Abstract
Many biologically active peptides found in nature exhibit a bicyclic structure wherein a head-to-tail cyclic backbone is further constrained by an intramolecular linkage connecting two side chains of the peptide. Accordingly, methods to access macrocyclic peptides sharing this overall topology could be of significant value toward the discovery of new functional entities and bioactive compounds. With this goal in mind, we recently developed a strategy for enabling the biosynthesis of thioether-bridged bicyclic peptides in living bacterial cells. This method involves a split intein-catalyzed head-to-tail cyclization of a ribosomally produced precursor peptide, combined with inter-sidechain crosslinking through a genetically encoded cysteine-reactive amino acid. This approach can be applied to direct the formation of structurally diverse bicyclic peptides with high efficiency and selectivity in living Escherichia coli cells and provides a platform for the generation of combinatorial libraries of genetically encoded bicyclic peptides for screening purposes.
1. Introduction
The structural diversity and breadth of biological activities exhibited by naturally occurring cyclic peptides have stimulated significant interest in this structural class for the development of chemical probes and therapeutics, in particular directed at challenging targets such as protein-protein and protein-nucleic acid interactions [1–3]. The growing interest in this area has translated into the development of various synthetic, semisynthetic, and biosynthetic strategies to obtain macrocyclic peptides and libraries thereof, including methods for obtaining head-to-tail, side-chain-to-tail, or side-chain-to-side-chain cyclic peptides that also comprise non-peptidic moieties and linkers [4–11]. Among the bioactive peptides found in nature, several exhibit a bicyclic structure wherein a head-to-tail cyclic structure is further constrained by an intramolecular linkage connecting two side chains of the peptide. Representative examples of these bicyclic peptides include α-amanitin, a potent inhibitor of eukaryotic RNA polymerases, the antimicrobial β-defensins, and cytotoxic plant-derived peptides belonging to the bouvardin and celogentin families. In these classes of molecules, the conformational rigidification imposed by the bicyclic backbone is often critical for their biological activity and beneficial toward increasing their cell permeability and proteolytic stability.
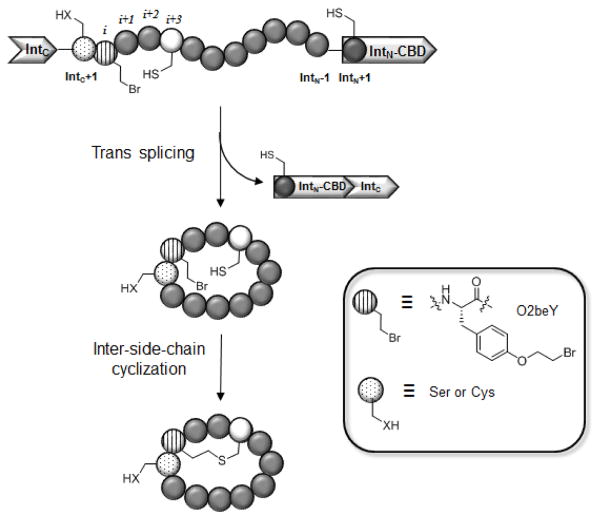
As part of our efforts toward developing versatile methodologies to access structurally diverse peptide-based macrocycles [12–16], we have recently devised a strategy to enable the ribosomal synthesis of ‘natural product-like’ bicyclic peptides, which feature a head-to-tail cyclic backbone along with an inter-side-chain thioether linkage [17,18]. As illustrated in Figure 1, this methodology couples split intein-mediated peptide circularization [19] with a spontaneous, intramolecular cross-linking reaction between a genetically encoded, cysteine-reactive unnatural amino acid (O-(2-bromoethyl)-tyrosine or O2beY) and a neighboring cysteine residue present in the peptide sequence [17,18]. Briefly, peptide head-to-tail circularization is achieving by framing a target peptide sequence between the C-terminal (IntC) and the N-terminal domain (IntN) of the naturally occurring split intein DnaE from Synechocystis sp. PCC6803 [19]. The overall mechanism of this reaction is described in Figure 2 and it involves the association of the two intein fragments to promote a N→S acyl shift at the level of the cysteine residue at the IntN+1 position. This thioester intermediate is intercepted by a nucleophilic residue at the IntC+1 position (either serine or cysteine) to undergo a trans(thio)esterification followed by the asparagine-mediated release of the side-chain-to-head linked cyclic peptide which upon O/S→N acyl shift gives the desired head-to-tail cyclic peptide. While this method can provide a means to generate genetically encoded libraries of head-to-tail cyclic peptides (e.g., via genetic randomization of residues within the peptide target sequence), its scope has remained restricted to the formation of monocyclic peptides [9]. To enable the biosynthesis of bicyclic peptides, we envisioned the possibility of incorporating, within the peptide target sequence, an electrophilic unnatural amino acid that could react with a nearby cysteine residue to form an hydrolytically and redox stable inter-side-chain thioether bond (Figure 1).
Figure 1.
Overall strategy for the ribosomal synthesis of thioether-bridged bicyclic peptides in E. coli. From the N- to C-terminus, the precursor protein comprises: (a) the C-terminal domain of DnaE split intein (IntC); (b) a variable target peptide sequence; (c) the N-terminal domain of DnaE split intein fused to a chitin binding domain (IntN-CBD). The target peptide sequence contains an initial Ser or Cys residue (dotted circle) at the ‘IntC+1’ site for mediating split intein-catalyzed head-to-tail cyclization and comprises the unnatural amino acid O-(2-bromoethyl)-tyrosine (O2beY, circle with vertical lines) and the reactive cysteine (white circle) for inter-side-chain crosslinking via thioether bond formation.
Figure 2.
Mechanism of split intein-catalyzed peptide circularization. IntN and IntC correspond to the N-domain and C-domain respectively, of DnaE split intein from Synechocystis sp. PCC6803. The IntN+1 cysteine and IntC+1 cysteine (or serine) residues are indicated.
