Abstract
Heightened and extended inflammation underlies the pathogenesis of many disorders, including inflammatory bowel disease, sepsis, and inflammatory arthritis. Ubiquitin networks help dictate the strength and duration of inflammatory signaling. In innate immunity, the itchy E3 ubiquitin protein ligase (ITCH)-A20 ubiquitin–editing complex inhibits receptor-interacting Ser/Thr kinase (RIPK) activation by removing Lys-63–linked polyubiquitinated chains from key proteins in the nuclear factor kappa B (NF-κB) signaling pathway. The complex then attaches polyubiquitinated chains to these proteins to target them for lysosomal or proteasomal destruction. ITCH is phosphorylated and thereby inhibited by inhibitor of nuclear factor kappa B kinase subunit beta (IKKβ) to fine-tune the inflammatory response to the strength of the offending signal. However, the biochemical mechanism by which E3 ubiquitination is impaired by IKK-driven phosphorylation remains unclear. Here, we report that this phosphorylation impedes ITCH binding to its cognate E2 ubiquitin ligase, UbcH7. Using CRISPR-Cas9 genetic knockout to mimic the ITCH-UbcH7–inhibited state, we further show that genetic UbcH7 deficiency phenocopies ITCH phosphorylation in regulating RIPK2 ubiquitination. We conclude that phosphorylation can disrupt the binding of an E3 ubiquitin ligase to an E2-conjugating enzyme, leading to prolonged inflammatory signaling. To our knowledge, this is the first report of E3 ubiquitin ligase phosphorylation inhibiting E3 ligase activity by impairing E2–E3 complex formation.
Keywords: E3 ubiquitin ligase, ubiquitin ligase, inflammation, phosphorylation, NF-kappa B, NF-κB, ubiquitination, ITCH, UbcH7, IKKβ
Introduction
As important as it is to initiate an inflammatory response, it is equally important to halt this response when it is no longer needed. A heightened and prolonged inflammatory response is implicated in the pathogenesis of diseases as diverse as inflammatory bowel disease, sepsis, inflammatory arthritis, and many others (1–5). Given the importance of a well-regulated inflammatory response, it is not surprising that the body has evolved mechanisms designed to maintain inflammatory homeostasis. For instance, among the first genes transcribed upon nuclear factor kappa B (NF-κB)2 activation are those designed to limit NF-κB activation (6–8). This sort of temporal self-limiting mechanism is crucial in tailoring the inflammation to the offending incident.
We have previously described a novel inflammatory feedback circuit involving a key protein kinase in the NF-κB pathway, inhibitor of NF-κB kinase subunit beta (IKKβ), and the itchy E3 ubiquitin ligase, ITCH, that inhibits many inflammatory signaling cascades, most notably TNF signaling and NOD2 signaling (9). ITCH acts in concert with the deubiquitinase A20 to perform ubiquitin-editing functions. In this complex, A20 deubiquitinates K63 polyubiquitinated proteins, and ITCH then attaches separate polyubiquitin chains that can subsequently target that protein for lysosomal or proteasomal degradation (5). The importance of this complex is manifest in vivo as A20−/− mice die within 3 weeks of birth of a massive, unregulated inflammatory response that includes severe colitis, severe ileitis, and severe hepatitis (10). ITCH−/− mice likewise die of an inflammatory disease as they develop severe pneumonitis and severe gastritis (11–13). Patients with ITCH mutations likewise develop inflammatory disease with lung inflammation, nephritis, and inflammatory bowel disease (14). Thus, not only is this pathway important in limiting acute signaling, but also in maintaining inflammatory homeostasis in vivo.
