Abstract
Recent discoveries of broadly neutralizing antibodies (bnAbs) in HIV-1–infected individuals have led to the identification of several major “vulnerable sites” on the HIV-1 envelope (Env) glycoprotein. These sites have provided precise targets for HIV-1 vaccine development, but identifying and utilizing many of these targets remain technically challenging. Using a yeast surface display–based approach, we sought to identify epitope-focused antigenic domains (EADs) containing one of the “vulnerable sites,” the CD4-binding site (CD4bs), through screening and selection of a combinatorial antigen library of the HIV-1 envelope glycoprotein with the CD4bs bnAb VRC01. We isolated multiple EADs and found that their trimeric forms have biochemical and structural features that preferentially bind and activate B cells that express VRC01 in vitro. More importantly, these EADs could induce detectable levels of neutralizing antibodies against genetically related autologous and heterologous subtype B viruses in guinea pigs. Our results demonstrate that an epitope-focused approach involving a screen of a combinatorial antigen library is feasible. The EADs identified here represent a promising collection of possible targets in the rational design of HIV-1 vaccines and lay the foundation for harnessing the specific antigenicity of CD4bs for protective immunogenicity in vivo.
Keywords: antibody, electron microscopy (EM), human immunodeficiency virus (HIV), vaccine, yeast, CD4bs, V3, epitope, guinea pig
Introduction
The global impact of a protective HIV-1 vaccine can hardly be overstated. Although both cell-mediated and humoral immune responses are believed to play critical roles in combating infection, it has been widely theorized that a successful vaccine should elicit a potent, sustainable neutralizing antibody response against a broad range of circulating HIV-1 strains (1, 2). Recent scientific advances have led to the identification of a growing number of broadly neutralizing antibodies (bnAbs)3 against HIV-1, which in turn have provided critical insights into the protective mechanisms of the human immune system and precise targets for immunogen design (3, 4). Broadly speaking, these bnAbs recognized five major “vulnerable sites” on the HIV-1 envelope glycoprotein gp160 as follows: 1) the CD4-binding site (CD4bs); 2) the glycan-associated V1V2; 3) V3 subdomains of gp120; 4) the membrane proximal external region (MPER) of gp41; and 5) the interface between gp120 and gp41 (5–7). Identifying antigens that can stimulate production of these bnAbs have become a major focus for HIV-1 vaccine researchers.
Traditionally, approaches to vaccine design have begun with the pathogen or its associated antigens aiming to manipulate their capacity to induce a protective antibody response in vivo. Although it has been remarkably successful against a large array of pathogens with little variability, it has become increasingly clear that a much more sophisticated approach is required to develop broadly protective vaccines against HIV-1 as it demonstrates continued sequence and structural evolution (8–10). Common mechanisms of HIV-1 immune evasion, such as sequence diversity, conformation multiplicity, and the glycan shield, enable viral escape from immune recognition and antiretroviral agents, and they pose major challenges for HIV-1 vaccine development (11–13). Recent progress in the HIV-1 vaccine field has resulted in the development of antibody ontogeny-based HIV-1 subunits or trimeric Env immunogens with emphasis on triggering the specific antibody germ line ancestors (14–19). These immunogens are able to trigger appreciable levels of autologous neutralizing antibodies in rabbits or to stimulate affinity maturation in the transgenic mouse models but failed to induce considerable heterologous bnAbs (20–30).
The idea of reverse vaccinology, in which vaccine design works backward from the host bnAbs and the antigenic domains they recognized, has recently become an exciting new avenue for HIV-1 research (31). But how exactly this idea will be translated into vaccine design has yet to be clearly defined and tested. Several methodologies are currently being investigated such as the epitope-focused scaffolding and engineering as well as stabilizing the envelope glycoprotein in the pre-fusion closed conformation using the structure-based design (17–20, 32–34). These approaches aim to characterize the antigenic regions closest to the bnAb epitope, thereby focusing the immune response on the key vulnerable sites while excluding regions that may elicit an ineffective or deleterious immune response. However, as many of the epitopes are conformational in nature, carving them out from the entire envelope glycoprotein while maintaining and stabilizing their native structures remains a technical challenge.
