Abstract
The retinoid X receptor α (RXRA) has been implicated in diverse hematological processes. However, it is unknown whether natural ligands of RXRA are present in hematopoietic cells. We adapted our upstream activation sequence-green fluorescent protein (UAS-GFP) reporter mouse to detect natural RXRA ligands in vivo. We observed reporter activity in diverse, primary mouse hematopoietic cells in vivo. We found that reporter activity increased during granulocyte colony stimulating factor (GCSF)–induced granulopoiesis and following phenylhydrazine (PHZ)–induced anemia, suggesting the presence of dynamically regulated natural RXRA ligands in hematopoietic cells. We found that mouse plasma could activate Gal4-UAS reporter cells in vitro, and plasma from mice treated with GCSF and PHZ recapitulated the patterns we observed in vivo. Severe vitamin A depletion had only a mild effect on RXRA reporter activity, whereas short-term fatty acid restriction reduced reporter activity, implicating fatty acids as plasma RXRA ligands. Through differential extraction, coupled with mass spectrometry, we identified the long chain fatty acid C24:5 as a natural RXRA ligand, which was dynamically increased in concentration in response to hematopoietic stress. Collectively, these data demonstrate that natural RXRA ligands are present and are dynamically increased in vivo in mouse hematopoietic cells.
Introduction
Retinoid X receptors (RXRs) are members of the superfamily of nuclear receptors (1). Like other nuclear receptors, the transcriptional activity of RXRs is ligand-dependent. Ligand binding results in a conformational shift of the terminal α helix (AF2 domain), which displaces bound co-repressors and facilitates the binding of co-activators.
Diverse molecules have been implicated as natural RXR ligands, including both retinoic acids and fatty acids (2). It is unknown whether any of these are present in hematopoietic cells in physiologically relevant quantities or whether they are dynamically increased during hematopoietic stress. 9-cis retinoic acid (9-cis-RA) has been described as a natural RXR ligand (3, 4). However, several groups reported 9-cis-RA to be absent or below detectable limits in testis, liver, heart, lung, and serum (5, 6). Another vitamin A metabolite, 9-cis-13,14-dihydroretinoic acid (9-cis-13,14-DHRA), was reported to be an endogenous RXR ligand in mouse serum, brain, and liver, although it is unclear if it is present in hematopoietic cells (7). Using a luciferase reporter assay, Lengqvist et al. showed that unsaturated fatty acids such as docosahexaenoic acid (DHA, C22:6), docosapentaenoic acid (DPA, C22:5) and arachidonic acid (AA, C20:4) activate RXRA in vitro, whereas saturated fatty acids like arachidic acid (C20:0) and stearic acid (C18:0) do not (8).
Three separate genes encode three RXR isoforms — RXRα (RXRA), RXRβ (RXRB), and RXRγ (RXRG) – at least one of which is present in every mammalian cell (9). These subtypes are highly conserved, with near-identical ligand binding pocket conformations (1). RXRs bind to DNA as either a homodimer (10) or a heterodimer (1); the heterodimeric partners of RXR include retinoic acid receptors (RARs), thyroid hormone receptors (TRs), the vitamin D receptor (VDR), peroxisome proliferator-activated receptors (PPARs), the liver X receptor (LXRs), the pregnane X receptor (PXR), and the constitutive androstane receptor (CAR) (11-15). By heterodimerizing with its partners, RXRs participates in diverse essential biological processes, such as development, metabolism, cell differentiation and cell death.
RXRs play important roles in hematopoiesis. RXRA abundance increases upon monocytic differentiation (16). Ectopic overexpression of RXRA in hematopoietic stem and progenitor cells inhibits granulopoiesis by impairing proliferation and differentiation, whereas functional genetic interference of Rxra promotes the generation of late-stage granulocytes in vitro (17). Rxra binds to the EPO promoter and promotes Epo transcription during E9.5-E11.5 phase of fetal liver erythropoiesis, prior to subsequently being supplanted by HNF4-dependent transcription of Epo from E11.5 onward (18). Rxrs also participate in osteoclast differentiation and postnatal bone remodeling (19). Conditional deletion of both Rxra and Rxrb in hematopoietic cells (Rxrg is not present in hematopoietic cells) generates giant, non-resorbing osteoclasts and increases bone mass in the male mice, and protected female mice from osteoporotic bone loss following ovariectomy. No information is available about the presence or distribution of natural ligands of RXRA in hematopoietic cells.
Here, we demonstrate that natural RXRA ligands are present in mouse hematopoietic cells and plasma in vivo, and are predominantly active in myeloid cells. We find that concentrations of RXRA ligands are dynamically increased in response to myeloid stress, such as exposure to GCSF and phenylhydrazine (PHZ). Moreover, we identify the long chain fatty acid C24:5 as an endogenous RXRA ligand that undergoes dynamic increases in concentrations during mouse hematopoietic stress.
Results
Gal4-UAS reporter adapted to detect RXRA ligands
To determine whether natural RXRA ligands are present in hematopoietic cells in vivo, we employed transgenic upstream activation sequence-green fluorescent protein (UAS-GFP) reporter mice (20). UAS promoter sequences are recognized by the yeast Gal4 transcription factor and are not activated by mammalian proteins. When the modular Gal4-DNA binding domain (Gal4-DBD) is fused to the RXRA-ligand binding domain (RXRA-LBD) and retrovirally expressed in UAS-GFP bone marrow Kit+ cells (Gal4-RXRA), the reporter should respond to intracellular ligands that bind and transactivate RXRA. We included an internal ribosomal entry site (IRES)-mCherry cassette in the Gal4-RXRA retroviral vectors to identify cells that had been transduced (fig. S1A). In Kit+ mouse bone marrow cells transduced with Gal4-RXRA retrovirus and cultured ex vivo, this system was sensitive to the natural RXRA agonist 9-cis-RA and to the synthetic RXRA agonist bexarotene (fig. S1B and C). The EC50 of 9-cis-RA was 1.3 nM and the EC50 of bexarotene was 5.1 nM, similar to previously reported results (3), thus validating the reporter assay.
Detection of RXRA ligands in mouse hematopoietic cells in vivo
To determine whether natural RXRA ligands are present in hematopoietic cells in vivo, bone marrow Kit+ cells from UAS-GFP transgenic mice were transduced with Gal4-RXRA retrovirus and then transplanted into lethally irradiated recipient mice (Fig. 1A). Six weeks after the transplantation, the recipient mice were sacrificed and bone marrow, peripheral blood, spleen, and peritoneal macrophages were collected and analyzed by flow cytometry. We found GFP+mCherry+ cells in all four tissue types, suggesting the presence of natural RXRA ligands in hematopoietic cells (Fig. 1B and 1C). The RXRA terminal helix (AF2 domain) is required for ligand-dependent transcription, and deletion of this domain acts as a negative control (21). We verified that the Gal4-RXRA and Gal4-RXRA-ΔAF2 fusion proteins were present in Kit+ bone marrow cells and NIH-3T3 cells following transduced with retrovirus (fig. S2A). Because long chain fatty acids have been implicated as natural ligands for both RXRs and PPARs, (8, 22) we also verified the specificity of the reporter and found that it was not activated by PPAR ligands when cells were transduced with Gal4-RXRA (fig. S2B and S2C). GFP+mCherry+ cells were not observed in UAS-GFP mice transplanted with Gal4-RXRA-ΔAF2, suggesting that the GFP+mCherry+ cells observed following Gal4-RXRA transplantation result from RXRA ligand binding, and not from heterodimeric cross-talk or non-specific reporter activity. Interestingly, reporter activity was not uniform across all hematopoietic populations. Subgate analysis using lineage cell markers revealed that GFP+mCherry+ cells are biased toward CD11b+ myeloid lineage cells (Fig. 1D).
Figure 1. Natural RXRA ligands in mouse hematopoietic cells in vivo.