Our previous work established that IKKβ, a key kinase in the NF-κB pathway, phosphorylates both A20 and ITCH. A20 phosphorylation limits its ability to function as a deubiquitinase (15), and ITCH phosphorylation inhibits its E3 ligase activity (9). This feed-forward inflammatory pathway allows the strength of IKKβ kinase activity to dictate the duration of inflammatory response, and as such, when TNF signaling is genetically removed from ITCH mice, the inflammatory phenotype of the ITCH mouse is diminished (9). Although we understand the consequences of ITCH phosphorylation at the cell biological level and on the organismal level, the biochemical mechanism by which IKKβ phosphorylation of ITCH inhibits its E3 ligase activity is unknown. In this work, we show that phosphorylation of ITCH causes a static interruption that diminishes its affinity for the E2 and this manifests in attenuated E3 ubiquitin ligase activity. Genetically removing the E2, UbcH7, phenocopies ITCH phosphorylation and therefore provides genetic evidence for the mechanism by which ITCH and phosphorylation by IKKβ alters its biochemical activity. To our knowledge, this is the first instance in which phosphorylation of an E3 ligase inhibits its ubiquitin ligase activity through impairment of the formation of an E2–E3 complex.
Results
ITCH phosphorylation impairs UbcH7 binding
We have previously shown that IKKβ phosphorylates ITCH on serine 687 within the HECT domain (9). Given the structural homology of HECT domains in general, we used the crystal structure of HECT domain–containing protein, E6AP, complexed with an E2 ubiquitin conjugating enzyme, UbcH7 (PDB ID: 1C4Z) (16), to model the effects of phosphorylation and mutagenesis on ITCH serine 687. Fig. 1 shows the structure of E6AP–UbcH7 complex and models for the ITCH–UbcH7 complex. The E6AP residue Ser-638 is located at the E2–E3 interface, where it forms a hydrogen bond with the main chain carbonyl of the UbcH7 residue Phe-63 from its L1 specificity loop (Fig. 1A, top left panel). Phe-63 in UbcH7 has been reported to make significant contribution to the E2–E3 interaction (16, 17). In the modeled structure of an ITCH–UbcH7 complex, residue Ser-687 of ITCH forms a similar hydrogen bond with the main chain carbonyl oxygen of Phe-63 from UbcH7 (Fig. 1A, top right panel). To examine the effects of phospho-ITCH and phosphomimetic S687D ITCH, we modeled each structure, which revealed significant clash between Phe-63 of UbcH7 and Asp-687 or pSer-687 from ITCH (Fig. 1A, lower panels). Because both models exhibit interruption of a main chain to side chain hydrogen bond, and significant steric hindrance at the E2-E3 interface, it is likely that the ITCH–UbcH7 interaction would be impaired upon Ser-687 phosphorylation (Fig. 1A).
Figure 1.
Phosphorylation disrupts UbcH7 binding. A, the structure of the E6AP–UbcH7 complex (1C4Z) is shown to the left of the panel series with the E6AP HECT domain colored cyan and UbcH7 colored yellow. The upper panel highlights the E6AP–UbcH7 interface near E6AP residue Ser-638 at the H7 helix, which forms a side chain to main chain hydrogen bond with UbcH7 residue Phe-63 at the L1 loop. To model the ITCH–UbcH7 interface, the structure of the ITCH HECT domain (3TUG, colored green) is superimposed onto the E6AP HECT domain within the E6AP–UbcH7 complex and shown at the upper right panel. Similar to the E6AP residue Ser-638, the ITCH residue Ser-687 is predicted to form the same side chain to main chain hydrogen bond with the UbcH7 residue Phe-63. Mutation of Ser-687 to Asp (lower left) or phosphorylation of Ser-687 (lower right) would lead to loss of the hydrogen bond and significant steric hindrance at the E2–E3 interface, thus severely impairing ITCH–UbcH7 interaction. B, HEK 293T cells were transfected with FLAG-ITCH and FLAG-ITCH S687D alone or with Omni-UbcH7 for co-precipitation. FLAG-ITCH was immunoprecipitated (IP) in each sample. After stringent washing of IP samples, lysate and IP samples were ran on SDS-PAGE, transferred to nitrocellulose, and probed as indicated. Similar results were obtained in three independent experiments. C, HEK 293T cells were stimulated with 1 μg/ml TNFα for the indicated time points and UbcH7 was immunoprecipitated. After gentle washing, lysate and IP samples were processed by Western blotting. Membranes were probed as indicated. Similar results were obtained in two independent experiments. D, Biacore SPR sensorgrams for binding of UbcH7 to wildtype or S687D mutant ITCH.