Our laboratory has recently developed a novel combinatorial antigen library approach for identifying antigenic domains containing the linear or conformational epitopes recognized by bnAbs (35–37). Using this system, we demonstrated here the successful isolation of the epitope-focused antigenic domain recognized by CD4bs bnAb VRC01 (EAD-VRC01). Structure and function analysis showed that EAD-VRC01 contains the large majority of CD4bs and is able to bind and attenuate the neutralizing activity of VRC01 in a pseudovirus-based assay. EAD-VRC01 is also able to trigger B cell activation through the formation of microclusters of the membrane-bound VRC01-BCRs and downstream signalings, the critical steps for B cells to differentiate into antibody-producing plasma cells. More importantly, immunization of guinea pigs with EAD-VRC01 induced detectable levels of neutralizing antibodies against genetically related autologous and heterologous subtype B viruses. Profiling of the neutralizing sera indicated the antibody specificity was largely directed to the CD4bs as well as V3 peptide on the envelope glycoprotein. We believe that the study reported here represents a novel and promising strategy for identifying EAD containing the desired bnAb epitopes. The EAD-VRC01 identified here could serve as a promising prototype for the future HIV-1 vaccines aiming to translate the specific antigenicity of CD4bs into the protective immunogenicity in vivo.
Results
Selection of the epitope-focused antigenic domain by VRC01
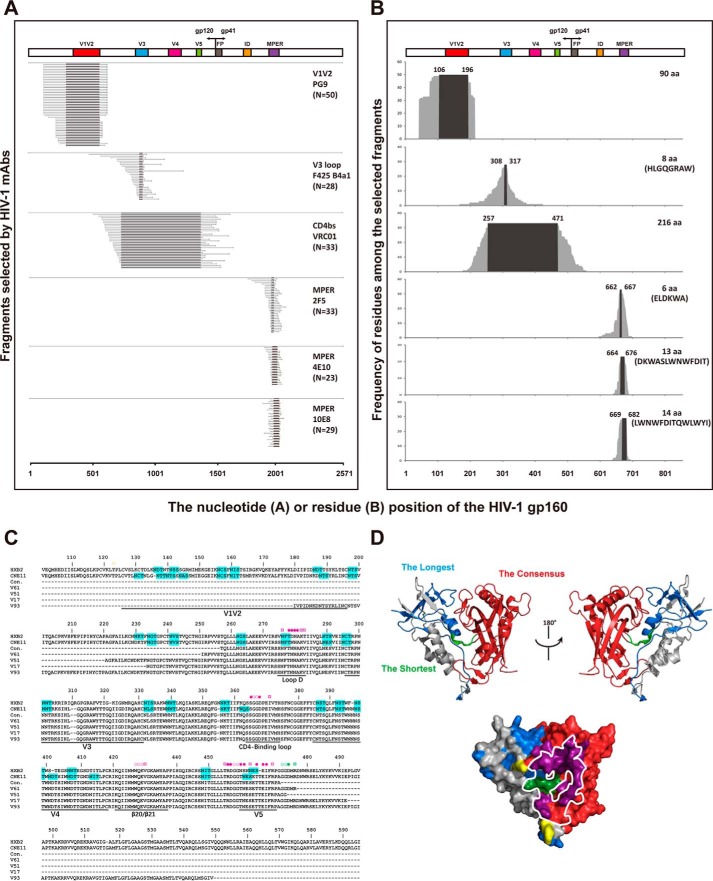
We have previously shown that the combinatorial antigen library displayed on the yeast surface was able to express both linear and conformational epitopes and was successfully applied to the profiling studies of antibody response during HIV-1 and influenza infection (35–37). Taking the advantage of the scale and diversity of displayed antigens, we sought to identify the epitope-focused antigenic domain (EAD) for HIV-1 immunogen design and development. To this end, we first constructed a combinatorial HIV-1 envelope antigen library based on the full-length envelope gene of CNE11 derived from an HIV-1 subtype B chronically infected patient in China (38). The pseudoviruses bearing the CNE11 envelope were able to infect the CCR5+CD4+TZM-bl cell line and demonstrated sensitivity to the majority of well-characterized bnAbs targeting the key vulnerable sites on the envelope (38). The actual protocol for construction, validation, and selection of the combinatorial antigen library has been illustrated in greater detail elsewhere (35–37). Specifically, through multiple rounds of fluorescence-activated cell sorting (FACS) and cloning, we were able to obtain 33 yeast clones that were highly reactive to VRC01, and a substantial number of clones to the control monoclonal antibodies (mAbs) such as PG9 (50 clones), 2F5 (33 clones), 4E10 (23 clones), and 10E8 (29 clones) (Fig. 1 and Fig. S1). Sequence analysis of the coding sequences revealed that they were variable in length but invariably contained the epitope specificity defined by each of the corresponding antibodies (Fig. 1B and Fig. S2) (5–7). This suggests that the selected EADs were correctly folded and displayed on the surface of the yeast. In particular, those selected by VRC01 (EAD-VRC01) largely spanned the V3, V4, and V5 regions of gp120, and those by PG9 (EAD-PG9) encompassed the V1V2 region of gp120, whereas those by 2F5 (EAD-2F5), 4E10 (EAD-4E10), and 10E8 (EAD-10E8) covered the MPER region of gp41 (Fig. 1, A–C). Of note, as PG9 is a glycan-dependent antibody, the EAD-PG9 must also contain an appropriate glycan moiety for specific recognition. The average length of the selected EAD-VRC01 is 282 aa, which is relatively longer than the EAD-PG9 (155 aa), EAD-2F5 (32 aa), EAD-4E10 (27 aa), and EAD-10E8 (29 aa). Furthermore, all of the 33 EAD-VRC01 sequences (Fig. S2), exemplified by the shortest (V61, 227 aa), the longest (V93, 372 aa), and the two intermediates (V51, 254 aa and V17, 260 aa), contained the critical residues as well as the conformational structure for VRC01 binding (Fig. 1, C and D). In fact, when superimposed onto the monomeric gp120, EAD-VRC01 significantly overlapped with the outer domain of gp120 and covered over 94% of the VRC01 footprint (Fig. 1D), in agreement with our initial design and expectation for EAD-VRC01.
Figure 1.
Identification and characterization of EAD-VRC01 from the HIV-1 gp160 combinatorial antigen library displayed on the yeast surface. A, EADs selected by the HIV-1 mAbs (PG9, F425 B4a1, VRC01, 2F5, 4E10, and 10E8) aligned to the original full-length CNE11 gp160 sequence used in the construction of the combinatorial yeast library. The target epitopes and the number (N) of sequences isolated by each mAb were indicated on the far right of each panel. The hypervariable regions V1–V5, interspersed conserved regions C1–C5 in gp120, fusion peptide (FP), immunodominant region (ID, and MPER in gp41 are indicated. B, frequency of each amino acid residue among the selected EADs along their corresponding positions in the CNE11 envelope glycoprotein. The consensus of amino acid sequences was numbered on the top of each graph according to HXB2 and highlighted by black background. The length and sequences of each consensus are shown on the far right wherever possible. C, sequence alignment of four EAD-VRC01 together with their consensus (con.) and corresponding sequences from CNE11 and HXB2. The EAD-VRC01 sequences represented the shortest (V61, 227 aa), the longest (V93, 372 aa), and the two intermediate ones (V51, 254 aa and V17, 260 aa). The contact residues with VRC01 were indicated with the symbol ○ for main-chain only contacts; ¤ for side-chain only contacts, and ● for both main- and side-chain contacts. Those colored in yellow were located outside the selected EAD-VRC01 fragments, in purple among the consensus EAD-VRC01 fragment, and in dark green in the shortest EAD-VRC01 V61 fragment. Potential glycosylation sites with the signature sequence NX(T/S) were highlighted in cyan. Dashes represented the gaps introduced to the preserve alignment. The hypervariable regions V1–V5, loop D, CD4-binding loop, and β20/β21 in gp120 were indicated. D, superimposing EAD-VRC01 sequences onto the monomeric gp120 (PDB code 3NGB) in ribbon and surface representations. The longest EAD-VRC01 was highlighted in light blue, the shortest in light green, and the consensus in red. The contact residues with VRC01 were colored as in C, and the contact surface of CD4 on the outer domain of gp120 was outlined by the white line.