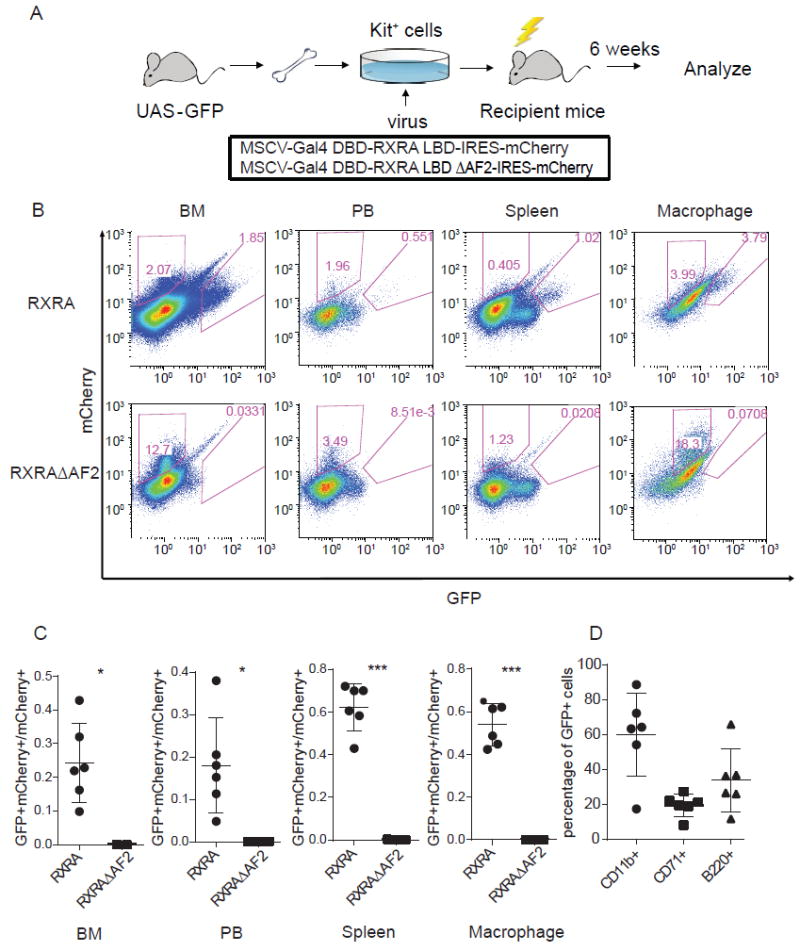
(A) Schema for bone marrow transplant procedure. Kit+ cells isolated from the bone marrow of UAS-GFP mice using MACS were transduced with one of the indicated retroviruses, then injected into lethally irradiated recipient mice. After 6 weeks of engraftment, recipient mice were sacrificed and their hematopoietic cells analyzed. (B) Representative FACS showing mCherry and GFP intensity in bone marrow cells (BM), peripheral blood (PB), spleen cells, and peritoneal macrophages from mice transplanted with UAS-GFP Kit+ bone marrow cells that were transduced with Gal4-RXRA or Gal4-RXRA-ΔAF2. (C) Ratio of GFP+mCherry+ cells relative to total mCherry+ cells in BM, PB, spleen, and peritoneal macrophages from mice transplanted with Kit+ bone marrow cells transduced with Gal4-RXRA (circles, N = 6 recipient mice) or RXRAΔAF2 (squares, N = 3 recipient mice). (D) The percentage of CD11b+ (circles), CD71+ (squares), and B220+ (triangles) cells in bone marrow GFP+mCherry+ from the recipient Gal4-RXRA mice (N = 6). Error bars represent standard deviation between individual mice. * P < 0.05, *** P < 0.001, T-test with Welch’s correction.
RXRA reporter output following GCSF and PHZ treatment
Given the roles of RXRA in myeloid maturation and the myeloid bias in GFP+mCherry+ cells observed in UAS-GFP transplants, we evaluated the effect of myeloid stress on natural RXR ligands in hematopoietic cells. We again transduced UAS-GFP bone Kit+ bone marrow cells with Gal4-RXRA retrovirus and transplanted these cells into recipient mice. After engraftment (~6 weeks), we treated transplanted mice with GCSF to induce granulopoiesis or with phenylhydrazine (PHZ) to induce hemolytic anemia and erythropoiesis (of note, following PHZ treatment we also observed granulopoiesis, with WBC peak at 18-22,000/μl, as has been noted elsewhere (23)). As a positive control, we also treated transplanted mice with bexarotene (a synthetic RXRA agonist). Compared to the mice without treatment, we found an increase in the proportion of GFP+mCherry+ cells in total bone marrow cells following GCSF or PHZ treatment (Fig. 2A and 2B). In addition, we found that the single cell abundance of GFP, indicated by mean fluorescence intensity (MFI), also increased (Fig. 2C). In comparison, GFP+mCherry+ cells were not detected in Gal4-RXRA-ΔAF2 transplanted mice following GCSF or PHZ treatment (Fig. 2A). These data suggest that intracellular concentrations of natural RXRA ligands are increased in hematopoietic cells during GCSF-induced granulopoiesis and PHZ-induced erythropoiesis, although concentrations of these natural ligands do not result in the maximum RXRA activation achieved by a potent pharmacologic agent (e.g. bexarotene).
Figure 2. UAS-GFP reporter output increased in hematopoietic cells following GCSF and PHZ treatment.
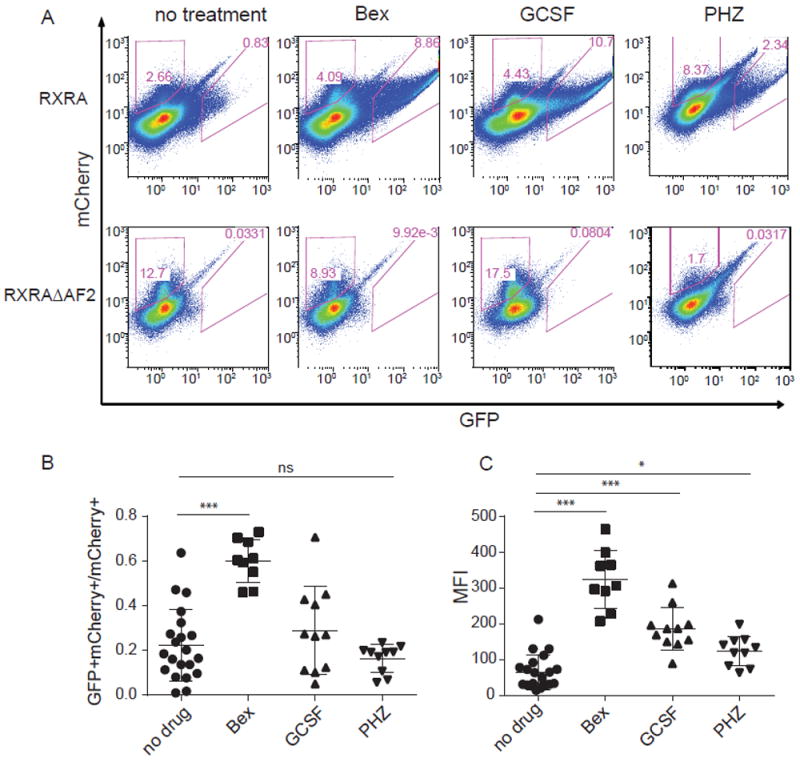
(A) mCherry and GFP intensity in whole bone marrow cells from recipient mice that were transplanted as in Fig. 1A and then treated with or without bexarotene, GCSF, or phenylhydrazine (PHZ). (B and C) Summary results of the ratio of GFP+mCherry+ cells to total mCherry+ cells (B) or mean fluorescence intensity (MFI) (C) in whole bone marrow cells from recipient mice transplanted with UAS-GFP Kit+ bone marrow cells transduced with Gal4-RXRA; N = 21 (Gal4-RXRA recipient mice without treatment), 9 (Gal4-RXRA recipient mice treated with bexarotene), 11 (Gal4-RXRA recipient mice treated with GCSF), 10 (Gal4-RXRA recipient mice treated with PHZ). Error bars represent standard deviation between individual recipient mice. * P < 0.05, *** P < 0.001, ANOVA with Tukey’s multiple comparison test.
Participation of RXRA in GCSF-induced granulopoiesis and hematopoietic stem cell mobilization
Because treatment of mice with bexarotene was able to induce greater activation of the UAS-GFP reporter than GSCF treatment (both in terms of the absolute proportion of GFP+mCherry+ cells, and the MFI of the GFP+mCherry+ cells), we determined if bexarotene could augment GCSF effects on granulopoiesis and HSCs mobilization. We found that bexarotene increased both the white blood cell (WBC) count and neutrophil (NE) count after GCSF treatment, compare to GCSF treatment alone (Fig. 3A and 3B). Bexarotene did not affect the absolute number of peripheral blood Kit+Lin-Sca-1+ (KLS) cells (Fig. 3C), or Kit+Lin-Sca-1- progenitor cells (Fig. 3D), but it did increase colony forming units (CFU, Fig. 3E) in peripheral blood following GCSF treatment.
Figure 3. RXRA in GCSF-induced granulopoiesis and hematopoietic stem cell mobilization.