To test this prediction, HEK 293T cells were transfected with FLAG-tagged ITCH and Omni-tagged UbcH7. Immunoprecipitation showed that whereas wildtype ITCH strongly bound UbcH7, the S687D phosphomimetic did not (Fig. 1B). We would then predict that upon IKK activation, the endogenous ITCH–UbcH7 interaction would be diminished. To test this, HEK 293T cells were stimulated with TNFα for the indicated time points corresponding to IKK activation. TNFα is a potent activator of IKKs (18), and has been shown to promote ITCH phosphorylation (9). UbcH7 was immunoprecipitated and analyzed for co-precipitating endogenous ITCH. We observed robust activation of IKKα/β, which was accompanied by a decrease in ITCH–UbcH7 binding (Fig. 1C). To then investigate difference in UbcH7 binding by wildtype or mutant ITCH, we purified UbcH7 and the recombinant HECT domains of ITCH and phosphomimetic S687D ITCH (Fig. S1) for surface plasmon resonance (SPR) experiments. Full-length ITCH and ITCHHECT bind UbcH7 with similar affinity (19). Wildtype ITCHHECT and S687D ITCHHECT were immobilized on an SPR chip and tested for binding to recombinant UbcH7. ITCHHECT was found to have a stronger binding affinity of 3.69 μm, whereas ITCH S687DHECT had a weaker 18.38 μm affinity (Fig. 1D, right panels). The affinity of the ITCHHECT-UbcH7 association was similar to those of E6APHECT-UbcH7 previously measured by fluorescence polarization and isothermal titration calorimetry (5.0 μm and 2.2 μm, respectively) (17). These experiments strongly suggest that unmodified serine 687 in ITCH is required for optimal binding to UbcH7, and that a phosphomimetic mutation at this site can impair affinity for UbcH7.
The Phe-63 residue of UbcH7 is responsible for phosphorylation-dependent coupling to ITCH
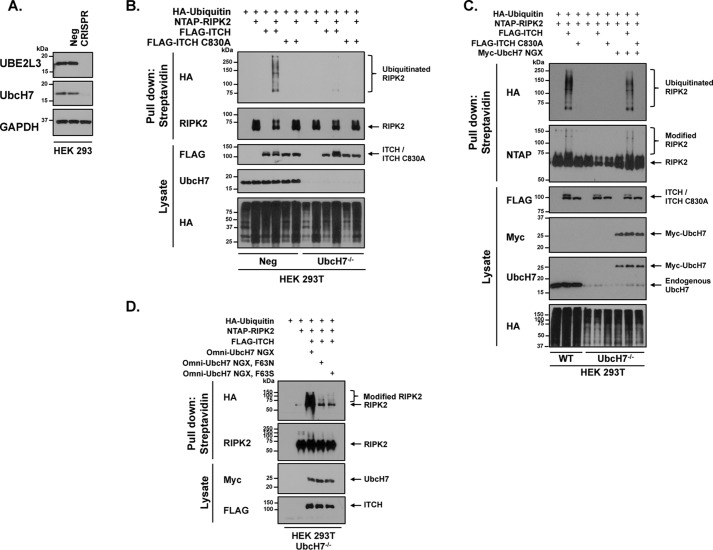
To then test the biochemistry of the model presented in Fig. 1A we measured the ability of ITCH to ubiquitinate an exogenous ligand, RIPK2. To this end, UbcH7 was genetically deleted from HEK 293T cells using CRISPR-Cas9 lentiviral transduction with a puromycin-resistant containing vector. Transduced cells were puromycin-selected and individual clones were isolated, tested for UbcH7 knockout, and pooled as the UbcH7−/− cell line. Because the same antibody often detects UbcH7 and UbcH8, lysates were probed with two antibodies confirmed to detect human UbcH7 (labeled as UBE2L3 and UbcH7; see supporting Materials and Methods). Both antibodies showed depletion of UbcH7 in the pooled clonal CRISPR cell line (Fig. 2A).