Binding kinetics of monomeric and trimeric EAD-VRC01 to VRC01 class mAbs
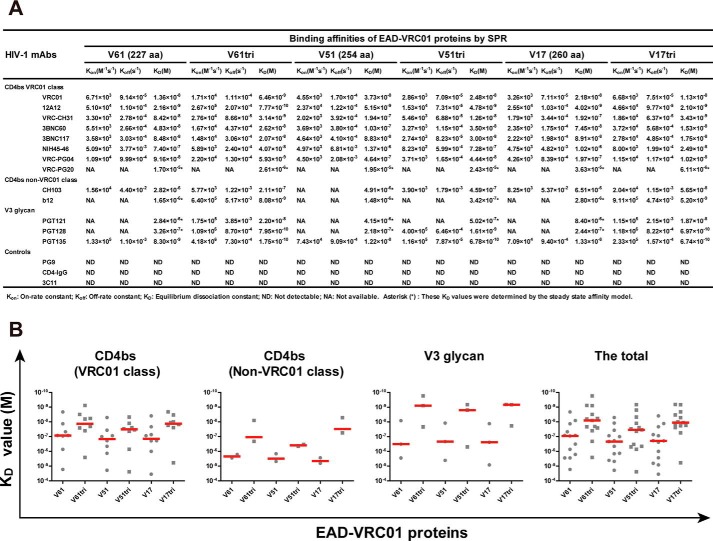
To further characterize the binding properties of EAD-VRC01 to a spectrum of mAbs with well-defined specificities, we expressed the monomeric and trimeric forms of three representatives EAD-VRC01 with different lengths (V61, 227 aa; V51, 254 aa; and V17, 260 aa) in Sf9 cells using the Bac-to-Bac baculovirus expression system. A foldon trimerization domain of T4 fibritin was fused to the C terminus of EAD-VRC01 for the production of trimers. The monomeric and trimeric V61, V51, and V17 in the cell supernatant were concentrated and purified by nickel-affinity chromatography and size-exclusion chromatography before being subjected to the binding kinetic studies by surface plasmon resonance (SPR) on a Biacore T200 (GE Healthcare). As shown in Fig. 2, the monomeric and trimeric V61, V51, and V17 were able to bind to the antigenic fragment (Fab) of all members of the VRC01 class mAbs, suggesting that the VRC01 epitope on the EAD-VRC01 was well-maintained and accessible. Furthermore, despite their differences in length, V61, V51, and V17 demonstrated comparable binding affinities (KD) to the Fab of VRC01 class mAbs, although the trimeric form demonstrated an overall average close to 10-fold stronger binding than the monomeric ones. Among all the VRC01 class mAbs tested, 12A12 bound the strongest whereas VRC-PG20 was the weakest (Fig. 2A). Furthermore, both the monomeric and trimeric forms of V61, V51, and V17 demonstrated similar levels of binding to the Fab of non-VRC01 class mAbs CH103 and b12, although the overall average was about 10-fold less compared with that of VRC01 class mAbs (Fig. 2, A and B). In addition, strong binding to the Fab of V3 glycan mAbs PGT121, PGT128, and PGT135 was also observed. In particular, the trimers demonstrated more than 2 log stronger binding than the monomers and also exceeded those of the VRC01 class mAbs (Fig. 2, A and B). This could be explained by the inclusion of the V3 region in the EAD-VRC01, which may become more exposed and accessible in the trimeric form. It is worthwhile to note that among the anti-V3 narrowly neutralizing mAbs tested (447–52D, 19b, and 39F), only 39F was able to bind to EAD-VRC01 (V61, V51, and V17) with significant preference for the trimers over the monomers (Fig. S3), suggesting that at least the N-terminal side of the V3 loop is exposed and accessible. These results clearly indicate that the three representatives EAD-VRC01 V61, V51, and V17 contain CD4bs and V3 glycan vulnerable sites identified on the full-length envelope glycoprotein of HIV-1, and the trimeric forms appear to adopt superior conformational structure than their monomeric counterparts for mAbs binding. Finally, none of the EAD-VRC01 V61, V51, and V17 had detectable bindings to the control mAbs PG9 and 3C11 or to soluble CD4-IgG (Fig. 2A). This is consistent with earlier reports where the outer domain of gp120 also failed to engage with the CD4 molecule despite its strong interaction with VRC01 (33).
