic factors, such as PRMT6, could contribute to more effective in vitro differentiation in the future.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to JL (SPP1463, DFG JL1389 5-2). We would like to thank Helge Hussong for assistance.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/1/18
References
- 1.Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11(5):513–519. [DOI] [PubMed] [Google Scholar]
- 2.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–3376. [DOI] [PubMed] [Google Scholar]
- 3.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1(1):57–64. [DOI] [PubMed] [Google Scholar]
- 4.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goode DK, Obier N, Vijayabaskar MS, et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev Cell. 2016;36(5):572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. [DOI] [PubMed] [Google Scholar]
- 7.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. [DOI] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. [DOI] [PubMed] [Google Scholar]
- 10.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. [DOI] [PubMed] [Google Scholar]
- 11.Riddell J, Gazit R, Garrison BS, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157(3):549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. [DOI] [PubMed] [Google Scholar]
- 13.Feng R, Desbordes SC, Xie H, et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA. 2008;105(16):6057–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ness SA, Kowenz-Leutz E, Casini T, Graf T, Leutz A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993;7(5):749–759. [DOI] [PubMed] [Google Scholar]
- 15.Batta K, Florkowska M, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 2014;9(5):1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zang C, Luyten A, Chen J, Liu XS, Shivdasani RA. NF-E2, FLI1 and RUNX1 collaborate at areas of dynamic chromatin to activate transcription in mature mouse megakaryocytes. Sci Rep. 2016;6:30255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tijssen MR, Cvejic A, Joshi A, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20(5):597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119(16):3724–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3): 299–304. [DOI] [PubMed] [Google Scholar]
- 20.Moreau T, Evans AL, Vasquez L, et al. Large-scale production of megakaryocytes from human pluripotent stem cells by chemically defined forward programming. Nat Commun. 2016;7:11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pimanda JE, Ottersbach K, Knezevic K, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA. 2007;104(45):17692–17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tober J, Yzaguirre AD, Piwarzyk E, Speck NA. Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development. 2013;140(18): 3765–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457(7231):892–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dore LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118(2):231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101(11):4333–4341. [DOI] [PubMed] [Google Scholar]
- 26.Goldfarb AN. Transcriptional control of megakaryocyte development. Oncogene. 2007;26(47):6795–6802. [DOI] [PubMed] [Google Scholar]
- 27.Kohrs N, Kolodziej S, Kuvardina ON, et al. MiR144/451 Expression is repressed by RUNX1 during megakaryopoiesis and disturbed by RUNX1/ETO. PLoS Genet. 2016;12(3):e1005946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuvardina ON, Herglotz J, Kolodziej S, et al. RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation. Blood. 2015;125(23):3570–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang G, Zhao X, Wang L, et al. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood. 2011;118 (25):6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh CP, Wang CQ, Ng CE, et al. RUNX1 meets MLL: epigenetic regulation of hematopoiesis by two leukemia genes. Leukemia. 2013;27(9):1793–1802. [DOI] [PubMed] [Google Scholar]
- 31.Ross K, Sedello AK, Todd GP, et al. Polycomb group ring finger 1 cooperates with Runx1 in regulating differentiation and self-renewal of hematopoietic cells. Blood. 2012;119(18):4152–4161. [DOI] [PubMed] [Google Scholar]
- 32.Yu M, Mazor T, Huang H, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45(3):330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herglotz J, Kuvardina ON, Kolodziej S, et al. Histone arginine methylation keeps RUNX1 target genes in an intermediate state. Oncogene. 2013;32(20):2565–2575. [DOI] [PubMed] [Google Scholar]
- 34.Lausen J. Contributions of the histone arginine methyltransferase PRMT6 to the epigenetic function of RUNX1. Crit Rev Eukaryot Gene Expr. 2013;23(3):265–274. [DOI] [PubMed] [Google Scholar]
- 35.Migliori V, Phalke S, Bezzi M, Guccione E. Arginine/lysine-methyl/methyl switches: biochemical role of histone arginine methylation in transcriptional regulation. Epigenomics. 2010;2(1):119–137. [DOI] [PubMed] [Google Scholar]
- 36.Guccione E, Bassi C, Casadio F, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449(7164):933–937. [DOI] [PubMed] [Google Scholar]
- 37.Hyllus D, Stein C, Schnabel K, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21(24):3369–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iberg AN, Espejo A, Cheng D, et al. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283 (6):3006–3010. [DOI] [PubMed] [Google Scholar]
- 39.Lee YH, Ma H, Tan TZ, et al. Protein arginine methyltransferase 6 regulates embryonic stem cell identity. Stem Cells Dev. 2012;21(14):2613–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolodziej S, Kuvardina ON, Oellerich T, et al. PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation. Nat Commun. 2014. May 29;5:3995. 10.1038/ncomms4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT, Zheng X, et al. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009;Chapter13:Unit 13.11. [DOI] [PubMed] [Google Scholar]
- 43.Ebert BL, Lee MM, Pretz JL, et al. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105(12):4620–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24(4):1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eram MS, Shen Y, Szewczyk MM, et al. A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem Biol. 2016;11(3):772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell LH, Drew AE, Ribich SA, et al. Aryl Pyrazoles as potent inhibitors of arginine methyltransferases: identification of the first PRMT6 tool compound. ACS Med Chem Lett. 2015;6(6):655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casadio F, Lu X, Pollock SB, et al. H3R42me2a is a histone modification with positive transcriptional effects. Proc Natl Acad Sci USA. 2013;110(37):14894–14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Jankovic V, Gural A, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22(5):640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H, Woo AJ, Waldon Z, et al. A Src family kinase-Shp2 axis controls RUNX1 activity in megakaryocyte and T-lymphocyte differentiation. Genes Dev. 2012;26(14):1587–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Huang G, Zhao X, et al. Post-translational modifications of Runx1 regulate its activity in the cell. Blood Cells Mol Dis. 2009;43(1):30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Draper JE, Sroczynska P, Tsoulaki O, et al. RUNX1B expression is highly heterogeneous and distinguishes megakaryocytic and erythroid lineage fate in adult mouse hematopoiesis. PLoS Genet. 2016;12(1): e1005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stein C, Notzold RR, Riedl S, Bouchard C, Bauer UM. The arginine methyltransferase PRMT6 cooperates with polycomb proteins in regulating HOXA gene expression. PLoS One. 2016;11(2):e0148892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein C, Riedl S, Ruthnick D, Notzold RR, Bauer UM. The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res. 2012;40(19):9522–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kleinschmidt MA, de Graaf P, van Teeffelen HA, Timmers HT. Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin-dependent kinase inhibitors. PLoS One. 2012;7(8): e41446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phalke S, Mzoughi S, Bezzi M, et al. p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 2012;40(19):9534–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Huang Y, Zhao J, Zhang Y, Lu J, Huang B. Suppression of PRMT6-mediated arginine methylation of p16 protein potentiates its ability to arrest A549 cell proliferation. Int J Biochem Cell Biol. 2012;44(12):2333–2341. [DOI] [PubMed] [Google Scholar]
- 57.Batta K, Menegatti S, Garcia-Alegria E, Florkowska M, Lacaud G, Kouskoff V. Concise review: recent advances in the in vitro derivation of blood cell populations. Stem Cells Transl Med. 2016;5(10):1330–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.