Figure 2.
Phe-63 mediates the phosphorylation-dependent coupling between UbcH7 and ITCH. A, HEK 293T cells were transduced with lentivirus containing CRISPR-Cas9 with either a negative control-RNA guide or UbcH7 Guide 1-RNA. Lysate from pooled select UbcH7−/− clones was run next to controls on SDS-PAGE and probed for UbcH7 and GAPDH as shown. B, negative control and UbcH7−/− HEK 293T cells were transiently transfected with HA-ubiquitin, NTAP-RIPK2, and ITCH or ITCH C830A to assess ITCH ubiquitination of RIPK2. Cellular lysates were collected and cleared. Streptavidin beads were used to pull down RIPK2 via SBP tag within the NTAP tag. Samples were analyzed by Western blotting. C, UbcH7−/− HEK 293T cells were transiently transfected with HA-ubiquitin, NTAP-RIPK2, FLAG-ITCH, FLAG-ITCH C830A, and Myc-UbcH7. NTAP was precipitated under stringent washing conditions (1% SDS, 2 m NaCl). Samples were run on SDS-PAGE next to WT samples that were similarly transfected. Samples were subjected to Western blot analysis as indicated. Results shown are representative of three independent experiments. D, HA-tagged ubiquitin, NTAP-tagged RIPK2, and FLAG-ITCH were transiently transfected into UbcH7−/− HEK 293T cells with CRISPR-resistant UbcH7 WT, F63N, or F63S. RIPK2 was pulled down with streptavidin beads. Samples were washed and subjected to SDS-PAGE and immunoblotting as indicated.
We then utilized this cell line by transiently transfecting HA-tagged ubiquitin, NTAP-RIPK2, and FLAG-ITCH, or FLAG-ITCH C830A. Streptavidin pulldown of NTAP-RIPK2 revealed more ITCH-ubiquitinated RIPK2 in the control cell line as compared with the UbcH7−/− cell line (Fig. 2B). In a similar experiment and to overcome potential CRISPR-mediated off-target effects, we replaced UbcH7 within the UbcH7−/− cell line using a CRISPR-resistant Myc-UbcH7 NGX construct to recover RIPK2 ubiquitination by ITCH (Fig. 2C). We then utilized this replacement strategy to test the requirement of Phe-63 on UbcH7 to mediate this interaction. In the molecular model presented in Fig. 1A, the carboxyl group of the peptide bond of Phe-63 mediates hydrogen bonding to ITCH. We know Phe-63 is critical as previous work using alanine-scanning mutations showed that mutation of Phe-63 to alanine limits E3 ubiquitin ligase activity (17). Because mutation of Phe-63 to alanine on UbcH7 is already known to inhibit E3 ligase activity, we scanned other, homologous E2 ligase sequences for alternative amino acids at this position. Many contain asparagine or serine at this position, and for this reason, Phe-63 was mutated to asparagine (F63N) or serine (F63S). We then used these mutants to test the amino acid side chain's ability to complement UbcH7 genetic loss. Although WT UbcH7 replacement into UbcH7−/−cells could clearly recover ITCH E3 ligase activity, F63N and F63S UbcH7 were significantly impaired in their ability to do so (Fig. 2D). These findings strongly support the phosphorylation-dependent UbcH7-ITCH biochemical coupling presented in the molecular model.