Figure 2.
Binding affinities of monomeric and trimeric EAD-VRC01 to HIV-1 mAbs measured by SPR. A, binding affinity between EAD-VRC01 and HIV-1 mAbs was measured by SPR and presented by Kon (on-rate constant), Koff (off-rate constant), and KD (equilibrium dissociation constant). The tested mAbs were CD4bs-targeted mAbs from multiple donors, including VRC01 class mAbs (VRC01, 12A12, VRC-CH31, 3BNC60, 3BNC117, NIH45–46, VRC-PG04, and VRC-PG20) and non-VRC01 class mAbs (CH103 and b12), V3 glycan-targeted mAbs (PGT121, PGT128, and PGT135), a V1V2-targeted mAb (PG9), CD4-IgG, and an irrelevant influenza hemagglutinin-targeted mAb (3C11) as the negative controls. The sensorgrams were fitted globally in 1:1 Langmuir model, and several KD values (denoted by asterisk symbols) were determined by the steady-state affinity model as the interactions were too fast to provide kinetic information. B, comparison of KD values within and between each mAb or combined group. The median value in each group was represented by the horizontal red line.
Attenuation of VRC01 Fab neutralization by monomeric and trimeric EAD-VRC01
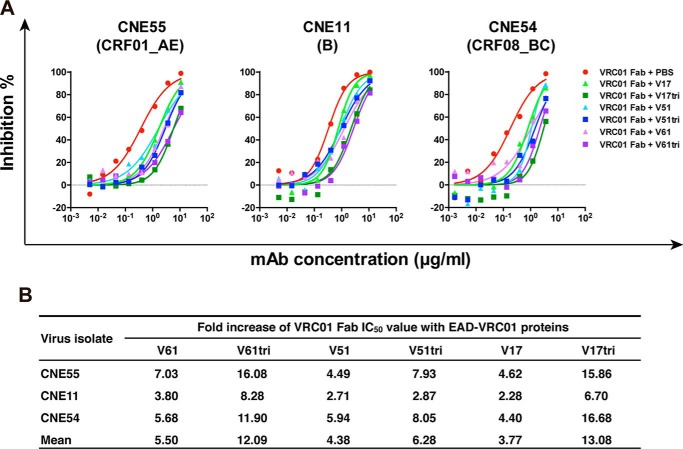
Although the CD4bs on EAD-VRC01 V61, V51, and V17 are detectable on the solid surface by SPR, it is uncertain whether they are well-preserved in solution. This question is critical for their engagement with the membrane-bound VRC01 in cell cultures and their capacity in inducing the preferred antibody specificities in experimental animals. To this end, we mixed the Fab of VRC01 with the monomeric and trimeric V61, V51, and V17 in solution before applying the pseudovirus-based neutralization assay. We hypothesized that if the CD4bs was indeed well-preserved in solution, such mixing would result in a spontaneous absorption and therefore a dramatic depletion of the neutralizing activity of VRC01 Fab. In fact, this is what we have found. As shown in Fig. 3, both monomeric and trimeric V61, V51, and V17 were able to attenuate neutralizing activity of VRC01 Fab in a dose-dependent manner against the three representative HIV-1 subtypes or circulating recombinant forms (CRFs) from China (38). In general, the trimer was more effective than the monomer in depleting the neutralizing activity but such effect appears to be EAD-VRC01 as well as viral strain-dependent. For instance, rather small differences were identified between EAD-VRC01 V51 and V51tri compared with that between EAD-VRC01 V61 and V61tri as well as between EAD-VRC01 V17 and V17tri (Fig. 3B). In contrast, the viral strain CNE11 was relatively insensitive to VRC01 Fab depletion than CNE55 and CNE54 (Fig. 3). Nevertheless, these results indicate that both the monomeric and trimeric forms of EAD-VRC01 V61, V51, and V17 preserved the CD4bs in solution, although the latter appeared to adopt superior conformational structure for VRC01 binding than their monomeric counterparts, consistent with the binding kinetic analysis conducted on the solid surface using SPR (see above).