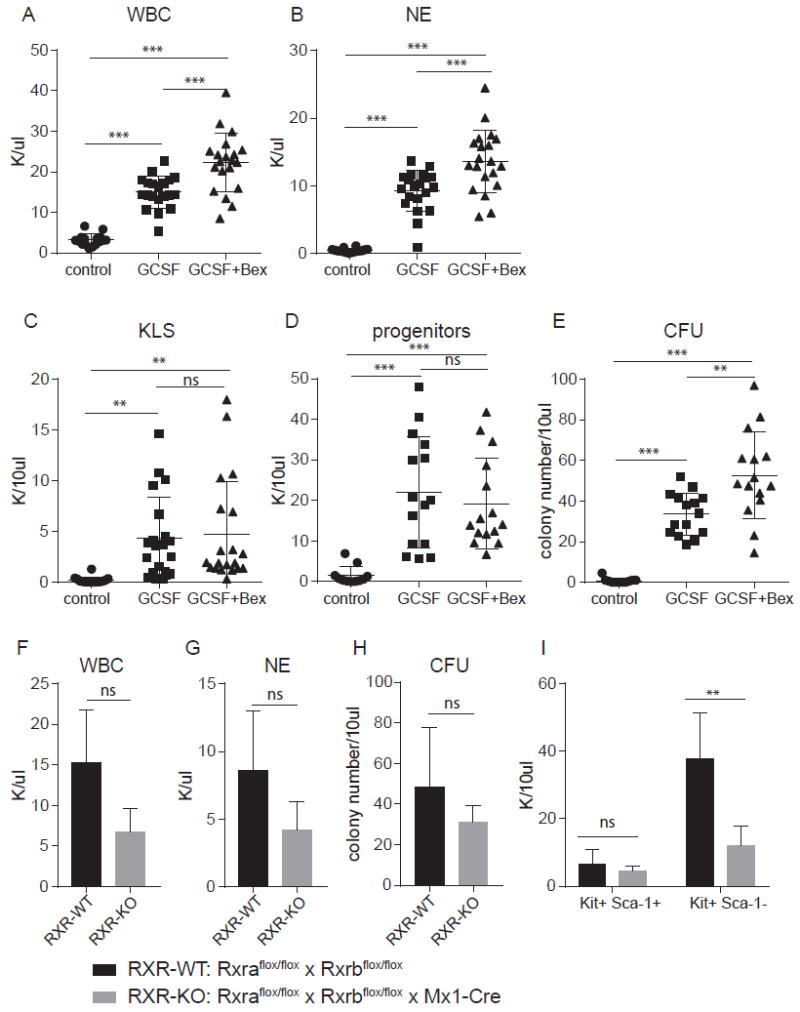
(A) White blood cell (WBC) count of C57BL6 mice treated with or without GCSF and bexarotene as indicated. (B) Neutrophil (NE) count of C57BL6 mice treated with or without GCSF and bexarotene as indicated. (C) Absolute number of Kit+Lin-Sca-1+ (KLS) cells in 10 μl peripheral blood of C57BL6 mice treated with or without GCSF and bexarotene as indicated. (D) Absolute number of Kit+Lin-Sca-1- (progenitors) cells in 10 μl peripheral blood of C57BL6 mice treated with or without GCSF and bexarotene as indicated. (E) Colony forming units in 10 μl peripheral blood of C57BL6 mice treated with or without GCSF and bexarotene as indicated. (F) WBC count of Rxraflox/flox × Rxrbflox/flox × Mx1-Cre (RXR-KO) mice and Rxraflox/flox × Rxrbflox/flox (RXR-WT) mice treated with GCSF. (G) NE count of RXR-KO mice and RXR-WT mice treated with GCSF. (H) Colony number generated from 10 μl peripheral blood of RXR-KO mice and RXR-WT mice. (I) Absolute number of Kit+Lin-Sca-1+ (KLS) cells and Kit+Lin-Sca-1- (progenitors) cells in 10 μl peripheral blood of RXR-KO mice and RXR-WT mice. A – B, control mice, N = 16; GCSF treated mice, N = 20; GCSF + bexarotene, N = 19 mice. C – E, control mice, N = 11; GCSF treated mice, N = 15; GCSF + bexarotene treated mice, N = 15. F – I, RXR-KO mice, n = 3; WT mice, n = 4. Error bars represent standard deviation between individual mice. * P < 0.05, ** P < 0.01, *** P < 0.001, ANOVA with Tukey’s multiple comparison test.
To determine whether RXR activity is essential for GCSF-induced granulopoiesis and mobilized HSCs’ function, we treated Rxraflox/flox × Rxrbflox/flox × Mx1-Cre (RXR-KO) mice and Rxraflox/flox × Rxrbflox/flox (RXR-WT) mice with GCSF (19). We observed a non-significant decrease in WBC and NE count, and peripheral blood colony forming units between RXR-KO and RXR-WT mice following GCSF treatment (Fig. 3F - 3H), but RXR-KO mice had fewer peripheral blood Kit+Lin-Sca-1- progenitor cells after GCSF treatment compared to RXR-WT mice (Fig. 3I). These data demonstrate that Rxra and Rxrb signaling contributes to GCSF-induced granulopoiesis and mobilized HSCs function, but is not essential.
Natural RXRA ligands in mouse plasma
To determine whether RXRA natural ligands are exclusively intracellular, or are more broadly available in the plasma, we transduced UAS-GFP bone marrow Kit+ cells with Gal4-RXRA retrovirus and treated the cultured cells ex vivo with mouse plasma. We found that plasma activated the reporter in a dose-dependent manner (Fig. 4A), and the ligands in 6 μl plasma (3%) activated the reporter as much as did 100 nM 9-cis-RA.
Figure 4. RXRA reporter activated by mouse plasma.
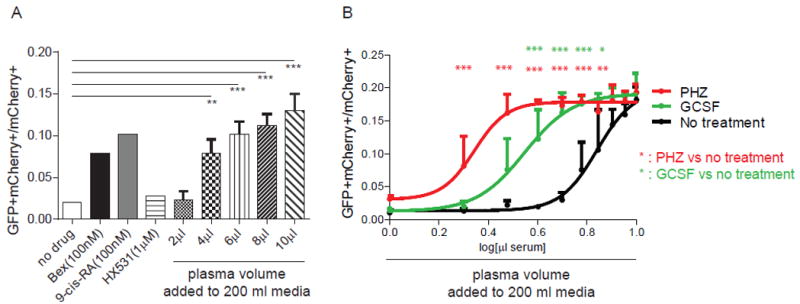
(A) UAS-GFP bone marrow Kit+ cells were transduced with Gal4-RXRA retrovirus and treated in culture with RXRA agonist (bexarotene and 9-cis-RA), antagonist (HX531), or mouse plasma as indicated, and the ratio of GFP+mCherry+ cells to total mCherry+ cells was determined. Cells were treated in a total volume of 200 μl media. Two way T-test compared against untreated control. (B) UAS-GFP bone marrow Kit+ cells were transduced with Gal4-RXRA retrovirus and treated in culture with plasma from indicated mice and the ratio of GFP+mCherry+ cells to total mCherry+ cells was determined. Error bars represent standard deviation between measurement of plasma obtained from individual mice (N = 3 mice per group). ANOVA with Tukey’s multiple comparisons compared results obtained at each plasma concentration. * P < 0.05, ** P < 0.01, *** P < 0.001.
Furthermore, we found that UAS-GFP reporter output increased when Kit+ cells were treated with plasma from mice exposed to PHZ or GSCF (Fig. 4B), consistent with our prior in vivo results (Fig. 2B), and we did not observe an increase following 5-fluoruracil (5-FU) treatment (fig. S3A). Again, reporter activity was absent in cells transduced with Gal4-RXRA-ΔAF2 (fig. S3B), and was not observed in cells transduced with Gal4-RARA (fig. S3C), again excluding the possibility of reporter activation via non-specific activation of the reporter or heterodimeric crosstalk.
Although both GCSF and PHZ increased UAS-GFP reporter activity and both induce granulopoiesis (23), the response following PHZ was modestly greater than following GCSF (Fig. 4B). Because of this, we focused ligand identification efforts primarily on plasma following PHZ treatment.
Dietary vitamin A deficiency and fatty acid restriction
To determine whether the reporter activity induced by mouse plasma might be due to vitamin A derivatives (e.g. 9-cis-RA or 9-cis-13,14-DHRA) (3, 4, 7), we analyzed plasma from vitamin A deficient mice (VAD) (24). Prior to PHZ treatment, we validated the presence of vitamin A deficiency using mass spectrometry detection of plasma retinol (Fig. 5A). Unexpectedly, activation of the GFP reporter by plasma from VAD mice was not statistically different from activation by plasma from control mice (VADC), suggesting that prolonged vitamin A deficiency resulted in a non-significant decrease in plasma RXR ligands. Furthermore, plasma from VAD mice treated with PHZ and plasma from VADC mice treated with PHZ activated the GFP reporter equally, suggesting that natural RXR ligands can be generated in mouse plasma in the absence of vitamin A (Fig. 5B).