Genetic deletion of UbcH7 phenocopies phosphorylation of ITCH
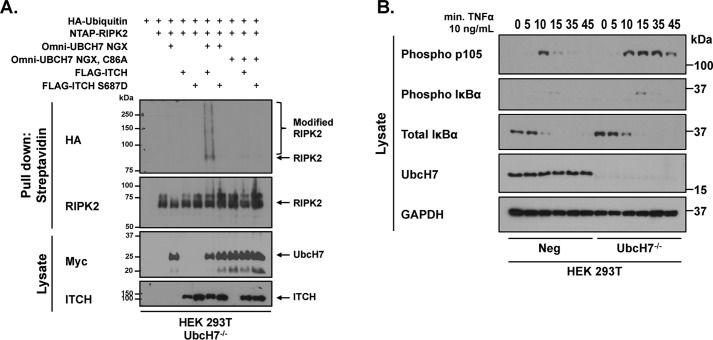
Given that phosphorylation of ITCH appears to alter UbcH7 binding and given that our previously published work established that ITCH phosphorylation prolonged NF-kB activity, we reasoned that genetic deletion of UbcH7 may mimic the phosphorylation of ITCH. To first establish functional similarity between UbcH7 loss and ITCH phosphorylation, UbcH7−/− cells were transfected with NTAP-tagged RIPK2 in the presence of HA-tagged ubiquitin before standard immunoprecipitated (IP) Western blotting was performed. Fig. 3A shows that both ITCH and S687D ITCH were inefficient at causing RIPK2 ubiquitination in UbcH7−/− cells. Replacement with CRISPR-resistant UbcH7 caused recovery of RIPK2 ubiquitination with WT ITCH, but not with S687D ITCH. Furthermore, reconstitution with a form of UbcH7 in which we have mutated the active cysteine to alanine (C86A UbcH7) does not allow recovery of ITCH-induced ubiquitination of RIPK2 (Fig. 3A). These findings strongly imply that UbcH7 genetic loss phenocopies ITCH phosphorylation.
Figure 3.
UbcH7 deficiency phenocopies ITCH phosphorylation. A, UbcH7−/− HEK 293T cells were transiently transfected with HA-ubiquitin, NTAP-RIPK2, FLAG-ITCH, FLAG-ITCH S687A, Myc-UbcH7, or Myc-UBCH7 C86A as indicated. NTAP was precipitated under stringent washing conditions (1% SDS, 2 m NaCl). Western blotting was then performed with the indicated antibodies. Samples were subjected to Western blot analysis as indicated. Results shown are representative of three independent experiments. B, negative control and UbcH7−/− HEK 293T cells were stimulated with 10 ng/ml TNFα for the indicated time points. Lysates were standardized for total protein and analyzed and subjected to SDS-PAGE and Western blot analysis.
Given this biochemical and functional similarity, we anticipated that, like genetic loss of ITCH or like ITCH phosphorylation, genetic UbcH7 deficiency would lead to prolonged inflammatory signaling. To test for prolonged inflammatory signaling, we stimulated both the Neg control cells and the UbcH7−/− cells with 10 ng/ml TNFα for the indicated times and queried lysates for downstream signaling. In Neg control cells, there was quick resolution of the phospho p105 signal in comparison to the prolonged phospho p105 signal in the UbcH7−/− cells (Fig. 2B). This signaling defect mimics the prolonged signaling in cells with ITCH S687D (9) and the prolonged signaling observed by other labs in ITCH−/− cells (5, 20, 21). Thus, genetic loss of UbcH7 mimics IKKβ phosphorylation of ITCH, and provides independent experimental verification that phosphorylation-mediated uncoupling of an E2-E3 interaction is physiologically relevant.
Discussion
The link between ubiquitination and phosphorylation is well known (22–24). The NF-κB inhibitor protein, IκBα, for instance, is phosphorylated on Ser-32 and Ser-36 such that it can be ubiquitinated and targeted to the proteasome (24, 25). Cell cycle re-entry and mitotic advancement proceed through similar systems (26, 27). Although it is well recognized that phosphorylation can recruit E3 ligases to a target (18, 27–29), it has yet to be shown that phosphorylation can alter E2-E3 binding. Coupled with our prior manuscript (9), our work shows that IKKβ phosphorylation of ITCH uncouples E2-E3 binding and can prolong inflammatory signaling. This work thus provides a new mechanism by which signal transduction output can be tailored to the stimulus.