Figure 3.
EAD-VRC01 attenuates the neutralizing activity of VRC01 Fab. A, neutralizing activity of VRC01 Fab in the presence of monomeric and trimeric EAD-VRC01 V61, V51, and V17 (30 μg/ml) against the three representative HIV-1 pseudoviruses bearing the envelope glycoprotein of CNE55 (clade CRF01_AE), CNE11 (clade B), and CNE54 (clade CRF08_BC) from China. B, fold increase in neutralizing IC50 of VRC01 Fab in the presence of EAD-VRC01 as compared with the mock controls in PBS.
Structural analysis of EAD-VRC01 trimer and its complex with VRC01 Fab by electron microscopy
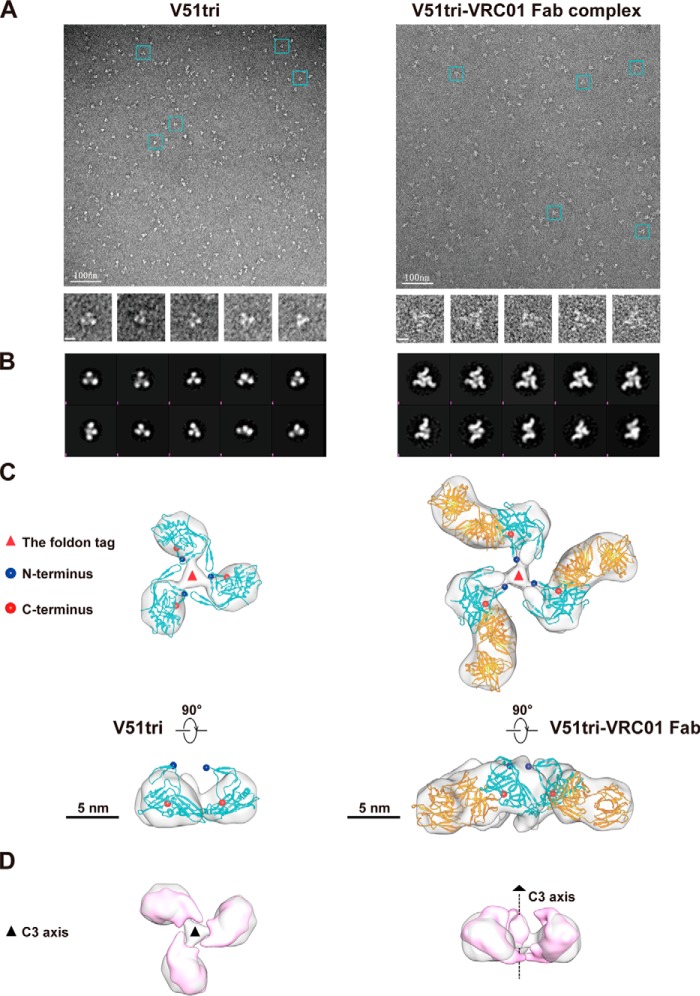
To further characterize the structural features of EAD-VRC01 and its binding with VRC01 Fab, we went further to analyze V51tri, one of the EAD-VRC01s, and its complex with VRC01 Fab using negative staining electron microscopy (EM). As shown in Fig. 4, trimeric features of the V51tri and V51tri–VRC01 Fab complex particles were clearly observed in the raw EM images (Fig. 4A). Reference-free 2D classification showed more details of the overall trimeric shapes of V51tri and V51tri–VRC01 Fab complex in both top and side views (Fig. 4B). In particular, the 2D classification of the V51tri–VRC01 Fab complex suggested that the majority of V51tri proteins were complexed with VRC01 Fab. Specifically, the reconstructed 3D EM maps of the V51tri and V51tri–VRC01 Fab complex showed the three monomeric subunits surrounding a cylindrical central core (Fig. 4C). Compared with V51tri, the V51tri–VRC01 Fab complex has additional densities protruding radially out from the lateral surface of each inner V51tri subunit. To further interpret the EM results, the V51tri structure model generated from the BG505 SOSIP.664 gp140 trimer (PDB code 4NCO) (39) and the VRC01 Fab crystal structure (PDB code 3NGB) (40) were fitted into the reconstructed 3D EM map (Fig. 4C). The fitted V51tri–VRC01 Fab complex model shows that the CD4bs was exposed to the outside and was spatially accessible to VRC01 Fab. Furthermore, the comparison between V51tri 3D EM map and its equivalent map generated from BG505 SOSIP.664 gp140 trimer (PDB code 4NCO) showed that the orientation and the overall arrangement of the monomers closely resemble those in the native gp140 trimers (Fig. 4D). Taken together, these results reinforced the notion that the trimeric form of V51, and most likely that of V61 and V17, contained the properly positioned and oriented epitope of VRC01 and are suitable for downstream biological and immunological studies.