Figure 5. Effect of vitamin A deficiency (VAD) and fatty acid deficiency (NF) on plasma RXRA ligands.
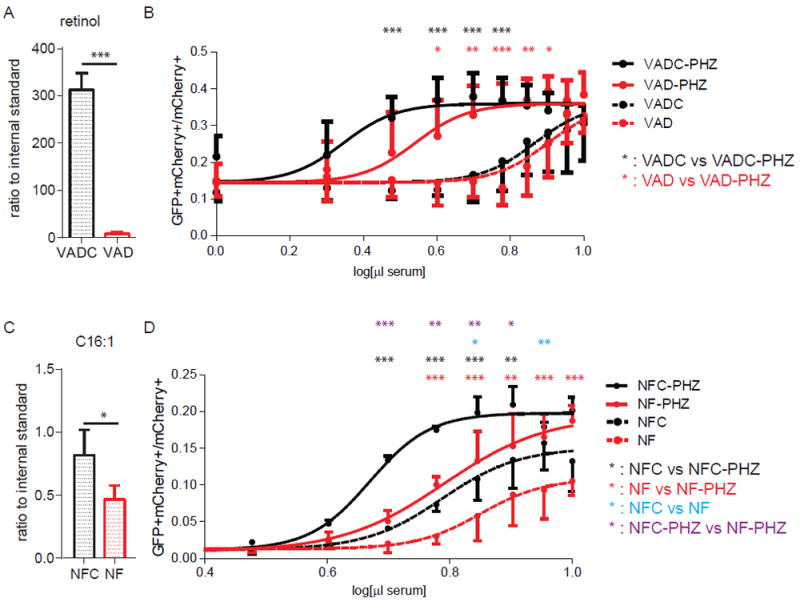
(A) Plasma concentration of retinol in VAD mice and control mice by mass spectrometry. Control mice, N = 5; VAD mice, N = 5. (B) UAS-GFP bone marrow Kit+ cells were transduced with Gal4-RXRA retrovirus and treated with plasma from VAD mice and control mice which were treated with or without PHZ. VAD: plasma from vitamin A deficient mice. VADC: plasma from mice treated with a control diet. VAD mice, N = 4; VAD + PHZ mice, N = 4; VADC mice, N = 5; VADC + PHZ mice, N = 5. (C) Plasma concentration of the fatty acid C16:1 in NF mice and control mice determined by mass spectrometry. NF mice, N = 5; NFC mice, N = 4. (D) The concentration of RXR ligands in fatty acid deficient mice was determined by treating reporter cells as in (B). NF: plasma obtained from mice treated with a fatty acid deficient diet. NFC: plasma from mice treated with a fatty acid control diet. NFC mice, N = 3; NFC + PHZ mice, N = 2; NF mice, N = 3; NF + PHZ mice, n = 4. Error bars represent standard deviation between measurements of plasma obtained from separate mice. * P < 0.05, ** P < 0.01, *** P < 0.001, ANOVA with Tukey’s multiple comparisons compared results obtained at each plasma concentration.
To determine whether the RXRA ligands that increase following PHZ treatment are fatty acids (e.g. DHA or AA) (8), we fed mice a no-fat (NF) diet or no-fat control (NFC) diet for 4 weeks, a period of time not associated with sequelae due to essential fatty acid deficiency (25). Unlike vitamin A, nonessential fatty acids can be synthesized in vivo. After 4 weeks with NF diet, the plasma concentration of palmitoleic acid C16:1 (a common glyceride constituent), was reduced by approximately 2 fold in mouse plasma (Fig. 5C). Unlike the VAD diet, plasma from mice fed a NF diet lead to decreased GFP activation, presumably reflecting reduced concentrations of RXR ligands in the plasma both at baseline, and following PHZ treatment (Fig. 5D). These data suggest that natural RXR ligands in mouse plasma are either essential fatty acids present in the diet or require essential fatty acid intake for their synthesis.
Identification of plasma RXRA ligands
To identify RXRA ligands by mass spectrometry (MS), we generated a stable 293T cell line that contains the UAS-GFP reporter and the fusion protein Flag-Gal4 DBD-RXRA LBD (293T-FXP). These cells produce GFP when treated with either plasma or serum and are amenable to greater experimental throughput of serum detection and subsequent pull-down studies. We tested commercially available serum from a wide range of mammals. We found that mouse serum and hamster serum have abundant RXRA ligands that activate the GFP reporter, rabbit serum and rat serum have moderate concentrations of RXRA ligands, whereas guinea pig serum and goat serum have undetectable concentrations of RXRA ligands (Fig. 6A), indicating that baseline RXRA activation may be different between different species. To characterize the serum-available RXRA ligands, we extracted mouse serum with different mixtures of immiscible solvents. We found the reporter-activating ligands could be maximally extracted in the organic phase at ratios of hexane:isopropanol (3:2) and methanol:H2O (9:1), consistent with mildly non-polar lipids (Fig. 6B and 6C).
Figure 6. Serum by extraction with immiscible solvents.
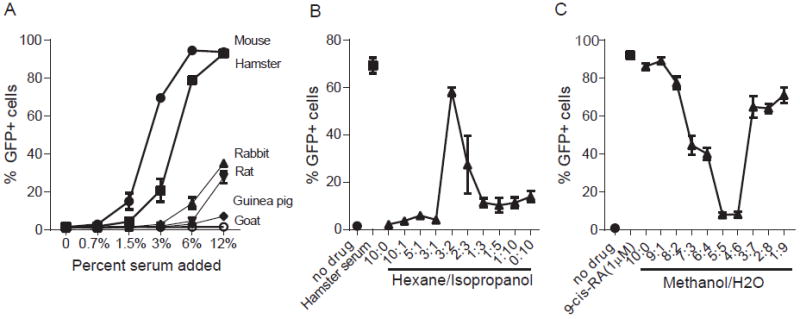
(A) Percentage of GFP+ cells in 293T-FXP reporter cells treated with or without serum from mouse, hamster, rabbit, rat, guinea pig or goat as indicated. (B) Percentage of GFP+ cells in 293T-FXP reporter cells treated with or without mouse serum extracted with hexane/isopropanol in different ratios. (C) Percentage of GFP+ cells in 293T-FXP cells reporter treated with or without mouse serum extracted with methanol/H2O in different ratios. Error bars indicate standard deviation between triplicate analysis.
By MS we first quantified the concentrations of diverse fatty acids under conditions we previously observed to have different UAS-Gal4 reporter activity. Based on our reporter results (Fig. 4-6), we anticipated that the RXRA ligands would meet the following requirements: at least 3 fold different between mouse serum vs. goat serum; 5 fold between mouse serum extracted with methanol:H20 9:1 vs. extracted with methanol:H2O 1:1; 1.5 fold different between VAD plasma vs. VAD-PHZ plasma; 1.5 fold different between NF plasma vs. NF-PHZ plasma; and 2 fold between NFC plasma vs. NFC-PHZ plasma. Several fatty acids fulfilled one or more of these criteria (Fig. 7A-F and table S1).
Figure 7. Mass spectrometry comparison of plasma and serum concentrations of long chain fatty acids C24:4 and C24:5.