Experimental procedures
Cell culture, transient transfection, and immunoprecipitation/pulldown
HEK 293T (CRL-1573; American Type Culture Collection) cells were grown in DMEM 10% SCS with 1% penicillin/streptomycin. HEK 293T cells were transfected with respective plasmids using a standard calcium phosphate transfection protocol and lysed 20–24 h post transfection. Cells were lysed in Triton lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm EGTA, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 5 mm iodoacetimide, 5 mm N-ethylmaleimide) with the following additives: calyculin, protease inhibitor mixture, 1 mm PMSF, 1 μm Na3VO4. Streptavidin or anti-FLAG Affinity gel beads (Sigma-Aldrich) were used in pulldown assays and co-immunoprecipitations and incubated at 4 °C overnight. Precipitated proteins on beads were washed in lysis buffer, under stringent conditions (1% SDS, 500 mm NaCl) four times before boiling in a 2× Laemmli sample buffer. Immunoprecipitation with anti-UBE2L3 (Cell Signaling Technology) was performed as per the manufacturer's protocol with Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology).
Western blotting
Samples were run through SDS-PAGE and transferred to nitrocellulose (see supporting Materials and Methods for antibody resources). Blocking and primary antibody dilution were performed as per the manufacturer's instruction.
Protein purification and SPR
The HECT domain of ITCH was Gibson-cloned into a bacterial expression vector encoding a GST-fusion tag and transformed into BL21 (DE3) codon plus RIPL cells (Agilent Technologies, Santa Clara, CA). UbcH7 was cloned into the pTrcHis bacterial expression vector and transformed into the same BL21 cells. The transformed BL21 cells were grown at 37 °C and protein expression was induced at 18 °C overnight with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG). Cells were harvested and lysed by sonication in a lysis buffer containing 25 mm Tris-HCl (pH 8.0), 150 mm NaCl. The ITCH HECT domains were purified using glutathione agarose and the UbcH7 was purified using nickel-nitrilotriacetic acid (Ni-NTA) beads. Buffer exchange was performed on the ITCH HECT domains for immobilization compatibility (10 mm HEPES, 150 mm NaCl, pH 7.4). The wildtype or mutant GST-HECT domain was immobilized through amine coupling using an Amine Coupling Kit (GE Healthcare Life Sciences), and the purified UbcH7 protein was flown over the immobilized HECT domain. The running buffer contained 25 mm Tris, pH 8.0, 100 mm NaCl, and 1 mm DTT. KD values were determined by kinetic fit with a heterogeneous ligand-binding model.
Lentiviral transduction, CRISPR-resistant UbcH7 construct
LentiCRISPR V2 (obtained from Addgene, Cambridge, MA) was customized with Guide 1 targeting UbcH7 (forward, 5′-CCGAAGCGGGTGCTCAGGCT-3′; reverse, 5′-AGCCTGAGCACCCGCTTCGG-3′). To make lentivirus, LentiCRISPR V2, psPAX, and pMD.2 were transfected into HEK 293T cells using a standard calcium phosphate transfection protocol. Media were harvested 2 days post transfection, cleared, and filtered through a .45-μm filter. Polybrene was added to the media prior to infecting target cells. Puromycin selection media were added 3 days after target cell infection with the CRISPR lentivirus and cells were split at a low concentration to facilitate selection of individual colonies. Six clones were pooled to create the Ubch7−/− cell line for experiments. To create a CRISPR-resistant UbcH7 construct (Myc-UbcH7 NGX), primers were designed to silently mutate the -NGG pam sequence (forward, 5′-GGTGAATGACCCACAGCCTGAGCAC-3′; reverse, 5′-GTGCTCAGGCTGTGGGTCATTCACC-3′). Linear DNA was transformed into Stbl3 cells for colony selection and sequencing confirmation.
Author contributions
J. M. P. and D. W. A. conceptualization; J. M. P., Y. C., and D. W. A. data curation; J. M. P., Y. C., T. S. X., and D. W. A. formal analysis; J. M. P. and D. W. A. investigation; J. M. P., Y. C., and T. S. X. methodology; J. M. P. and D. W. A. writing-original draft; J. M. P. and D. W. A. project administration; T. S. X. software; D. W. A. supervision; D. W. A. funding acquisition; D. W. A. writing-review and editing.