The design and realization of this strategy involved a series of important considerations. First, the reactivity of the unnatural amino acid has to be properly tuned in order to support the desired intramolecular thioether bond formation, while avoiding undesirable reactions with other nucleophiles present in the intracellular milieu (e.g., glutathione). Another important requirement was the availability of an efficient system for the ribosomal incorporation of such unnatural amino acid into the precursor polypeptide expressed in E. coli. To this end, we established that O-(2-bromoethyl)-tyrosine (O2beY) has the desired reactivity profile and we engineered an orthogonal aminoacyl-tRNA synthetase (AARS)/tRNA pair for incorporation of O2beY in response to an amber stop codon (TAG), respectively [17]. The O2beY-specific AARS was derived from the Methanocaldococcus jannaschii tyrosyl-tRNA synthetase, which had been previously evolved for encoding a variety of tyrosine derivatives via the amber stop codon suppression technology [20]. The O2beY-specific AARS was found to offer an excellent amber stop codon suppression efficiency (85%), enabling the expression of O2beY-containing proteins in high yields (e.g., ~40 mg/L for O2beY-containing yellow fluorescent protein).
Using a series of model polypeptide constructs produced in E. coli, it was established that O2beY is able to form a thioether linkage with a neighboring cysteine residue that is located from two to eight residues apart from O2beY with high efficiency (80–100% cyclization yield) [17]. Furthermore, the efficiency of the cross-linking reaction was found to be independent of the composition of the target peptide sequence and relative orientation of the O2beY/Cys pair (i.e. O2beY/Cys vs. Cys/O2beY) across the various constructs tested.
By combining split intein-mediated peptide circularization with the O2beY/Cys cross-linking strategy (Figure 1), it has been possible to direct the spontaneous biosynthesis of natural product-like bicyclic peptides inside living E. coli cells [17,18]. To illustrate such capability, a streptavidin binding motif (His-Pro-Gln) was introduced within the target peptide sequence of the precursor polypeptide, thus enabling the isolation and identification of the desired bicyclic products, along with potential acyclic or monocyclic byproducts, via streptavidin affinity capture followed by LC-ESI-MS and MS/MS analyses. Using this approach, bicyclic peptides comprising between 12 and 18 amino acid residues and featuring different O2beY/Cys connectivities (e.g., i / i+3 and i / i+8 linkages) and thus varying ring sizes could be produced in E. coli in good to excellent yields (70–97%) [17,18]. Mechanistic studies showed that these bicyclic peptides are generated at the post-translational level primarily through head-to-tail cyclization followed by inter-side-chain crosslinking. In the presence of target sequences less prone to split intein-mediated circularization, however, the thioether bridging reaction was found to increase the overall yield of the bicyclization reaction, presumably by facilitating the trans splicing process [18].
The protocols provided here describe procedures for the cloning, production in E. coli, and isolation of two representative bicyclic peptides featuring a i / i+3 and a i / i+8 thioether linkage and comprising a streptavidin-binding motif for isolation from E. coli cells via affinity capturing. This general methodology can be readily adapted to generate libraries of genetically encoded bicyclic peptides which can be screened using in vitro assays [18] or, alternatively, using high throughput methods based on intracellular reporter [21] or selection systems [22], in order to identify bicyclic peptides with novel or improved functional properties.
2. Materials
2.1 Reagents
Chemically competent BL21(DE3) cells.
Ampicillin stock solution (1000×): Dissolve ampicillin sodium salt in water at 100 mg/mL. Filter sterilized and store in aliquots at −80°C for up to 6 months.
Chloramphenicol stock solution (1000×): Dissolve chloramphenicol in ethanol at 30 mg/mL. Store in aliquots at −80°C for up to 6 months.
Sodium hydrogen phosphate (Na2HPO4).
Potassium dihydrogen phosphate (KH2PO4).
Sodium chloride (NaCl).
Ammonium chloride (NH4Cl).
Lysogeny Broth (LB) medium: Dissolve 10 g tryptone, 5 g yeast extract, and 10 g NaCl in 1 L of deionized water. Autoclave the solution and store at room temperature.
M9 salts (5×) solution: Dissolve 64 g Na2HPO4, 15 g KH2PO4, 2.5 g NaCl, and 5 g NH4Cl in 1 L of deionized water. Autoclave the solution and store at room temperature.
M9 medium: To a 1 L sterile flask, add 370 mL of distilled water, 100 mL of 5× M9 salt solution, 500 μL of 2 M MgSO4, 50 μL 1 M CaCl2, 5 mL glycerol, and 50 mL of 10% yeast extract (Note 1).
Magnesium sulfate (MgSO4) solution: Dissolve 24 g MgSO4 in 100 mL of deionized water to make 2 M solution, autoclave and store at room temperature.
Calcium chloride (CaCl2) solution. Dissolve 1.1 g CaCl2 in 10 mL of deionized water to make 1 M solution, filter sterilize and store at room temperature.
Sterile glycerol: Autoclave 100 mL glycerol and store at room temperature.
Arabinose stock solution (10%): Dissolve 1 g of arabinose in 10 mL of deionized water, filter sterilize and store at −20°C.
Chitin beads (New England Biolabs).
Streptavidin-coated agarose beads.
(L)-N-tert-butoxycarbonyl-tyrosine.
Dibromoethane.
Potassium carbonate (K2CO3).
pSFBAD09 and pJJDuet30 plasmids (Addgene #11963 and #11962, respectively).
Agarose.
Deoxynucleotide (dNTPs) mix solution.
High fidelity DNA Polymerase (e.g., Phusion HF DNA polymerase from NEB) and appropriate buffer solution (e.g. Phusion buffer HF 5× from NEB).
PCR Purification Kit (e.g., QIAquick from Qiagen).
Gel Extraction Kit (e.g., QIAquick from Qiagen).
Restriction enzymes Dpn I, Nde I, and Kpn I HF and appropriate buffer (e.g. CutSmart buffer 10× from NEB).
Calf intestinal alkaline phosphatase (CIP).
T4 DNA ligase and T4 DNA ligase reaction buffer.
Stock solution of 200 mM O-2-bromoethyl-tyrosine (O2beY) in water. The synthesis of O2beY is described below.
Phosphate buffer: 50 mM potassium phosphate solution, pH 8.0.
2.2 Solvents
Acetic acid (HOAc).
Trifluoroacetic acid (TFA).
Formic acid (FA).
Acetonitrile.
Hexanes.
Dimethylsulfoxide (DMSO).
Deionized water.
N-N-Dimethylformamide (DMF).
Dichloromethane (DCM).
Ethyl Acetate (EtOAc).
3. Methods
3.1 Synthesis of O-(2-bromoethyl)tyrosine (O2beY)
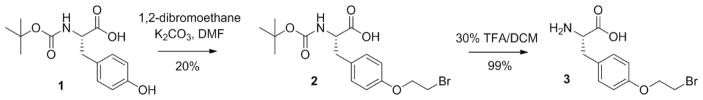
As described in the scheme reported in Figure 3, the unnatural amino acid O-(2-bromoethyl)tyrosine (O2beY) can be prepared in two steps starting from commercially available N-Boc-tyrosine.