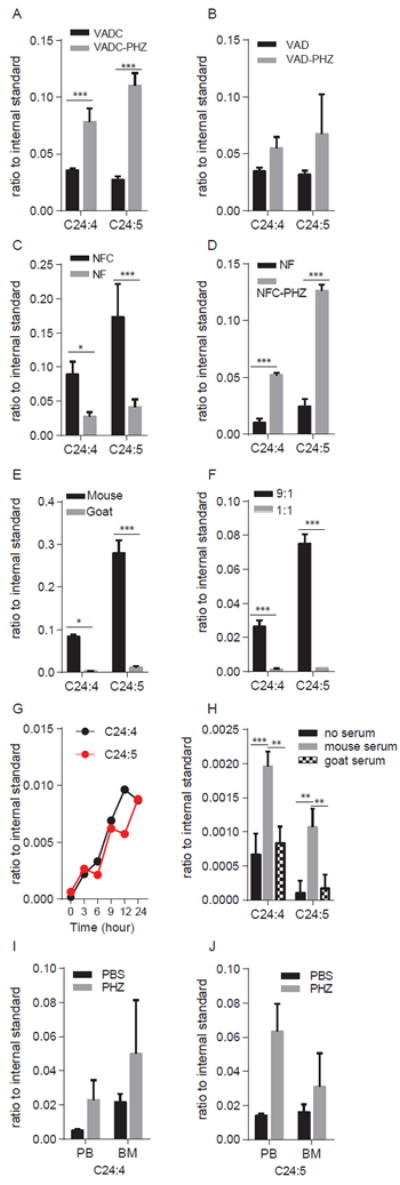
The concentration of C24:4 and C24:5 were determined in: (A) plasma from VADC mice treated with or without PHZ; VADC mice, N = 4; VADC + PHZ mice, N = 5; (B) plasma from VAD mice treated with or without PHZ; VAD mice, N = 4; VAD + PHZ mice, N = 4; (C) plasma from NF mice and NFC mice; NFC mice, N = 4; NF mice, N = 5; (D) plasma from NF mice and NFC mice treated with PHZ; NF mice, N = 3; NFC + PHZ, N = 2. (E) serum from mouse and goat analyzed in triplicate; (F) mouse serum extracted by methanol:H2O in 9:1 and 1:1 ratio and analyzed in triplicate. (G) Concentrations of C24:4 and C24:5 extracted from Flag-Gal4-RXRA following immunoprecipitation from 293T-FXP cell extracts treated with mouse serum for 0, 3, 6, 9, 12, or 24 hours, single determination performed at each time point. (H) Concentrations of C24:4 and C24:5 extracted from Flag-Gal4-RXRA following immunoprecipitation of 293T-FXP cells treated with or without mouse serum or goat serum for 12 hrs, analyzed in triplicate (I). Quantification of the fatty acid C24:4 in peripheral blood (PB) and bone marrow (BM) from mice treated with or without PHZ. PBS treated mice, N = 2; PHZ treated mice, N = 2. (J) Quantification of the fatty acid C24:5 in peripheral blood (PB) and bone marrow (BM) from mice treated with or without PHZ by mass spectrometry. PBS treated mice, N = 2; PHZ treated mice, N = 2. Error bars represent standard deviation of individual mice or biological triplicate experiments as indicated. * P < 0.05, ** P < 0.01, *** P < 0.001, T-test.
Second, we assessed fatty acids that bound to Flag-Gal4-RXRA during immunoprecipitation. We treated 293T-FXP cells with mouse or goat serum up to 24 hours, lysed cells, incubated with anti-Flag beads overnight, extracted the beads with hexane:isopropanol (3:2), and analyzed the extraction by MS. Only 6 fatty acids bound to RXRA in a time-dependent manner: C22:3, C22:4, C22:6, C24:4, C24:5, and C24:6 (Fig. 7G and table S1).
Third, we compared concentrations of lipids from mouse vs. goat serum that could be immunoprecipitated with Flag-Gal4-RXRA, and found that C24:4 and C24:5 were three times higher in mouse serum pull-down samples vs. goat serum pull-down samples (Fig. 7H).
Fourth, we analyzed peripheral blood cells and bone marrow cells from mice treated with or without PHZ by MS. Again, we found concentrations of fatty acids C24:4 and C24:5 increased three-fold in peripheral blood cells and increased two-fold in bone marrow cells after PHZ treatment (Fig. 7I-J). These data suggest that in mouse hematopoietic cells and in mouse serum, the long chain fatty acids C24:4 and C24:5 could be candidate RXRA ligands, with concentrations that increase in response to hematopoietic stress induced by PHZ.
Binding of long chain fatty acids C24:4 and C24:5 to RXRA
First, we evaluated the possibility that C24:4 and C24:5 could potentially dock within the RXRA ligand-binding pocket. We modeled lipid docking using Surflex-dock(26) and crystal structures containing oleic acid (PDB 1DFK, which has the AF2 domain in an inactive configuration) or 9-cis-RA (PDB 1XDK, which has the AF2 domain in an active configuration)(27-29). Results suggested a number of plausible docked poses within the RXRA ligand-binding domain result from the torsional mobility of these lipids, and both could be docked within either configuration of the AF2 domain (Fig. 8A-B and fig. S4 and S5). In all predicted configurations, the carboxyl group interacted with the R321 side-chain. Furthermore, previously reported ligand-contacting side chains could be modeled with close (< 3 angstroms) distances of C24:4 and C24:5 (e.g. L331, A332, V347, I350, C437, H440, L441, F444) (27-29).
Figure 8. Transactivation of RXRA by C24:5.
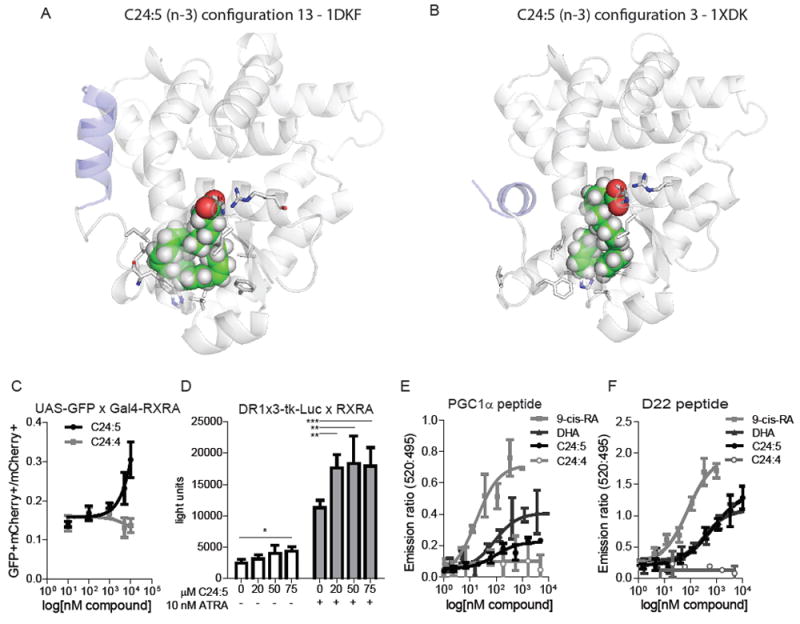
(A-B) In silico models of C24:5 docked in RXRA in an active configuration (A: PDB 1DKF) or and inactive configuration (B: PDB 1XDK) (C) Transactivation of UAS-GFP Kit+ bone marrow cells transduced with Gal4-RXRA and treated in culture with synthesized fatty acid C24:5 or C24:4. Error bars indicate standard deviation of independent triplicates. (D) 293T cells were transfected with DR1x3-tk-Luc and CMX-RXRA plasmids and treated as indicated. Error bars indicate standard deviation of independent triplicates compared by T-test. (E and F) TR-FRET of RXRA binding to indicated co-activator peptides and indicated concentrations of ligands. Error bars represent standard deviation of independent triplicate experiments. EC50 determined by fitting the data to a log(agonist) variable slope with four parameters. * P < 0.05, ** P < 0.01, *** P < 0.001.
Second, we examined the function of synthesized C24:4 and C24:5 on primary UAS-GFP Kit+ cells when transduced with Gal4-RXRA retrovirus. In repeat MS studies, the peak of the synthetic C24:5 compound was identical to the inferred C24:5 plasma peak (fig. 6). The fatty acid C24:4 did not activate the GFP reporter, whereas C24:5 activated the GFP reporter in a dose-dependent manner (Fig. 8C). We also observed that C24:5 could activate a DR1-luciferase reporter (containing 3 multimerized repeats of the RXRA direct repeat-1 sequence) when cells were transduced with full-length RXRA, and that low dose ATRA augmented the response to the lipid, as has been observed with DHA (Fig. 8D) (8).
Third, to determine whether the long chain fatty acid C24:5 can bind and activate RXRA directly, we performed FRET-based co-activator binding assays. C24:5 induced dose-dependent binding of both PGC1 and D22 peptides, with an EC50 of 94 and 712 nM, comparable to the results observed with DHA (86 and 366 nM), but lower than the results observed with 9-cis-RA (17 and 79 nM) (Fig. 8G-H). C24:4 did not lead to binding of either PGC1 or D22 (Fig. 8E and F). These data implicate the long chain fatty acid C24:5 as a natural ligand of RXRA in mouse which undergoes dynamic increases in concentrations in response to hematopoietic stress.