Supplementary Material
Acknowledgments
We thank George Dubyak, Theresa Pizarro, Fabio Cominelli, and Pamela Wearsch (at Case Western Reserve University) and XiaoXia Li (Cleveland Clinic Foundation) for their helpful critiques. We thank Jie Yang for help with recombinant protein expression and purification. We thank Yinghua Chen at the Protein Expression Purification and Crystallography Core Facility from the Department of Physiology and Biophysics for help with Biacore studies.
This work was supported by National Institutes of Health Grants R01GM086550 (to D. W. A.), P01DK091222 (to D. W. A.), and F31GM108403A (to J. M. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1 and supporting Materials and Methods.
- NF-κB
- nuclear factor kappa B
- ITCH
- itchy E3 ubiquitin protein ligase
- RIPK
- receptor-interacting Ser/Thr kinase
- CRISPR
- clustered regularly interspaced short palindromic repeats
- SPR
- surface plasmon resonance
- IP
- immunoprecipitated.
References
- 1. Podolsky D. K. (2002) Inflammatory bowel disease. N. Engl. J. Med. 347, 417–429 10.1056/NEJMra020831 [DOI] [PubMed] [Google Scholar]
- 2. Wiersinga W. J., Leopold S. J., Cranendonk D. R., and van der Poll T. (2014) Host innate immune responses to sepsis. Virulence 5, 36–44 10.4161/viru.25436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker R. G., Hayden M. S., and Ghosh S. (2011) NF-κB, inflammation, and metabolic disease. Cell Metab. 13, 11–22 10.1016/j.cmet.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arend W. P. (2001) Cytokine imbalance in the pathogenesis of rheumatoid arthritis: the role of interleukin-1 receptor antagonist. Semin. Arthritis Rheum. 30, 1–6 10.1053/sarh.2001.23693 [DOI] [PubMed] [Google Scholar]
- 5. Shembade N., Harhaj N. S., Parvatiyar K., Copeland N. G., Jenkins N. A., Matesic L. E., and Harhaj E. W. (2008) The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat. Immun. 9, 254–262 10.1038/ni1563 [DOI] [PubMed] [Google Scholar]
- 6. Krikos A., Laherty C. D., and Dixit V. M. (1992) Transcriptional activation of the tumor necrosis factor α-inducible zinc finger protein, A20, is mediated by κB elements. J. Biol. Chem. 267, 17971–17976 [PubMed] [Google Scholar]
- 7. Smith M. F. Jr, Eidlen D., Arend W. P., and Gutierrez-Hartmann A. (1994) LPS-induced expression of the human IL-1 receptor antagonist gene is controlled by multiple interacting promoter elements. J. Immunol. 153, 3584–3593 [PubMed] [Google Scholar]
- 8. Sun S. C., Ganchi P. A., Ballard D. W., and Greene W. C. (1993) NF-kappa B controls expression of inhibitor I kappa B alpha: Evidence for an inducible autoregulatory pathway. Science 259, 1912–1915 10.1126/science.8096091 [DOI] [PubMed] [Google Scholar]
- 9. Perez J. M., Chirieleison S. M., and Abbott D. W. (2015) An IkappaB kinase-regulated feedforward circuit prolongs inflammation. Cell Rep. 12, 537–544 10.1016/j.celrep.2015.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee E. G., Boone D. L., Chai S., Libby S. L., Chien M., Lodolce J. P., and Ma A. (2000) Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289, 2350–2354 10.1126/science.289.5488.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perry W. L., Hustad C. M., Swing D. A., O'Sullivan T. N., Jenkins N. A., and Copeland N. G. (1998) The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 18, 143–146 10.1038/ng0298-143 [DOI] [PubMed] [Google Scholar]
- 12. Matesic L. E., Copeland N. G., and Jenkins N. A. (2008) Itchy mice: The identification of a new pathway for the development of autoimmunity. In Immunology, Phenotype First: How Mutations Have Established New Principles and Pathways in Immunology (Beutler B., ed.), pp. 185–200. Springer, Dordrecht, The Netherlands: 10.1007/978-3-540-75203-5 [DOI] [PubMed] [Google Scholar]