Figure 3.
Synthetic route for preparation of O-(2-bromoethyl)tyrosine (O2beY).
Transfer 20 mL of dry DMF to a 125-mL round-bottom flask equipped with a stirring bar.
To the DMF solution, add N-tert-butoxycarbonyl-tyrosine 1 (2 g, 7.1 mmol) and potassium carbonate (2.94 g, 21.3 mmol) under argon.
To the DMF solution of step 2, add dibromoethane (1.83 mL, 21.3 mmol) dropwise over 20 min. Stir the reaction mixture at room temperature.
Monitor the progress of the alkylation reaction by thin-layer chromatography (TLC) using hexane/ethyl acetate (7:3) as the mobile phase (Rf for 1: 0.05; Rf for 2: 0.5).
Upon completion of the reaction (approximately 18 hours), filter the reaction mixture using a Buchner funnel.
Dilute the filtrate with 60 mL of water (Note 2).
Acidify the water/DMF solution with HCl 0.1 M to pH 4.
Extract the acidified water/DMF solution with 2 × 100 mL of ethyl acetate. Combine the organic layers and dry them over sodium sulfate. Remove the organic solvent under reduced pressure in rotary evaporator to obtain 2 as a crude product (yellow oil).
Purify O-2-bromoethyl-N-tert-butoxycarbonyl-tyrosine 2 by flash column chromatography using a 10:9:1 mixture of hexanes : ethyl acetate : acetic acid as the solvent system.
Identify the fractions containing O-2-bromoethyl-N-tert-butoxycarbonyl-tyrosine 2 by TLC, combine them, and remove the solvent under reduced pressure in a rotary evaporator to yield 2 as an off-white powder (0.54 g, 20% yield).
Dissolve 10–15 mg of isolated 2 in deuterated chloroform (CDCl3) and confirm its identity by 1H NMR and mass spectrometry. 1H NMR (400 MHz, CDCl3) δ 1.38 (s, 9H), 2.99–3.04 (m, 2H), 3.43 (t, 2H, J = 6 Hz), 3.58 (t, 1H; J = 6 Hz), 4.22 (t, 2H, J = 6.4 Hz), 6.80 (d, 2H, J = 8.4 Hz), 7.05 (d, 2H, J = 8.4). MS (ESI) calculated for C14H19NO5 [M]+: m/z 387.07, found 387.17.
For deprotection of the Boc protecting group (2 → 3 in Figure 3), add 2 g of purified 2 to 20 mL of 30% (v/v) trifluoroacetic acid in dichloromethane in a round-bottom flask equipped with a stirring bar. Stir the solution at 0°C.
Monitor the reaction by TLC using hexane/ethyl acetate (7:3) as the mobile phase (Rf for 2: 0.5; Rf for 3: 0).
Upon completion of the reaction (approximately 2 hours), remove the solvent under reduced pressure in a rotary evaporator.
Dissolve the crude residue in 10 mL of acetic acid and remove the solvent under reduced pressure in a rotary evaporator (Note 3). Repeat this step one more time to yield the final product, O-2-bromoethyl-tyrosine 3, as an off-white solid (1.7 g, 99% yield).
Dissolve 10–15 mg of isolated 3 in deuterated methanol (CD3OD) and confirm its identity by 1H and 13C NMR and mass spectrometry. 1H NMR (400 MHz, CD3OD) δ 3.02 – 3.23 (2H, m), 3.65 (2H, t, J = 5.6 Hz), 4.10 (1H, t, J= 5.8 Hz), 4.26 (2H, t, J = 5.6 Hz), 6.90 (2H, d, J = 8.4), 7.18 (2H, d, J = 8); 13C NMR (125 MHz, CD3OD) δ 28.9, 35.3, 54.1, 67.8, 114.8, 126.7, 130.2, 157.9, 167.6. MS (ESI) calculated for C11H14BrNO3 [M+H]+: m/z 288.02, found 288.51.
3.2 Cloning of the expression vectors for the precursor protein
The methodology described in this protocol involves the use of two plasmid vectors; a first one for directing the expression of the split intein-containing precursor protein and a second one encoding for the orthogonal AARS/tRNA pair for amber stop codon suppression with the unnatural amino acid O2beY. Various combinations of commercially available plasmid vectors for recombinant protein expression can be used for this purpose but the choice of the vectors must satisfy certain requirements. In general, the two plasmids need to carry two different and compatible origins of replication to allow for their stable co-existence in the bacterial cell. In addition, they need to carry two different antibiotic resistance markers to enable the selection of transformant E. coli cells containing both plasmids. E. coli cells that contain only one of the two vectors would either produce only the AARS/tRNA system and no precursor protein, or produce a truncated form of the precursor protein due to failed suppression of the amber stop codon introduced within the peptide target sequence.
In the present protocol, the plasmid encoding for the precursor protein is based on a pBAD vector system (Thermo Fischer Scientific), which contains a ColE1 origin of replication and an ampicillin resistance marker. The gene encoding for the precursor protein is under an arabinose-inducible promoter (AraBAD promoter) and is composed of: (a) IntC-domain of DnaE split intein from Synechocystis sp., (b) a desired peptide target sequence containing a codon for encoding of the cysteine residue (TGT or TGC) and an amber stop codon (TAG) for encoding of the cysteine-reactive O2beY; (c) IntN-domain of DnaE split intein from Synechocystis sp.; and (d) the chitin-binding domain (CBD) of chitinase A1 from Bacillus circulans.
The plasmid encoding for the amber stop codon suppression system is based on a pEVOL vector [23], which carries a p15A origin of replication and a chloramphenicol ampicillin resistance marker. This vector contains a first copy of the AARS gene under an arabinose-inducible promoter (AraBAD promoter), a second copy of the AARS gene under an under a constitutive glnS promoter, and a single copy of the cognate amber stop codon suppressor tRNA under a constitutive proK promoter. The Obe2Y-specific AARS gene is an engineered variant of M. jannaschii tyrosyl-tRNA synthetase carrying the following mutations: Y32G, E107P, D158A, L162A. The cognate tRNA is an engineered variant of M. jannaschii tyrosyl tRNA, in which the anticodon has been replaced with the triplet CUA for recognition of an amber stop codon.