Discussion
RXRA is a ligand-dependent transcription factor that participates in several essential biological processes, including development, metabolism, and hematopoiesis. Although multiple RXRA deficient mouse models have been employed to study its function, these studies have not addressed whether endogenous RXRA ligands are present or whether their concentrations are altered in response to stimuli in vivo in hematopoietic cells. Other groups have employed a UAS-Gal4 reporter system to explore natural RXRA ligands in the developing mouse spinal cord, although none have examined the presence of natural RXRA ligands in hematopoiesis (30). By using a similar in vivo reporter system, we found that natural RXRA ligands are present and are dynamically increased in concentration in mouse hematopoietic cells and plasma following hematopoietic stress induced by GCSF and PHZ. We identified the long chain fatty acid C24:5 as a natural RXRA ligand in mice.
RARA-RXRA heterodimers play important roles in hematopoiesis and leukemogenesis (31, 32). At least ten X-RARA fusion proteins have been found in acute promyelocytic leukemia (APL) (33). Using the same in vivo reporter strategy, we did not detect RARA natural ligands in vivo (20), or in mouse plasma (fig. S3C). Instead, we observed evidence of natural ligands of RXRA, the silent partner of RARA, in hematopoietic cells and plasma, and found that reporter activity increased during myeloid stress, such as GCSF and PHZ (Fig. 2 and 4), but not following stem cell stress, such as 5-FU (fig. S3A). As a control, we observed no reporter activity when bone marrow cells were transduced with Gal4-RXRA-ΔAF2. Deletion of the AF2 domain prevents ligand-independent activation of the reporter, and thus the results suggest that the increased reporter activity correlated with increased concentrations of natural RXRA ligands. Because this deletion prevents ligand-dependent activation of the reporter, it does not control for alternative, ligand-independent effects on RXRA that could result from GCSF or PHZ treatment, and this limitation must be acknowledged, although increased concentrations of RXRA ligands DHA and C24:5 were observed in parallel by mass spectrometry.
RXRs are the central members of the nuclear receptor superfamily (1) and can function as homodimers (10) or heterodimers (11-15). PPAR-RXRs are permissive heterodimers, which can be activated by either PPAR ligands or RXR ligands (34). In contrast, TR-RXR, VDR-RXR are non-permissive heterodimers, which only can be activated by TR ligands and VDR ligand (35). PPARs have been shown to participate in hematopoietic stem and progenitor cells self-renewal (36, 37); TRs influence erythroid progenitors proliferation and differentiation (38); and VDR deficiency has been shown to promote hematopoietic stem and progenitor cells survival in spleen (39). Both RXRs and PPARs can be activated by long-chain fatty acids. However, when we treated our reporter cells with PPAR ligands, we did not observe activation,(8, 22) suggesting that the UAS-Gal4 system was specifically responding to RXRA ligands (fig. S2B and C).
We found that RXRA reporter activity increased during GSCF-induced granulopoiesis. Through studies with RXR-KO mice, we found that RXR activation contributed to GCSF induced mobilization of HSCs (although it was not absolutely required) and that GCSF induced mobilization of HSCs was augmented by the addition of pharmacologic concentrations of RXR ligands (Fig. 3A – E).
9-cis-RA and 9-cis-13,14-DHRA have been proposed as natural RXRA ligands in mouse serum, brain, and liver (3, 4, 7). However, when we examined plasma in a vitamin A deficient (VAD) mouse model, in which the retinol concentrations were undetectable (Fig. 5A), we still detected activation of our RXRA reporter, consistent with the presence of RXRA ligands (Fig. 5B). These data suggest the predominant plasma RXRA ligands under baseline conditions and following PHZ treatment are unlikely to be vitamin A derivatives.
In contrast, multiple groups have suggested that fatty acids might serve as activating RXR ligands. The crystallographic analysis of constitutively active mouse RXRA mutant RXRA F318A revealed a continuous U-shaped electron density in the ligand-binding cavity, suggesting the presence of a bound ligand, consistent with a fatty acid of 16 or 18 carbon atoms (27). C18:1 can activate RXRA reporter ex vivo but with low efficiency (8). We detected fatty acid C18:1 in mouse serum, more abundant than in goat serum, but its concentration was not affected by PHZ treatment (table S1). Moreover, we found that C18:1 could be extracted by methanol:H2O (1:1), but the products extracted by methanol:H2O (1:1) did not activate the RXRA reporter (Fig. 6C). Unsaturated fatty acids C20:4, C22:5, and C22:6 were identified as Rxr ligands in mouse brain (8). We observed C22:5 in both mouse and goat serum, although goat serum did not activate the RXRA reporter (Fig. 6A and Table S1). We also observed that C22:5 was not pulled down when reporter cells were incubated with serum, although C22:6 was (table S1). However, PHZ treatment did not consistently affect C22:6 concentrations in VAD mice and NF mice (table S1), suggesting that C22:6 may contribute to basal RXR activation, but does not appear to increase in concentration in response to PHZ. In contrast, C24:5 was consistently detected by MS under conditions that lead to activation of the RXRA reporter, was dynamically increased in plasma concentration by PHZ, and synthetic C24:5 could activate the RXRA reporter, activate a DR1-luciferase reporter when co-expressed with full-length RXRA, and induced dose-dependent co-activator binding to purified RXRA (Fig. 7 and 8), suggesting that this lipid is a natural RXRA ligand that increases in plasma concentration in response to hematopoietic signals.
In summary, we observed evidence of activating natural ligands for RXRA in mouse hematopoietic cells, and in plasma and serum. Concentrations of these ligands were increased following GCSF- and PHZ-induced myeloid stress, and we identified C24:5 as a natural RXRA ligand that undergoes dynamic increases in concentration during myeloid stress.
Materials and Methods
Reagents
Bexarotene was from LC laboratories. Phenylhydrazine, 5- Fluorouracil, corn oil, hexane, isopropanol, were from Sigma. GCSF was from Amgen. GW6471, GW7647, pioglitazone, and tesaglitazar were from Tocris. Anti-mouse CD11b (M1/70)-BV421, anti-mouse c-Kit (2B8)-BV421, anti-mouse B220 (RA3-6B2)-PE-Cy7 were from BD Bioscience. Anti-mouse CD8 (53-6.7)-eFluor450, anti-mouse CD71 (R17217) –eFluor450, anti-mouse Sca-1 (D7)-APC, anti-mouse Gr-1 (RB6-8C5) –APC, anti-mouse CD4 (GK1.5) –APC, anti-mouse Ter119 (TER119) – APC, anti-mouse c-Kit (2B8) –PE-Cy7, anti-mouse CD19 (eBio1D3) –PE-Cy7, anti-mouse Ter119 (TER119)-PE-Cy7, anti-mouse CD127 (ATR34) PE-Cy7, anti-mouse CD8 (53-6.7) –PE-Cy7, anti-mouse CD4 (RM4-5) –PE-Cy7, anti-mouse CD3e (145-2C11) –PE-Cy7 were from eBioscience. Sera from Mouse, Hamster, Rabbit, Rat, Guinea pig, and Goat were obtained from Equitech-Bio, Inc., and sera was obtained while animals were maintained on a standard diet. C24:4 ((9Z, 12Z, 15Z, 18Z, 21Z)-tetracosa-9,12,15,18,21-tetraenoic acid) and C24:5 ((9Z, 12Z, 15Z, 18Z, 21Z)-tetracosa-9,12,15,18,21-pentaenoic acid) were synthesized from Avanti Polar Lipids. Inc. The DR1-Luc reporter and pBABE-RXRA plasmids were gifts from Vivek Arora, Washington University.
Mice
UAS-GFP and Rxraflox/flox × Rxrbflox/flox × Mx1-Cre mice were bred as described (19, 20). RXR deletion was induced by intraperitoneal injecting Rxraflox/flox × Rxrbflox/flox × Mx1-Cre mice with pI:pC 300μg/mouse; 4 doses were given, every other day. And RXR deletion was confirmed by PCR 4 weeks after mice were treated with pI:pC. Bexarotene was administrated by oral gavage, suspended in sterile corn oil, 1 mg/mouse/day for 3 days, and mice were sacrificed at day 4. GCSF was administrated by subcutaneous injection, 125 μg/kg every 12 hours for 9 injections, and mice were sacrificed 2 hours after the last injection. All mice were cared for in the experimental animal center of Washington University School of Medicine. The Washington University Animal Studies Committee approved all animal experiments.
Vitamin A deficient mouse model
Vitamin A deficient mice were generated as described (24). Briefly, FVB female mice were fed with vitamin A deficient diet (Teklad, TD.86143) or vitamin A control diet (Teklad, TD.91280) during pregnancy. The offspring received the same diet for at least 12 weeks. Plasma concentrations of retinol were undetectable by mass spectrometry prior to experimental intervention.
