Although the frequency of TP53 mutations in hematologic malignancies is low, these mutations have a high clinical relevance and are usually associated with poor prognosis. Somatic TP53 mutations have been detected in up to 73.3% of cases of acute myeloid leukemia (AML) with complex karyotype and 18.9% of AML with other unfavorable cytogenetic risk factors.1 AML with TP53 mutations, and/or chromosomal aneuploidy, has been defined as a distinct AML subtype. In low-risk myelodysplastic syndromes (MDS), TP53 mutations occur at an early disease stage and predict disease progression.2 TP53 mutation diagnosis is now part of the revised European LeukemiaNet (ELN) guidelines.3,4
The use of next generation sequencing (NGS), particularly ultra-deep sequencing, has led to the discovery that patients with either MDS or AML (either de novo, secondary or therapy-related) present multiple TP53 mutations, suggesting that several TP53 independent clones may co-exist.5 Patient follow up also reveals a highly dynamic evolution of these mutations during disease progression in treated and untreated patients.6,7 This observation is in line with the recent recognition that human tumors harbor an extensive genetic intratumoral heterogeneity.8 These findings will likely have implications for therapy and biomarker discovery, and determination of genetic complexity is becoming part of clinical decision-making processes in the age of precision medicine.
In this report, in silico analysis of the UMD_TP53 database showed that TP53 variants detected in patients with multiple TP53 alterations are fully oncogenic. Furthermore, using long-range single-molecule real-time (SMRT) sequencing on AML and MDS patients harboring multiple TP53 mutations, we showed that all of these variants are localized on different subclones, emphasizing the considerable tumor heterogeneity in these patients.
The 2017 release of the UMD_TP53 database contains the mutation status of 75,448 patients, including 922 cases of AML and 899 cases of MDS (Online Supplementary Table S1).9,10 Among these patients, 158 MDS cases (22.3%) and 99 AML cases (13%) harbor more than one TP53 variant in their tumors, higher than the rate observed in solid tumors (Online Supplementary Table S1). Chronic Lymphocytic Leukemia (CLL) patients also harbor a high frequency of tumors with multiple TP53 mutations. This feature has been observed with increasing frequency over recent years with the advent of deep sequencing techniques. Whether or not all of the multiple variants identified in these patients are truly deleterious or comprise a mix of driving and passenger mutations has never been addressed. The UMD_TP53 database includes quantitative functional data for all TP53 missense variants and can therefore be used to determine whether patients with multiple TP53 mutations frequently harbor non-deleterious TP53 variants. Analysis of the 257 AML and MDS cases with more than one TP53 variant showed that the majority (98%) of these variants are true deleterious TP53 mutations with complete loss of function and not simply random passenger mutations co-selected during tumor progression (Online Supplementary Table S2 and Online Supplementary Figure S1 a to e). A few non-deleterious variants have been identified, but they are likely very rare non-somatic polymorphisms.
To further demonstrate the presence and the dynamics of multiple independent tumor clones in AML and MDS, we have developed a novel, third-generation single-molecule real-time (SMRT) sequencing assay using the Pacific Biosciences platform with long-read lengths that span the most frequently mutated region of the TP53 gene. Sanger sequencing cannot be used to define the allelic distribution of multiple TP53 variants. This is also true for standard NGS if the two variants are more than 200 base pairs apart. On the other hand, SMRT analysis can be used to phase mutations located multiple kilobases apart directly from sequencing reads.
Eleven patients harboring multiple TP53 mutations in their tumors were enrolled. For 3 patients, sequential samples were available to assess the evolution of the various variants. The TP53 status of these patients was already defined according to stringent clinical criteria using either Sanger sequencing or standard NGS (Online Supplementary Material and Online Supplementary Table S3). In silico analysis of all these variants using the UMD_TP53 database showed that they were true deleterious TP53 mutations that have already been described in various types of cancer (Online Supplementary Table S1).