While the plasmid/promoter configuration described above has proven effective toward producing bicyclic peptides in E. coli in good yields (> 0.5 mg/L culture) according to the methods described herein, alternative configurations are likely to be equally viable provided the key elements are included and the plasmid compatibility issues are properly addressed. For example, O2beY-containing proteins have been successfully expressed using a different vector (e.g. pET vector) and from a gene under a different inducible promoter (e.g. IPTG-inducible T7 promoter) [17].
The following procedure was utilized to prepare pBAD-based vectors for the expression of the representative precursor proteins Z3C_O2beY and Z8C_O2beY described in Table 1. The oligonucleotide primers used in this protocol are described in Table 2 and were obtained by custom synthesis from IDT Technologies.
Table 1.
Name and target sequences of the precursor protein constructs described in the protocols.
| Construct name | Target sequence[a] | Cys position[b] |
|---|---|---|
| Z3C_O2beY | S(O2beY)TNCHPQFANA | i+3 |
| Z8C_O2beY | S(O2beY)TNVHPQFCNA | i+8 |
Full-length precursor protein corresponds to IntC-(target sequence)-IntN-(Chitin Binding Domain).
Position of reactive cysteine residue (underlined) in relation to O2beY (= i position).
Table 2.
Oligonucleotide primers.
| Primer | Sequence |
|---|---|
| SICLOPPS_for | 5′-CAGGTCATATGGTTAAAGTTATCGGTCGTCGATCC-3′ |
| SICLOPPS_rev | 5′-CAACAGGTACCTTTAATTGTACCTGCGTCAAGTAATGGAAAG-3′ |
| Z3C_for | 5′-CGCAGTTCGCGAACGCGTGCTTAAGTTTTGGCACCGAAATT-3′ |
| Z3C_1/2_rev | 5′-GGATGGCAGTTGGTCTAGCTATTGTGGGCGATAGCACCATTAGC-3′ |
| Z3C_2/2_rev | 5′-CGCGTTCGCGAACTGCGGATGGCAGTTGGTCTAGCTATTGTG-3′ |
| Z8C_1/2_rev | 5′-GATGCACGTTGGTCTAAGAATTGTGGGCGATAGCACCATTAGC-3′ |
| Z8C_2/2_rev | 5′-CGCGTTGCAGAACTGCGGATGCACGTTGGTCTAAGAATTG-3′ |
| pBAD_seq_for | 5′-GGATCCTACCTGACGCTTTTTATCG-3′ |
The genes encoding the N-terminal and C-terminal DnaE inteins from Synechocystis sp. were extracted from pSFBAD09 and pJJDuet30 plasmids [24] (Addgene #11963 and #11962, respectively). The pSFBAD09 vector also served as the host vector for insertion of the precursor protein gene.
For preparation of the Z3C_O2beY construct, PCR amplify the DnaE-IntC-containing segment of the precursor protein gene using pSFBAD09 vector as the template, primer SICLOPPS_for as the forward primer, and primer Z3C_1/2_rev as the reverse primer. PCR solution: 10 μL Phusion buffer HF 5×, 36 μL deionized water, 0.5 μL template plasmid (= 50 ng/μL), 1 μL SICLOPPS_for primer (10 μM), 1 μL Z3C_1/2_rev primer (10 μM), 1 μL dNTP mix, 0.5 μL Phusion polymerase (Note 4). PCR method: 98°C for 2 min, then 30 cycles: 98°C for 15 sec, 61°C for 15 sec, 72°C for 30 sec; then 72°C for 2 min, then 4°C. Analyze the in vitro synthesized dsDNA by resolving on 1.5% agarose gel.
Add 0.5 μL of Dpn I to the reaction mixture of step 2 (to digest the plasmidic DNA) and incubate at 37°C for 2 hours. Purify the product of the PCR reaction (0.12 Kbp) using QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s instructions.
Using the purified PCR product from step 3 as the template, perform second PCR reaction using primer SICLOPPS_for as the forward primer and primer Z3C_2/2_rev as the reverse primer. PCR solution: 10 μL Phusion buffer HF 5×, 36 μL deionized water, 1 μL PCR product from step 3 (50–100 ng/μL), 1 μL SICLOPPS_for primer (10 μM), 1 μL Z3C_2/2_rev primer (10 μM), 1 μL dNTP mix, 0.5 μL Phusion polymerase. PCR method: 98°C for 2 min, then 30 cycles: 98°C for 15 sec, 58°C for 15 sec, 72°C for 30 sec; then 72°C for 2 min, then 4°C. Analyze the in vitro synthesized dsDNA by resolving on 1.5% agarose gel.
Purify the product of the PCR reaction (0.13 Kbp) in step 4 using a PCR purification kit according to the manufacturer’s instructions.
PCR amplify the DnaE-IntN-containing segment of the precursor protein gene using pJJDuet30 vector as the template, primer Z3C_for as the forward primer and SICLOPPS_rev as the reverse primer. PCR solution: same as in step 2 with the use of appropriate primers and template as described here. PCR method: 98°C for 2 min, then 30 cycles: 98°C for 15 sec, 61°C for 15 sec, 72°C for 30 sec; then 72°C for 2 min, then 4°C. Analyze the in vitro synthesized dsDNA by resolving on 1.5% agarose gel.
Add 0.5 μL of Dpn I to the PCR reaction of step 6 (to digest the plasmidic DNA) and incubate at 37°C for 2 hours. Purify the product of the PCR reaction (0.38 Kbp) using a PCR purification kit according to the manufacturer’s instructions.
Merge the genes isolated in steps 5 and 6 by PCR Overlap Extension using SICLOPPS_for and SICLOPPS_rev as superprimers. PCR solution: 10 μL Phusion buffer HF 5×, 28 μL deionized water, 5 μL each of the gene segments (150–200 ng each in 1:1 ratio), 1 μL dNTP mix, 0.5 μL Phusion polymerase. PCR method: 98°C for 2 min, then 6 cycles: 98°C for 15 sec, 55°C for 15 sec, 72°C for 30 sec; then add 1 μL of 1:1 mixture of SICLOPPS_for and SICLOPPS_rev primers; then 30 cycles: 98°C for 15 sec, 63°C for 15 sec, 72°C for 30 sec; then 72°C for 2 min, then 4°C. Analyze the in vitro synthesized dsDNA by resolving on 1.5% agarose gel. (Note 5)
Purify the product of the PCR Overlap Extension reaction (0.51 Kbp) in step 8 using a PCR purification kit according to the manufacturer’s instructions.