Diet-restricted fatty acid mouse model
A diet-restricted fatty acid mouse model was generated as described (25). Briefly, FVB mice were fed with No fat diet (Teklad, TD.03314) and control diet (Teklad, TD.130321) for 4 weeks prior to experimental intervention. Plasma concentrations of palmitoleic acid (C16:1) were evaluated by mass spectrometry prior to experimental intervention.
Retrovirus production
Retrovirus production was performed as described (40). Calcium chloride transfection and low-passage 293T cells were used for virus packaging. 5 × 106 293T cells were seeded in 10cm dishes in DMEM (high glucose) + 10% FBS, 18 ~ 24 hours before transfection and grow to 80% confluence. 12 μg MSCV-Gal4 DBD-RXRA LBD-IRES-mCherry (Gal4-RXRA), MSCV-Gal4 DBD-RXRA LBD ΔAF2-IRES-mCherry (Gal4-RXRA ΔAF2), or MSCV-Gal4 DBD-RARA LBD-IRES-mCherry (Gal4-RARA), 8 μg Ecopak, and 155 μl 2M CaCl2 were mixed, adjust the volume to 1.25 ml by adding H2O. 1.25 ml 2 × HEPES Buffer was drop-wise added to the mixture. The mixture was incubated at room temperature for 20 min and then drop-wise added onto 293T cells. Fresh medium was changed after 12 hours transfection. Virus was collected at 48 hours and 72 hours and stored at -80°C.
Bone marrow transplantation
Femurs, tibias, and pelvises were isolated from 6-8 weeks old UAS-GFP mice. Bone marrow cells were collected by centrifuging bones at 6,000 rpm for 2 min. Red blood cells were lysed by ACK Buffer (NH4Cl 150mM, KHCO3 10mM, Na2EDTA 0.1mM) on ice for 5 min. Kit+ cells were isolated by magnetic-activated cell sorting (MACS) using Automacs Pro (Miltenyl Biotec) per manufacture’s protocol. Kit+ cells were cultured in transplant media (RPMI 1640 + 15% FBS + 20ng/ml mSCF + 25ng/ml mFlt3L + 10ng/ml mIL3 + 10ng/ml Tpo + 50μM β-mercaptoethanol) overnight, and transduced with Gal4-RXRA or Gal4-RXRA ΔAF2 retrovirus by spinfection with 10 μg/ml polybrene and 10mM HEPES at 2,400 rpm, 30°C for 90 min in an Eppendorf 5810R centrifuge. Kit+ cells were spinfected with same virus second time after 24 hours. Transduced cells were injected to lethally irradiated recipient mice 2 hours after the second spinfection. After 6 weeks engraftment, mice were sacrificed and analyzed.
Flow cytometry
Bone marrow, peripheral blood, spleen, and peritoneal macrophages were collected from the bone marrow transplant mice following engraftment (typically ~6 weeks). For hematopoietic stem and progenitor cell analysis, cells were stained with Lineage-PE-Cy7, c-Kit-BV421, Sca-1-APC. For myeloid lineage analysis, cells were stained with c-Kit-PE-Cy7, CD11b-BV421, Gr-1-APC. For erythroid lineage analysis, cells were stained with c-Kit-PE-Cy7, CD71-eFluor450, Ter119-APC. For lymphoid lineage analysis, cells were stained with B220-PE-Cy7, CD4-APC, CD8-eFluor450. Fluorescence was detected using a Gallios Flow Cytometer (Beckman Coulter).
Plasma or serum extraction
50 μl mouse plasma or serum was mixed with 500 μl hexane:isopropanol or methanol:H2O and vortexed vigorously for 30 seconds. Following centrifugation at 12,000 rpm for 5 minutes, the upper phase (organic phase) was collected into a new tube and stored at -80 °C until analyzed.
Pull-down assays
We generated stable 293T cells expressing UAS-GFP and 3×Flag-Gal4 DBD-RXRA LBD IRES - Puro (293T-FXP). 1 × 106 293T-FXP cells per well were seeded in 6-well-plates, and treated with mouse serum or goat serum. Cells were collected and lysed with RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1% Deoxycholate, 0.1% SDS, 1mM EDTA, proteinase inhibitor) at 0, 3, 6, 9, 12, and 24 hrs. 293T-FXP cell lysate was incubated with pre-washed anti-Flag magnetic beads and rotated at 4 °C overnight. Beads were washed with TBST three times. Washed beads were resuspended in 50 μl TBST, and then extracted with hexane:isopropanol (3:2). Organic phase was collected and stored in -80 °C until analysis.
Mass spectrometry
For retinol analysis, 100 μl of plasma extraction sample was used. Prior to MS analysis, 50 ng of retinol-d6 as the internal standard was added to sample. Then, the solvents in the sample were dried under a stream of nitrogen. The dried sample was re-dissolved in 1 ml ethanol:H2O (1:1) for MS analyses.
For fatty acids analysis, 100 μl of plasma extraction sample or 400 μl pull down sample was used for DMAPA (dimethylaminopropylamine) derivatization for improving MS sensitivity of fatty acids. Prior to the derivatization, 50 ng (10 ng for pull-down sample) of AA-d8 (arachidonic acid-d8) as the internal standard was added to each sample. The solvents in the sample were dried under a stream of nitrogen. To derivatize the sample, 50 μl of 100 mM EDC (N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride) and 50 μl of 50 mM DMAPA and 50 mM DMAP (4-dimethylaminopyridine) were added to the dried fatty acid samples and heated at 50 °C for 30 minutes. The samples were dried under nitrogen and then dissolved in 1ml ethanol:H2O (1:1) for MS analysis.
In silico ligand docking analysis
Docking of C24:5 and C24:4 against crystal structures of RXRA (PDB 1DKF and 1XDK) was performed with Surflex-dock (26). MOL2 files for each compound were generated from SMILES strings using Open Babel.(41) Surflex-Dock receptor protomols were generated with a threshold of 0.25 and a bloat of 2.0 and subsequently docked using the default ‘-pgeom’ docking accuracy parameter set. Residues lining the binding pocket were defined as those within 4 Å of oleic acid (PDB 1DKF) or 9-cis-RA (PDB 1XDK). Images were generated in Pymol (v.1.8.6.0, Schrodinger).
Luciferase detection
293T cells were transfected with pBABE-RXRA in combination with DR1x3-TK-Luc using Lipofectamine 2000 (Invitrogen). Six hours after transfection, the cells were collected and plated into a 48 well plate in 1% BSA media in triplicate. DHA, fatty acid 24:5, Bex, or 9-cis RA were added to the cells as a BSA complex (BSA-Albumin, Fraction V, Fatty Acid Free, Calbiochem), with equal amounts of BSA added to all samples.(42) After 40 hour incubation, the cells were harvested and assayed for luciferase (Luc Assay System with Reporter Lysis Buffer, Promega) in a Beckman Coulter LD400 plate reader.
Time Resolution-FRET assay
The assay was performed using LanthaScreen TR-FRET Retinoic X Receptor alpha Coactivator assay (Invitrogen). Reaction mixture contained RXR alpha LBD-GST 10 nM, TB anti GST antibody 5 nM, Fluorescein-PGC1α or D22 peptide 500 nM, DTT 5mM, DMSO 1% and testing compounds. 20 μl of the reaction mixture was analyzed after 1 hour incubation at room temperature and analyzed using a Synergy2 Plate Reader (BioTek Instruments), using a 340 nm filter with a 30 nm bandwidth, and an emission filter was centered at 520 nm with a 25 nm bandwidth and 495 nm with a 10 nm bandwidth). The TR-FRET ratio was calculated by dividing the emission signal at 520 by the emission signal at 495.
Data analysis
Flow cytometry data was analyzed with FlowJo software version 10. Statistical analysis was performed using Prism (Graphpad). T-test and ANOVA tests were performed, as appropriate. FRET studies were analyzed using a sigmoidal dose response with variable slope. All studies were performed in triplicate, unless otherwise indicated. Error bars represent standard deviation. Data points without error bars have standard deviations below Graphpad’s limit to display.
Supplementary Material
Acknowledgments
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO. for the use of the Flow Cytometry Core. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant P30 CA91842. We thank High-Throughput Screening Center at Washington University School of Medicine in St. Louis, MO. We thank Deborah Laflamme for technical assistance and Feng Gao for statistical assistance. This work was supported by NIH R01 HL128447 (JS Welch), NIH P50 CA171963 (Project 1, JS Welch), and by grants from the Spanish Ministry of Economy and Competitiveness (SAF2015-64287R, SAF2015-71878-REDT) (M Ricote). The mass spectrometry facility at Washington University is supported by NIH P30 DK020579, Daniel Ory. J.S.W., H.N. and M.R. designed experiments, performed experiments, and wrote the manuscript. H.F., O.M., G.H., M.P.M, T.E.F., G.R.B. designed and performed experiments.