- 13. Melino G., Gallagher E., Aqeilan R. I., Knight R., Peschiaroli A., Rossi M., Scialpi F., Malatesta M., Zocchi L., Browne G., Ciechanover A., and Bernassola F. (2008) Itch: A HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 15, 1103–1112 10.1038/cdd.2008.60 [DOI] [PubMed] [Google Scholar]
- 14. Lohr N. J., Molleston J. P., Strauss K. A., Torres-Martinez W., Sherman E. A., Squires R. H., Rider N. L., Chikwava K. R., Cummings O. W., Morton D. H., and Puffenberger E. G. (2010) Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am. J. Hum. Genet. 86, 447–453 10.1016/j.ajhg.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hutti J. E., Turk B. E., Asara J. M., Ma A., Cantley L. C., and Abbott D. W. (2007) IkappaB kinase beta phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-kappaB pathway. Mol. Cell. Biol. 27, 7451–7461 10.1128/MCB.01101-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., and Pavletich N. P. (1999) Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 10.1126/science.286.5443.1321 [DOI] [PubMed] [Google Scholar]
- 17. Eletr Z. M., and Kuhlman B. (2007) Sequence determinants of E2-E6AP binding affinity and specificity. J. Mol. Biol. 369, 419–428 10.1016/j.jmb.2007.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mercurio F., Zhu H., Murray B. W., Shevchenko A., Bennett B. L., Li J., Young D. B., Barbosa M., Mann M., Manning A., and Rao A. (1997) IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278, 860–866 10.1126/science.278.5339.860 [DOI] [PubMed] [Google Scholar]
- 19. Riling C., Kamadurai H., Kumar S., O'Leary C. E., Wu K. P., Manion E. E., Ying M., Schulman B. A., and Oliver P. M. (2015) Itch WW domains inhibit its E3 ubiquitin ligase activity by blocking E2-E3 ligase trans-thiolation. J. Biol. Chem. 290, 23875–23887 10.1074/jbc.M115.649269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tao M., Scacheri P. C., Marinis J. M., Harhaj E. W., Matesic L. E., and Abbott D. W. (2009) ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr. Biol. 19, 1255–1263 10.1016/j.cub.2009.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. You F., Sun H., Zhou X., Sun W., Liang S., Zhai Z., and Jiang Z. (2009) PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immun. 10, 1300–1308 10.1038/ni.1815 [DOI] [PubMed] [Google Scholar]
- 22. Hunter T. (2007) The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 23. Tigno-Aranjuez J. T., and Abbott D. W. (2012) Ubiquitination and phosphorylation in the regulation of NOD2 signaling and NOD2-mediated disease. Biochim. Biophys. Acta 1823, 2022–2028 10.1016/j.bbamcr.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karin M., and Ben-Neriah Y. (2000) Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu. Rev. Immunol. 18, 621–663 10.1146/annurev.immunol.18.1.621 [DOI] [PubMed] [Google Scholar]
- 25. Perkins N. D. (2006) Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene 25, 6717–6730 10.1038/sj.onc.1209937 [DOI] [PubMed] [Google Scholar]
- 26. King R. W., Deshaies R. J., Peters J.-M., and Kirschner M. W. (1996) How proteolysis drives the cell cycle. Science 274, 1652–1659 10.1126/science.274.5293.1652 [DOI] [PubMed] [Google Scholar]
- 27. Fraser J. A., Vojtesek B., and Hupp T. R. (2010) A novel p53 phosphorylation site within the MDM2 ubiquitination signal: I. Phosphorylation at SER269 in vivo is linked to inactivation of p53 function. J. Biol. Chem. 285, 37762–37772 10.1074/jbc.M110.143099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skowyra D., Craig K. L., Tyers M., Elledge S. J., and Harper J. W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219 10.1016/S0092-8674(00)80403-1 [DOI] [PubMed] [Google Scholar]
- 29. Wei W., Li M., Wang J., Nie F., and Li L. (2012) The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting disheveled protein. Mol. Cell. Biol. 32, 3903–3912 10.1128/MCB.00251-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.