Digest the purified product of the PCR Overlap Extension reaction from step 9 using Nde I and Kpn I HF restriction enzymes. Double digest mixture: 100–300 ng DNA, 1 μL Nde I, 1 μL Kpn I-HF, 5 μL 10× CutSmart buffer, deionized water to final volume of 50 μL. Leave the reaction in a water bath for 45 min.
Digest the pSFBAD09 vector using Nde I and Kpn I restriction enzymes. Double digest mixture: 300–500 ng vector DNA, 1 μL Nde I, 1 μL Kpn I-HF, 5 μL 10× CutSmart buffer, deionized water to final volume of 50 μL. Leave the reaction in a water bath for 45 min.
To the double digest reaction mixture from step 11 add of 1 μL Calf Intestinal Alkaline Phosphatase (CIP) in order to dephosphorylate the 5′ and 3′ end of the digested vector and prevent circularization of the vector without the insert. Incubate the reaction mixture for additional 30 min in a water bath at 37°C.
Load the digested insert (step 10) and digested vector (step 12) into separate wells of a 1.2% agarose gel. Resolve the dsDNA products by electrophoresis and excise the bands corresponding to the insert and linearized vector. Combine the excised bands and extract the dsDNA by a gel extraction kit according to the manufacturer’s instructions. Elute the DNA with 30 μL of the elution buffer provided with the kit.
Ligate the insert and vector together using T4 ligase. Ligation solution mixture: 12 μL deionized water, 1 μL T4 ligase, 2 μL 10× T4 ligase buffer, 5 μL DNA mixture from step 13. Leave the ligation reaction for 2 h at room temperature. Cloning of the insert into the Nde I / Kpn I cassette in the pSFBAD09 vector result in the fusion of the CBD protein to the C-terminus of the DnaE-IntC-(target sequence)-DnaE-IntN gene (Figure 4).
Transform the purified ligation mixture (step 14) into chemically competent DH5α cells or equivalent E. coli cells for general cloning purposes. Typical procedure: add 5 μL of the ligation mixture to a 50 μL aliquot of chemically competent DH5α cells. Gently tap the tube and incubate the cells on ice. After 30 min, place the tube in a water bath pre-warmed at 42°C for 30 seconds followed by transfer to ice for additional 5 min.
Add 500 μL of LB medium and incubate the cells with shaking for 45 min at 37°C. Plate 100 μL of the cell culture onto LB agar plates containing ampicillin at 100 mg/L and incubate the plate at 37°C for 14–18 hours.
Pick a colony from the plate with a sterile toothpick and inoculate a culture tube containing 5 mL of LB medium supplemented with ampicillin at 100 mg/L. After overnight incubation at 37°C, pellet the cells by centrifugation at 5,000 × g, and extract the plasmidic DNA from the cells using a plasmid miniprep kit according to the manufacturer’s instruction.
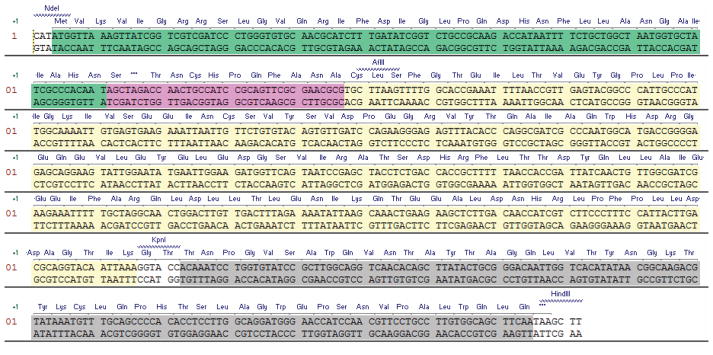
Confirm the identity of the clone construct by DNA sequencing using pBAD_seq_for as sequencing primer. The DNA and protein sequence of the gene for expression of the precursor protein Z3C_O2beY is provided in Figure 4.
The Z8C_O2beY construct can be cloned using an identical procedure (steps 2 through 18) with the exception that primer Z8C_rev_1/2 and Z8C_rev_2/2 are to be used instead in step 6.
Figure 4.
DNA and protein sequence for the precursor protein Z3C_O2beY (Table 1). The target peptide sequence is highlighed in pink and is preceeded by the IntC domain of DnaE split intein (green) and C-terminally fused to DnaE IntN domain (yellow) and the chitin binding domain affinity tag (grey).
3.2 Expression of the O2beY-containing precursor proteins
The following section describes a typical procedure for the expression of the precursor protein constructs Z3C_O2beY and Z8C_O2beY described in Table 1. An important consideration concerning the choice of the E. coli strain for recombinant expression of these constructs is the use of a non-suppressor strain such as BL21 or BL21(DE3). E. coli strains characterized by genotypic signatures such as supD, supH, supE, supF, supP, or supZ contain mutated tRNAs that cause the incorporation of a natural amino acid (Ser, Gln, Tyr, etc.) in response to an amber stop codon, thus interfering with the incorporation of O2beY into the target sequence during the expression of the precursor protein. As illustrated by the model constructs described in Table 1, the precursor proteins should consist of, from the N- to the C-terminus, the DnaE IntC-domain, followed by an arbitrary peptide target sequence containing the O2beY and cysteine residues spaced by one to seven interjecting residues, followed by the DnaE IntN-domain fused to a chitin-binding domain (CBD). While the CBD domain is not essential for production of the bicyclic peptide, it is useful for isolation of the precursor protein and trans splicing product via chitin-affinity chromatography and subsequent analysis of the extent of split intein-mediated splicing of the precursor protein.
In terms of design principles, target peptide sequences comprising between 12 and 18 amino acid residues have been successfully cyclized in living E. coli cells yielding the desired bicyclic peptides in high yields (70–99%) [18]. It is reasonable to assume that shorter and longer sequences would be also amenable to bicyclization. As mentioned earlier, the inter-side-chain cross-linking reaction proceeds most efficiently (80–100%) when the O2beY and cysteine residues are separated by 2 up to 7–8 residues. Constructs with the O2beY/Cys pair in i/i+10 to i/i+12 relationships are still able to form the thioether bridge but with reduced efficiency (20–50%) [17]. The first residue of the peptide target sequence (i.e., ‘IntC+1 position’, Figure 1) is involved in the mechanism of split intein-catalyzed trans splicing (Figure 2) and must be a serine or cysteine residue. In a side-by-side comparison experiment, precursor proteins containing a serine or cysteine residue at this site were found to lead to the desired bicyclic peptide with equal efficiency [18], suggesting that the bicyclization process is not affected by the inherently different nucleophilicity of these amino acids. If a cysteine residue occupies the IntC+1 site, however, O2beY should be ideally positioned in the IntC+2 position so to prevent alkylation of the mechanistically important cysteine by means of O2beY-borne bromoethyl group. In this i/i+1 arrangement, the O2beY/Cys cross-linking reaction was indeed found to be very inefficient (<5%), possibly due to the formation of a strained 14-membered macrocycle [17]. This structural requirement is eliminated when a serine residue is used as IntC+1 residue.