Footnotes
The authors declare no conflicts of interest.
References and Notes
- 1.Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson MI, Xia Z. The retinoid X receptors and their ligands. Biochim Biophys Acta. 2012;1821:21–56. doi: 10.1016/j.bbalip.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, Chambon P. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci U S A. 1993;90:30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 5.Arnold SL, Amory JK, Walsh TJ, Isoherranen N. A sensitive and specific method for measurement of multiple retinoids in human serum with UHPLC-MS/MS. Journal of lipid research. 2012;53:587–598. doi: 10.1194/jlr.D019745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones JW, Pierzchalski K, Yu J, Kane MA. Use of fast HPLC multiple reaction monitoring cubed for endogenous retinoic acid quantification in complex matrices. Anal Chem. 2015;87:3222–3230. doi: 10.1021/ac504597q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruhl R, Krzyzosiak A, Niewiadomska-Cimicka A, Rochel N, Szeles L, Vaz B, Wietrzych-Schindler M, Alvarez S, Szklenar M, Nagy L, de Lera AR, Krezel W. 9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice. PLoS Genet. 2015;11:e1005213. doi: 10.1371/journal.pgen.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lengqvist J, Mata De Urquiza A, Bergman AC, Willson TM, Sjovall J, Perlmann T, Griffiths WJ. Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor alpha ligand-binding domain. Mol Cell Proteomics. 2004;3:692–703. doi: 10.1074/mcp.M400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Nohara A, Kobayashi J, Mabuchi H. Retinoid X receptor heterodimer variants and cardiovascular risk factors. J Atheroscler Thromb. 2009;16:303–318. doi: 10.5551/jat.no786. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XK, Lehmann J, Hoffmann B, Dawson MI, Cameron J, Graupner G, Hermann T, Tran P, Pfahl M. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature. 1992;358:587–591. doi: 10.1038/358587a0. [DOI] [PubMed] [Google Scholar]
- 11.Bugge TH, Pohl J, Lonnoy O, Stunnenberg HG. RXR alpha, a promiscuous partner of retinoic acid and thyroid hormone receptors. The EMBO journal. 1992;11:1409–1418. doi: 10.1002/j.1460-2075.1992.tb05186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature. 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Bio. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 16.Defacque H, Commes T, Legouffe E, Sevilla C, Rossi JF, Rochette-Egly C, Marti J. Expression of Retinoid X Receptor alpha is increased upon monocytic cell differentiation. Biochemical and biophysical research communications. 1996;220:315–322. doi: 10.1006/bbrc.1996.0403. [DOI] [PubMed] [Google Scholar]
- 17.Taschner S, Koesters C, Platzer B, Jorgl A, Ellmeier W, Benesch T, Strobl H. Down-regulation of RXRalpha expression is essential for neutrophil development from granulocyte/monocyte progenitors. Blood. 2007;109:971–979. doi: 10.1182/blood-2006-04-020552. [DOI] [PubMed] [Google Scholar]
- 18.Makita T, Hernandez-Hoyos G, Chen TH, Wu H, Rothenberg EV, Sucov HM. A developmental transition in definitive erythropoiesis: erythropoietin expression is sequentially regulated by retinoic acid receptors and HNF4. Genes Dev. 2001;15:889–901. doi: 10.1101/gad.871601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menendez-Gutierrez MP, Roszer T, Fuentes L, Nunez V, Escolano A, Redondo JM, De Clerck N, Metzger D, Valledor AF, Ricote M. Retinoid X receptors orchestrate osteoclast differentiation and postnatal bone remodeling. J Clin Invest. 2015;125:809–823. doi: 10.1172/JCI77186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu H, Chacko J, Hadwiger G, Welch JS. Absence of natural intracellular retinoids in mouse bone marrow cells and implications for PML-RARA transformation. Blood Cancer J. 2015;5:e284. doi: 10.1038/bcj.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mascrez B, Mark M, Dierich A, Ghyselinck NB, Kastner P, Chambon P. The RXRalpha ligand-dependent activation function 2 (AF-2) is important for mouse development. Development. 1998;125:4691–4707. doi: 10.1242/dev.125.23.4691. [DOI] [PubMed] [Google Scholar]
- 22.Huang JT, Welch JS, Ricote M, Binder CJ, Willson TM, Kelly C, Witztum JL, Funk CD, Conrad D, Glass CK. Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 23.Dornfest BS, Lapin DM, Naughton BA, Adu S, Korn L, Gordon AS. Phenylhydrazine-induced leukocytosis in the rat. J Leukoc Biol. 1986;39:37–48. doi: 10.1002/jlb.39.1.37. [DOI] [PubMed] [Google Scholar]
- 24.Ikami K, Tokue M, Sugimoto R, Noda C, Kobayashi S, Hara K, Yoshida S. Hierarchical differentiation competence in response to retinoic acid ensures stem cell maintenance during mouse spermatogenesis. Development. 2015;142:1582–1592. doi: 10.1242/dev.118695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Jain AN. Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 27.Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 28.Pogenberg V, Guichou JF, Vivat-Hannah V, Kammerer S, Perez E, Germain P, de Lera AR, Gronemeyer H, Royer CA, Bourguet W. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J Biol Chem. 2005;280:1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- 29.Egea PF, Mitschler A, Moras D. Molecular recognition of agonist ligands by RXRs. Mol Endocrinol. 2002;16:987–997. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- 30.Solomin L, Johansson CB, Zetterstrom RH, Bissonnette RP, Heyman RA, Olson L, Lendahl U, Frisen J, Perlmann T. Retinoid-X receptor signalling in the developing spinal cord. Nature. 1998;395:398–402. doi: 10.1038/26515. [DOI] [PubMed] [Google Scholar]
- 31.Johnson BS, Chandraratna RA, Heyman RA, Allegretto EA, Mueller L, Collins SJ. Retinoid X receptor (RXR) agonist-induced activation of dominant-negative RXR-retinoic acid receptor alpha403 heterodimers is developmentally regulated during myeloid differentiation. Mol Cell Biol. 1999;19:3372–3382. doi: 10.1128/mcb.19.5.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang YA, Shen K, Ishida Y, Wang Y, Kakizuka A, Brooks SC. Induction of murine leukemia and lymphoma by dominant negative retinoic acid receptor alpha. Mol Carcinog. 2005;44:252–261. doi: 10.1002/mc.20144. [DOI] [PubMed] [Google Scholar]
- 33.De Braekeleer E, Douet-Guilbert N, De Braekeleer M. RARA fusion genes in acute promyelocytic leukemia: a review. Expert Rev Hematol. 2014;7:347–357. doi: 10.1586/17474086.2014.903794. [DOI] [PubMed] [Google Scholar]
- 34.Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 35.Shulman AI, Mangelsdorf DJ. Retinoid x receptor heterodimers in the metabolic syndrome. N Engl J Med. 2005;353:604–615. doi: 10.1056/NEJMra043590. [DOI] [PubMed] [Google Scholar]
- 36.Ito K, Turcotte R, Cui J, Zimmerman SE, Pinho S, Mizoguchi T, Arai F, Runnels JM, Alt C, Teruya-Feldstein J, Mar JC, Singh R, Suda T, Lin CP, Frenette PS, Ito K. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354:1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HY, Gao X, Barrasa MI, Li H, Elmes RR, Peters LL, Lodish HF. PPAR-alpha and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature. 2015;522:474–477. doi: 10.1038/nature14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer A, Mikulits W, Lagger G, Stengl G, Brosch G, Beug H. The thyroid hormone receptor functions as a ligand-operated developmental switch between proliferation and differentiation of erythroid progenitors. The EMBO journal. 1998;17:4291–4303. doi: 10.1093/emboj/17.15.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeanson NT, Scadden DT. Vitamin D receptor deletion leads to increased hematopoietic stem and progenitor cells residing in the spleen. Blood. 2010;116:4126–4129. doi: 10.1182/blood-2010-04-280552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu H, Hadwiger G, Fujiwara H, Welch JS. Pathways of retinoid synthesis in mouse macrophages and bone marrow cells. Journal of leukocyte biology. 2016 doi: 10.1189/jlb.2HI0415-146RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang PK, Khatchadourian A, McKinney RA, Maysinger D. Docosahexaenoic acid (DHA): a modulator of microglia activity and dendritic spine morphology. J Neuroinflammation. 2015;12:34. doi: 10.1186/s12974-015-0244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