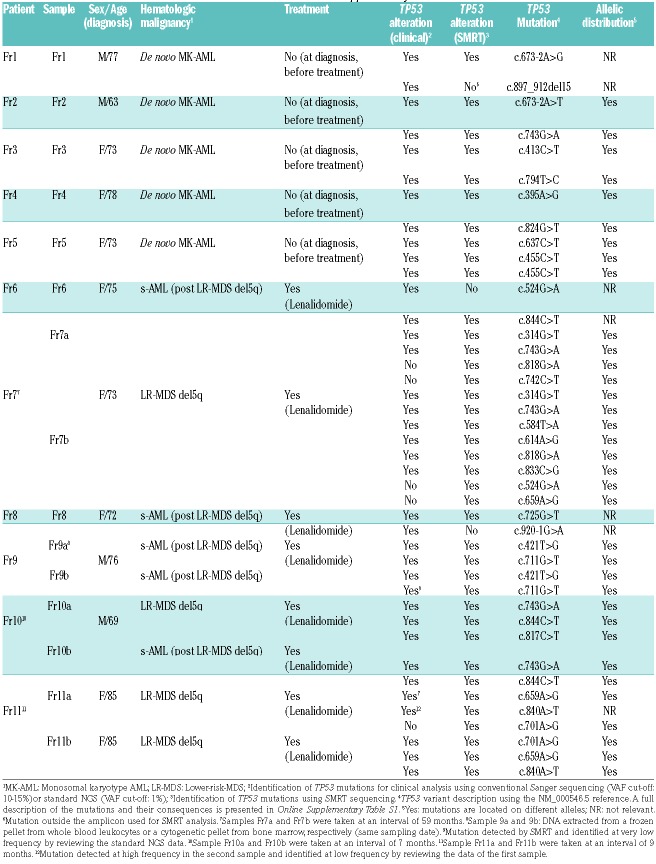
The majority of mutations detected for clinical evaluation were readily identified by SMRT, except for 2 variants that were not included in the amplicon used for analysis. SMRT identified 5 mutations that were not identified by clinical analysis (Online Supplementary Table S3 and Online Supplementary Figure S2). Manual examination of the sequencing data performed for clinical analysis confirmed that 2 of these mutations were detected at a frequency below the cut-off used for the analysis (Table 1). Most of the remaining mutations detected by SMRT were present at a very low frequency (Online Supplementary Figure S2). The variant allele frequency (VAF) observed for each variant detected by the two analyses was remarkably similar, despite being performed in different centers according to very different methodologies (Online Supplementary Figure S3).
Table 1.
Patient characteristics. More information is available in Online Supplementary Table S1.
Our analysis shows that all oncogenic TP53 variants were located in different alleles (Table 1 and Online Supplementary Figures S4a to S4k). For two samples, patient Fr7, sample 7b and patient Fr2, the close proximity of two TP53 variants allowed analysis of the alignment obtained after standard NGS and confirmed that these mutations were carried by different alleles (Online Supplementary Figures S5 and S6). For 2 samples, the allelic distribution was also confirmed by the observation that the different TP53 variants were associated with different TP53 haplotypes (Online Supplementary Figure S4 j and k, patients Fr10 and Fr11).
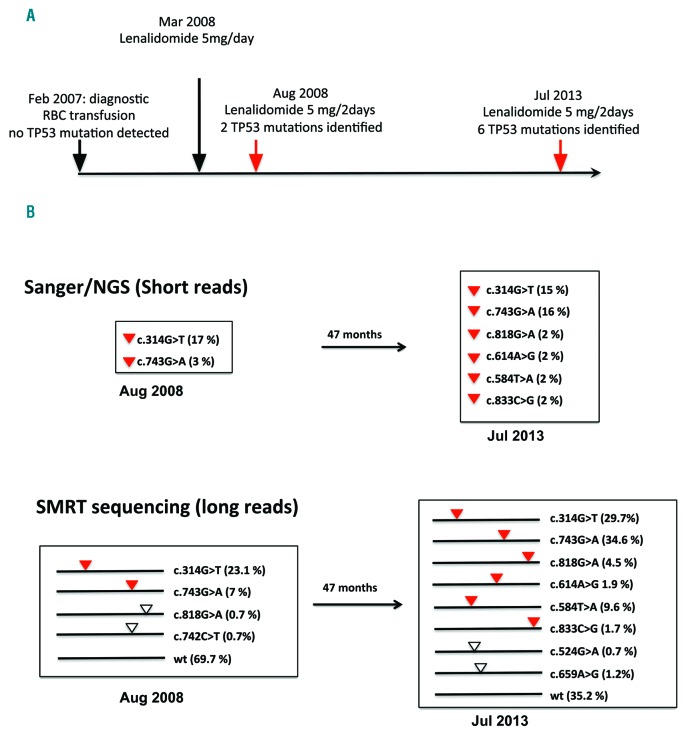
Figure 1 shows a typical result observed for two samples collected 5 years apart from a patient with multiple TP53 mutations. Of note, the diagnostic sample was negative with standard NGS and was therefore not used for SMRT analysis. In the first sample analysed by SMRT, clinical analysis identified two pathogenic TP53 mutations confirmed by SMRT. SMRT analysis also identified two novel TP53 mutations at very low frequency and showed that the 4 variants were distributed in different TP53 molecules. The two novel variants were readily identified in the second sample collected 5 years later. New TP53 variants were also identified by both methodologies (2 variants) and 2 additional variants were found at low frequency by SMRT (Figure 1). All these variants, carried by different TP53 molecules, were true driver mutations already identified in multiple tumor types, as shown by their high frequency in the UMD_TP53 database. This dynamic evolution of the various subclones can also lead to the elimination of certain subclones, as shown for patient Fr10 with the disappearance of TP53 variants (Online Supplementary Figure S4J).