Add 50 ng of pBAD_Z3C (or pBAD_Z8C) plasmid and 50 ng μL of pEVOL_O2beY plasmid to a 50 μL aliquot of chemically competent BL21(DE3) E. coli cells. Gently tap the tube to mix the DNA and incubate the cells on ice for 30 minutes.
Place the cells in a heat block at 42°C for 30 seconds followed by a quick transfer to ice for additional 5 minutes.
Add 500 μL of LB medium and incubate the cells with shaking for 45 min at 37°C.
Plate 100 μL of the LB culture onto an LB agar plate containing 50 mg/L ampicillin and 30 mg/L chloramphenicol. Place the plate in an incubator at 37°C for 14–18 hours until visible colony growth is observed.
Pick a single colony from the plate with a sterile toothpick and inoculate a culture tube containing 5 mL of LB medium supplemented with 50 mg/L ampicillin and 30 mg/L chloramphenicol. Incubate the culture tube overnight with shaking at 37°C.
Prepare 500 mL of M9 medium using presterilized solutions. Add ampicillin to 50 mg/L and chloramphenicol to 30 mg/L concentration. Mix well (Note 6).
Inoculate 500 mL of M9 medium with the overnight cell culture from step 5. Incubate the flask at 37°C with shaking (150–200 rpm) until OD600 reaches 0.6 (~ 3–4 hours).
Once the desired cell density is reached, transfer the culture flask to room temperature. After 30 min, add L-arabinose to a final concentration of 0.1% (w/v) and O2beY to a final concentration of 2 mM from a stock solution of 0.2 M in water (Note 7).
Grow the culture in a shaking incubator at 27°C for 12 hours followed by incubation at 37°C for 3 hours (Note 8).
Harvest the cells by centrifugation at 5,000 × g for 10 min. Resuspend the cells in 50 mL of phosphate buffered saline solution (100 mM sodium phosphate, 150 mM NaCl, pH 7.5).
Lyse the cells by sonication. Method: amplitude 50, process time 30 s, pulse on 1 s, pulse off 10 s. Repeat this method two times.
Centrifuge the cell lysate at 18,000 × g for 30 min at 4 °C. Use the clarified lysate immediately or flash freeze it in dry ice and store it at −80°C. If frozen, use within one week.
3.4 Isolation and characterization of the DnaE trans splicing products
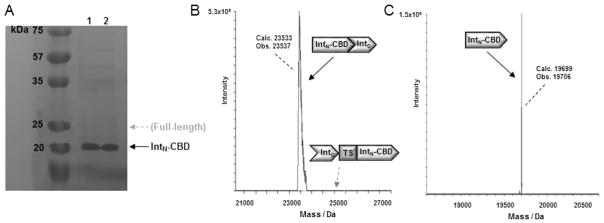
A key step in the formation of the bicyclic peptide products according to the general methodology of Figure 1 is the trans splicing event occurring upon association of the N- and C-domain of DnaE split intein, which results in the head-to-tail cyclization of the target peptide sequence (Figure 2). The presence of the CBD domain fused to the C-terminal end of the DnaE IntN domain allows for the isolation of both the full-length precursor protein (IntC-(target sequence)-IntN-CBD) and spliced protein (IntN-CBD/IntC complex) via chitin-affinity capturing using chitin-coated agarose beads. Upon elution of these proteins from the chitin beads, the extent of trans splicing can be estimated by measuring the relative amount of full-length protein and spliced intein by SDS-PAGE analysis followed by gel band densitometry (Figure 5A). The occurrence and extent of DnaE-catalyzed trans splicing can be assessed and further confirmed by LC-ESI-MS analysis of the chitin-affinity purified proteins (Figure 5B).
Figure 5.
Analysis of DnaE trans splicing products. A) SDS-PAGE analysis of the protein construct Z3C_O2beY (lane 1) and Z8C_O2beY (lane 2) isolated using chitin beads. The bands corresponding to IntN-CBD and full-length precursor protein are labeled. The latter is not visible due to complete splicing of the precursor protein. B) Deconvoluted MS spectrum of chitin-affinity purified IntN-CBD/IntC complex from Z3C_O2beY sample. The position of the full-length precursor protein (if present) is also indicated. (C) Deconvoluted MS spectrum corresponding to the dissociated IntN-CBD fragment upon treatment of the chitin-affinity purified IntN-CBD/IntC complex (from Z3C_O2beY sample) with 6 M guanidine hydrochloride.
For the constructs described in Table 1, the molecular weights for IntN-CBD and the IntN-CBD/IntC complex are 19,699 Da and 23,533 Da, respectively, whereas the molecular weights for the full-length precursor proteins Z3C_O2beY and Z8C_O2beY are 24,892 Da and 24,920 Da, respectively. It should be noted that under standard LC-ESI-MS conditions using C18 reverse-phase chromatography in water/acetonitrile systems with 0.1% formic acid and electrospray ionization (ESI), the non-covalent IntN-CBD/IntC complex is sufficiently stable to be observed in the associated form (Figure 5B). In contrast, under denaturing conditions such as in the presence of sodium dodecyl sulfate (Figure 5A) or guanidinium chloride (Figure 5C), the IntN-CBD/IntC complex gives rise to a large (19.7 KDa) and a small (3.8 KDa) protein fragment corresponding to the dissociated IntN-CBD and IntC polypeptides.
Transfer 0.5 mL of chitin beads to a 15 mL tube and spin them down at 1,400 × g for 1 min. Carefully remove the solvent, resuspend the beads in 2 mL of phosphate buffer and spin them down using the same conditions, repeat this process two more times.
To the chitin beads add 3 mL aliquot of lysed cells and incubate for 1 h on ice. Wash the beads two times with 2 mL of phosphate buffer, as described in step 1, each time remove as much of the buffer as possible without disturbing the beads. Avoid prolonged incubation of beads in the phosphate buffer to circumvent potential release of the proteins from the chitin beads.