Figure 4.
Negative staining EM and image reconstruction of the purified V51tri and V51tri–VRC01 Fab complex. A, negative staining EM micrographs of the V51tri (left) and V51tri–VRC01 Fab complex (right). Raw micrographs were shown at the top. Representative particles were highlighted by the cyan boxes and shown at the bottom. The scale bars, 100 nm in the raw micrographs and represented 10 nm in the boxed particle images. B, reference-free class averages of the V51tri (left) and V51tri–VRC01 Fab complex (right). C, three-dimensional (3D) reconstructions of the V51tri (left) and V51tri–VRC01 Fab complex (right) were shown in the top (top) and side (bottom) views with the fitted models. The V51tri and V51tri–VRC01 Fab models were generated from the crystal structures of BG505 SOSIP.664 gp140 trimer (PDB code 4NCO) and gp120–VRC01 Fab complex (PDB code 3NGB). The fitted V51 monomers were indicated by the cyan ribbon and the fitted VRC01 Fabs were indicated by the orange ribbon. The N and C termini of the fitted V51 monomers were marked with the blue and red balls, respectively. Positions of the foldon tags were indicated by the red triangles and the scale bars, 5 nm. D, comparison of the V51tri EM map (in gray) and its equivalent map (in magenta) generated from the crystal structure of BG505 SOSIP.664 gp140 (left, top view; right, side view). The position of the 3-fold axis was indicated by a black dotted line.
EAD-VRC01 triggers surface BCR clustering and downstream signalings in B cells
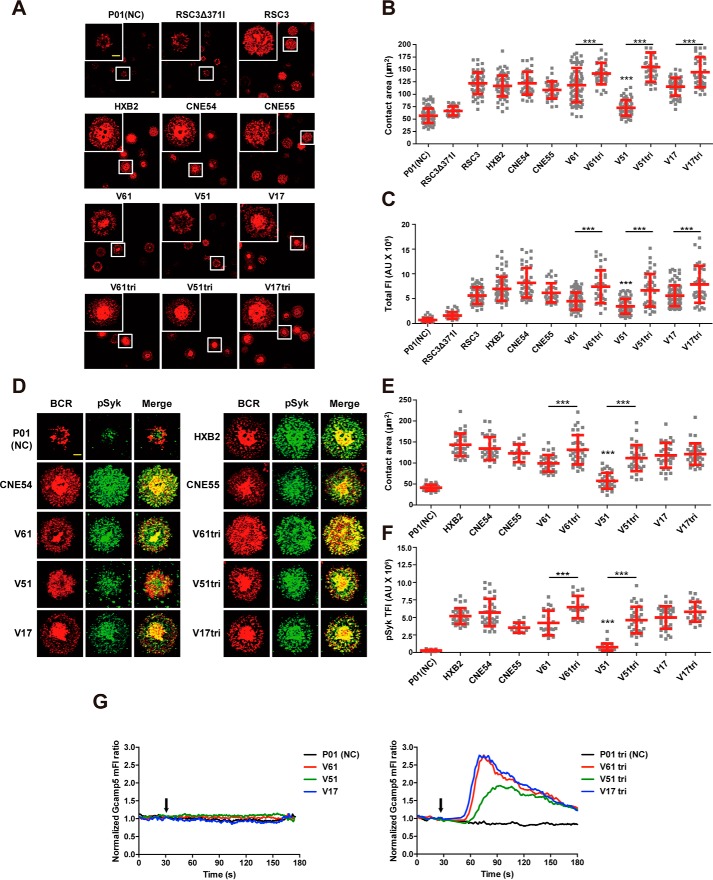
Although EAD-VRC01 V61, V51, and V17 are able to bind to the soluble VRC01, it is uncertain whether they could activate B cells by triggering surface VRC01-BCR clustering and downstream signalings, the critical steps for B cells to differentiate into antibody-producing plasma cells. To address this question, we constructed the human Ramos B cells to express human IgM-BCRs with the variable regions of the heavy and light chain from VRC01 (Ramos-VRC01 B cells) and loaded onto a planar lipid bilayer containing the monomeric or trimeric V61, V51, and V17 for 10 min at 37 °C. The formation and accumulation of BCR microclusters on the antigen contact area were examined using total internal reflection fluorescence microscopy (TIRFM) as reported previously (41–43). As shown in Fig. 5, A–C, V61, V51, and V17 were all able to trigger the formation of VRC01-BCR microclusters within B cell immunological synapses, but the trimeric forms were clearly more potent than the monomeric ones. Among the monomeric forms, V51 was the weakest, whereas V61 and V17 were comparable in terms of the contact area on the antigen-containing surface (Fig. 5B) and the total fluorescence intensity of the VRC01-BCR