Figure 1.
Clinical course and TP53 mutation analysis on patient Fr7. A. Patient Fr7 was diagnosed in February 2007 with lower-risk MDS with del5q with no TP53 mutations detected at a cut-off of 1%. One year after initiation of red blood cell (RBC) transfusion for anemia, patient Fr7 was treated with lenalidomide at a dosage of 5 mg/day, but rapidly experienced major adverse effects, leading to reduction of the dosage to 5 mg every other day (or less in a context of poor adherence). Five months after onset of therapy, 2 TP53 mutations were identified together with improvement of anemia. Five years later, in July 2013 (and until the present time), patient Fr7 was still alive and with no disease progression to secondary AML despite growth of at least 6 TP53-mutant clones, suggesting possible clonal equilibrium due to competition between the numerous mutant clones. B. Sanger sequencing and/or standard NGS analysis is shown in the upper part with 2 and 6 mutations in sample 7a and 7b, respectively. No allelic distribution can be inferred from this type of analysis. SMRT sequencing (lower part) provides an accurate picture of the allelic distribution of each TP53 variant, as well as the remaining wt allele. The frequencies of the 9 different alleles are shown in brackets. Red triangle: TP53 variants identified by both types of analysis. White triangle: TP53 variants detected only by SMRT sequencing.
Using both in silico analysis and SMRT sequencing, we demonstrate that the presence of multiple subclones with different TP53 variants is a common feature in AML and MDS. All TP53 variants detected in MDS and AML patients by SMRT sequencing are true, physically independent TP53 variants, confirming the results of indirect computational studies currently used to infer cancer heterogeneity. It is highly likely that each TP53 variant belongs to an independent subclone arising from a wild-type TP53 founder clone. The observation of multiple subclones with different TP53 variants in these patients suggests the occurrence of a specific genetic background in the founder clones that requires TP53 inactivation for further progression. All of these subclones present a highly dynamic evolution, but it remains to be determined whether this evolution is driven by treatment, a natural characteristic of the tumor or both. A recent study on 1,514 MDS patients after stem-cell transplantation showed that 283 patients (19%) had at least one oncogenic TP53 mutation and a poor overall survival.11 One hundred and two (36%) of these patients had more than one TP53 variant (range 2–6). It is likely that the use of a sensitive methodology for DNA sequencing will reveal that tumors with multiple TP53 variants constitute a general feature raising potential problems for treatment options. Finally, in this report, we demonstrate the efficiency of SMRT sequencing for the analysis of complex samples. The rapid progress in NGS, combining longer reads, increased sensitivity and decreased costs, will allow investigation of the whole sequence of clinically relevant genes in a single analysis. Long-read RNA-seq analysis could also be used to address the issue of TP53 alternative spliced transcripts that have already been described to be of clinical interest in AML.12
Supplementary Material
Footnotes
Funding: this work was supported by Radiumhemmets Forskningsfonder and the Swedish Cancer Society (Cancerfonden) to TS. SMRT sequencing was performed by the National Genomics Infrastructure (NGI) hosted by SciLifeLab Uppsala. The authors are most grateful to the Molecular Hematology team and to the IRCNA Tumor Bank (CHU de Nantes, Institut de Cancérologie de l’Ouest, Saint-Herblain F44800, France) for their assistance.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016; 374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jadersten M, Saft L, Smith A, et al. TP53 Mutations in low-risk myelodysplastic syndromes with del(5q) predict disease Progression. J Clin Oncol. 2011;29(15):1971–1979. [DOI] [PubMed] [Google Scholar]
- 3.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malcovati L, Hellström-Lindberg E, Bowen D, et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European LeukemiaNet. Blood. 2013; 122(17):2943–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31(3):705–711. [DOI] [PubMed] [Google Scholar]
- 6.Mossner M, Jann JC, Nowak D, et al. Prevalence, clonal dynamics and clinical impact of TP53 mutations in patients with myelodysplastic syndrome with isolated deletion (5q) treated with lenalidomide: results from a prospective multicenter study of the german MDS study group (GMDS). Leukemia. 2016;30(9):1956–1959. [DOI] [PubMed] [Google Scholar]
- 7.da Silva-Coelho P, Kroeze LI, Yoshida K, et al. Clonal evolution in myelodysplastic syndromes. Nat Commun. 2017;815099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. [DOI] [PubMed] [Google Scholar]
- 9.Leroy B, Ballinger ML, Baran-Marszak F, et al. Recommended guidelines for validation, quality control, and reporting of TP53 variants in clinical practice. Cancer Res. 2017;77(6):1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35(6):672–688. [DOI] [PubMed] [Google Scholar]
- 11.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anensen N, Hjelle SM, Van Belle W, et al. Correlation analysis of p53 protein isoforms with NPM1/FLT3 mutations and therapy response in acute myeloid leukemia. Oncogene. 2012;31(12):1533–1545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.