Add 0.2 mL of acetonitrile:H2O mixture (70:30 v/v) to the chitin beads and incubate at room temperature for one minute to release any chitin-bound protein (Note 9).
Carefully separate the eluate from the beads and lyophilize the collected eluate.
The identity of the proteins can be determined by LC-ESI-MS and/or SDS-PAGE analysis, Figure 5. For LC-ESI-MS analysis dissolve the lyophilized eluates in a minimal amount (i.e. 100–200 μL) of water or desired buffer (i.e. 6 M guanidinium hydrochloride) with gentle mixing. For SDS-PAGE analysis dissolve the lyophilized eluates in a minimal amount of water.
3.3 Isolation and characterization of the macrocyclic peptide products
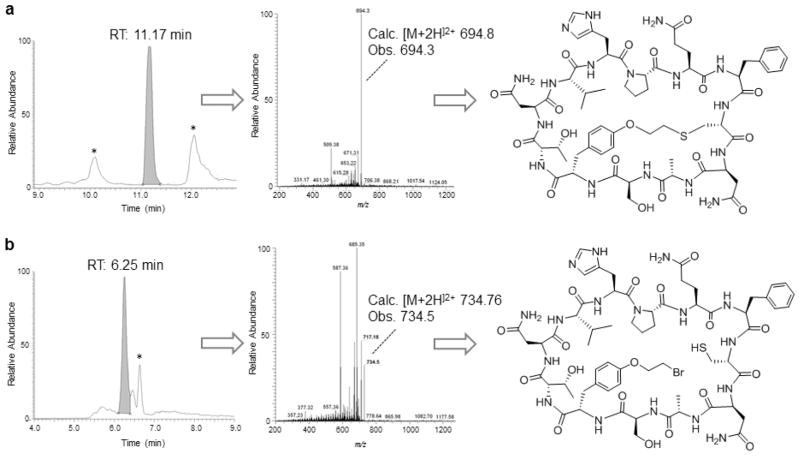
The precursor proteins described in Table 1 were designed to contain a His-Pro-Gln (HPQ) motif which is known to bind to streptavidin [25,26] and which can be exploited to facilitate the isolation of the macrocyclic peptide products via capturing with streptavidin-coated agarose beads. In addition to the desired bicyclic peptide product (Figure 1), a possible product of the reaction of Figure 1 is the head-to-tail monocyclic peptide, which occurs whenever the O2beY/Cys cross-linking reaction does not proceed in a quantitative manner. Both the bicyclic and monocyclic peptide products (along with any undesired adduct, Note 10) incorporate the HPQ motif and are captured during the streptavidin-binding procedure, while remaining distinguishable based on their different masses and behavior to MS/MS fragmentation. Accordingly, after elution from the streptavidin beads, these peptides products can be resolved and quantified by LC-ESI-MS analysis under standard analytical conditions (Figure 6). In the following, a protocol for the isolation, identification, and quantification of the bicyclic and monocyclic peptide products generated upon in vivo cyclization of the protein precursor Z8C_O2beY is described.
Figure 6.
LC–MS analysis of macrocyclic products obtained from in vivo cyclization of Z8C_O2beY precursor protein. (A) LC-MS extracted-ion chromatogram (left panel), MS/MS fragmentation spectrum (middle panel), and chemical structure (right panel) of bicyclic peptide isolated via streptavidin affinity from cells expressing the contruct Z8C_O2beY. (B) Same as (A) for the head-to-tail cyclic peptide byproduct. Peaks labeled with * correspond to unrelated multicharged ions from adventitious proteins.
Transfer 0.1 mL of the streptavidin-coated agarose slurry to a 15 mL tube and spin them down at 1,400 × g for 1 min. Carefully remove the solvent, resuspend the beads in 2 mL of phosphate buffer and spin them down using the same conditions, repeat this process four more times.
Transfer 3 mL aliquot of lysed cells to the tube containing streptavidin-coated agarose beads and incubate for 3 hours under gentle shaking on ice (Note 11). Wash the beads two times with 2 mL of the phosphate buffer as described in step 1. Carefully remove the solvent above the beads without disturbing them.
Incubate the agarose beads with 0.2 mL of acetonitrile:H2O mixture (70:30 v/v) for one minute to release any streptavidin-bound peptides and lyophilize the eluates (Note 11).
Dissolve the lyophilized eluates in minimal amount of DMSO (i.e. 10–20 μL) and dilute with water to final concentration of DMSO of 10%. The identity of the eluted peptides can be determined by LC-ESI-MS analysis, Figure 6. For measuring the relative amount of the mono- and bicyclic peptide products all of the potential peptide products need to be taken into consideration (Note 10).
Acknowledgments
This work was supported by the U.S. National Institutes of Health (grant R21 CA187502). MS instrumentation was supported by the U.S. National Science Foundation (grants CHE-0840410 and CHE-0946653).
Footnotes
M9 medium needs to be freshly prepared each time using presterilized components. If the M9 medium is made from non-sterile components and then autoclaved, the salts will precipitate.
The DMF reaction should be diluted with a minimum of 3× volumes of water to ensure successful product extraction.
Substitution of the TFA with acetic acid lowers the toxicity of the unnatural amino acid to bacterial cells during expression of the precursor proteins.
Keep all of the reagents for the PCR reactions at −20°C and only take out just prior to use. Always add polymerase last, just prior to starting the PCR reaction.
The success of the PCR Overlap Extension reaction is highly dependent upon the quality of the PCR products that are to be joined together. If the gene segments are of low quality (e.g., as apparent from smearing of the corresponding band on the agarose gel), the PCR Overlap Extension reaction may not work or may lead to mixture of products. If necessary, higher quality gene segments can be prepared by optimizing the PCR conditions, e.g. by varying the annealing time and temperature or by re-designing the primers. Alternatively, the PCR product can be purified by gel extraction using commercial kits such as QIAquick Gel Extraction Kit (Qiagen).
Upon addition of CaCl2, some precipitate may be observed. Mix well until it is completely dissolved.
The O2beY stock solution is a suspension, mix thoroughly prior to addition to bacterial cells. Store the O2beY stock solution at −20°C.
The additional 3-hour incubation step was found to promote DnaE-mediated trans splicing and thus head-to-tail cyclization of the peptide.
Chitin beads can be re-used up to five times without significant loss of CBD binding capacity. To preserve their binding properties, the beads should be washed 3× with 5 mL of phosphate buffer immediately after treatment with the acetonitrile:H2O mixture and can be stored in the same buffer at 4°C for several weeks.
Other possible byproducts may consist of modified adducts of the head-to-tail monocyclic peptide resulting from nucleophilic displacement of O2beY side-chain 2-bromoethyl group with abundant cellular nucleophiles (i.e., cysteine, glutathione) or water. While the possible occurrence of these side-reactions should not be ignored, these adducts were never observed across several different target peptide sequences [17,18], supporting the high chemo- and regioselectivity of the O2beY-mediated cross-linking reaction.
Streptavidin-coated agarose beads can be re-used up to three times without significant loss of binding capacity. To preserve their binding properties, the beads should be washed 5× with 5 mL of phosphate buffer immediately after treatment with the acetonitrile:H2O mixture and can be stored in the same buffer at 4°C for several weeks.
References
- 1.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery--an underexploited structural class. Nat Rev Drug Discov. 2008;7(7):608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 2.London N, Raveh B, Schueler-Furman O. Druggable protein-protein interactions--from hot spots to hot segments. Curr Opin Chem Biol. 2013;17(6):952–959. doi: 10.1016/j.cbpa.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Bionda N, Fasan R. Peptidomimetics of α-helical and β-strand protein binding epitopes. In: Czechtizky W, Hamley P, editors. Small Molecule Medicinal Chemistry. Strategies and technologies. Wiley & Sons, Inc; Hoboken, NJ: 2015. pp. 431–464. [Google Scholar]
- 4.White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat Chem. 2011;3(7):509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 5.Hipolito CJ, Suga H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: the FIT and RaPID systems. Curr Opin Chem Biol. 2012;16(1–2):196–203. doi: 10.1016/j.cbpa.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Smith JM, Frost JR, Fasan R. Emerging strategies to access peptide macrocycles from genetically encoded polypeptides. J Org Chem. 2013;78(8):3525–3531. doi: 10.1021/jo400119s. [DOI] [PubMed] [Google Scholar]
- 7.Frost JR, Smith JM, Fasan R. Design, synthesis, and diversification of ribosomally derived peptide macrocycles. Curr Opin Struct Biol. 2013;23(4):571–580. doi: 10.1016/j.sbi.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Baeriswyl V, Heinis C. Polycyclic peptide therapeutics. ChemMedChem. 2013;8(3):377–384. doi: 10.1002/cmdc.201200513. [DOI] [PubMed] [Google Scholar]
- 9.Lennard KR, Tavassoli A. Peptides come round: using SICLOPPS libraries for early stage drug discovery. Chemistry. 2014;20(34):10608–10614. doi: 10.1002/chem.201403117. [DOI] [PubMed] [Google Scholar]
- 10.Bowers AA. Biochemical and biosynthetic preparation of natural product-like cyclic peptide libraries. Medchemcomm. 2012;3(8):905–915. [Google Scholar]
- 11.Josephson K, Ricardo A, Szostak JW. mRNA display: from basic principles to macrocycle drug discovery. Drug Discovery Today. 2014;19(4):388–399. doi: 10.1016/j.drudis.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Smith JM, Vitali F, Archer SA, Fasan R. Modular Assembly of Macrocyclic Organo-Peptide Hybrids Using Synthetic and Genetically Encoded Precursors. Angew Chem Int Ed Engl. 2011;50(22):5075–5080. doi: 10.1002/anie.201101331. [DOI] [PubMed] [Google Scholar]
- 13.Satyanarayana M, Vitali F, Frost JR, Fasan R. Diverse organo-peptide macrocycles via a fast and catalyst-free oxime/intein-mediated dual ligation. Chem Commun (Camb) 2012;48(10):1461–1463. doi: 10.1039/c1cc13533c. [DOI] [PubMed] [Google Scholar]
- 14.Frost JR, Vitali F, Jacob NT, Brown MD, Fasan R. Macrocyclization of Organo-Peptide Hybrids through a Dual Bio-orthogonal Ligation: Insights from Structure-Reactivity Studies. Chembiochem. 2013;14(1):147–160. doi: 10.1002/cbic.201200579. [DOI] [PubMed] [Google Scholar]
- 15.Smith JM, Hill NC, Krasniak PJ, Fasan R. Synthesis of bicyclic organo-peptide hybrids via oxime/intein-mediated macrocyclization followed by disulfide bond formation. Org Biomol Chem. 2014;12(7):1135–1142. doi: 10.1039/c3ob42222d. [DOI] [PubMed] [Google Scholar]
- 16.Frost JR, Jacob NT, Papa LJ, Owens AE, Fasan R. Ribosomal Synthesis of Macrocyclic Peptides in Vitro and in Vivo Mediated by Genetically Encoded Aminothiol Unnatural Amino Acids. ACS Chem Biol. 2015;10(8):1805–1816. doi: 10.1021/acschembio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bionda N, Cryan AL, Fasan R. Bioinspired strategy for the ribosomal synthesis of thioether-bridged macrocyclic peptides in bacteria. ACS Chem Biol. 2014;9(9):2008–2013. doi: 10.1021/cb500311k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bionda N, Fasan R. Ribosomal Synthesis of Natural-Product-Like Bicyclic Peptides in Escherichia coli. Chembiochem. 2015;16(14):2011–2016. doi: 10.1002/cbic.201500179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci U S A. 1999;96(24):13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 21.Nordgren IK, Tavassoli A. A bidirectional fluorescent two-hybrid system for monitoring protein-protein interactions. Mol Biosyst. 2014;10(3):485–490. doi: 10.1039/c3mb70438f. [DOI] [PubMed] [Google Scholar]
- 22.Horswill AR, Savinov SN, Benkovic SJ. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc Natl Acad Sci U S A. 2004;101(44):15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol. 2010;395(2):361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 24.Zuger S, Iwai H. Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat Biotechnol. 2005;23(6):736–740. doi: 10.1038/nbt1097. [DOI] [PubMed] [Google Scholar]
- 25.Katz BA. Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence. Biochemistry. 1995;34(47):15421–15429. doi: 10.1021/bi00047a005. [DOI] [PubMed] [Google Scholar]
- 26.Naumann TA, Savinov SN, Benkovic SJ. Engineering an affinity tag for genetically encoded cyclic peptides. Biotechnol Bioeng. 2005;92(7):820–830. doi: 10.1002/bit.20644. [DOI] [PubMed] [Google Scholar]