microclusters (Fig. 5C). The differences among the three trimeric forms were quite minimum but significantly more potent than their respective monomeric counterparts. In fact, they were equivalent if not more potent than the positive controls such as the monomeric full-length gp120 glycoprotein (HXB2, CNE54, and CNE55) and resurfaced stabilized core 3 of gp120 (RSC3) (Fig. 5, B and C). The negative control P01 (a EAD-PG9 from the same library) and RSC3Δ371I had either negligible or barely detectable activity in inducing the formation of VRC01-BCR microclusters.
Figure 5.
Triggering VRC01-BCR signalings and calcium mobilization by EAD-VRC01. A, synaptic accumulation of BCRs triggered by EAD-VRC01 in the human Ramos B cells expressing VRC01-IgM-BCRs in TIRFM images. The EAD-VRC01 as well as control proteins, such as P01 (a EAD-PG9 as the negative control), RSC3 (the bait protein for isolating VRC01 mAb), RSC3Δ371I (lacked a single amino acid in RSC3 at position 371), HXB2 gp120 (clade B), CNE54 gp120 (clade CRF08_BC), and CNE55 gp120 (clade CRF01_AE), were used to trigger VRC01-BCR clustering. The magnified insets (upper left corner) show the highlighted cells in the original image with the white boxes. The BCR molecules are labeled in red, and the scale bar in the inset window represents 1.5 μm. B and C, statistical analysis for the contact area (B) and total fluorescence intensity (TFI) (C) of accumulated BCRs in the immunological synapses of the Ramos-VRC01 B cells. Each dot in the plot represents the data of one cell. Two-tailed t tests were used for the statistical analysis between the monomer and trimer of the same EAD-VRC01. Bars indicate the mean values and standard deviations. ***, p < 0.001. D, synaptic recruitment of phosphorylated Syk (pSyk) to the contact area of the Ramos B cells after BCR accumulation by EAD-VRC01 in TIRFM images. The BCRs and pSyk molecules are labeled in red and green. The scale bar, 1.5 μm. E and F, statistical analysis for the contact area (E) and TFI (F) of recruited pSyk in the immunological synapses. Each dot in the plot represents the data of one cell. Two-tailed t tests were used for the statistical analysis between the monomer and trimer of the same EAD-VRC01. Bars indicate the mean values and standard deviations. ***, p < 0.001. G, EAD-VRC01 induces the calcium flux in the Ramos B cells. Gcamp5 biosensor reported the Ca2+ flux by flow cytometry in the Ramos B cells after the stimulation of EAD-VRC01 monomers (left) or trimers (right) at the indicated time points (black arrow). P01 and P01tri stated above were negative controls.
The ability of V61, V51, and V17 to activate BCR downstream signalings in the Ramos-VRC01 B cells was further analyzed using intracellular immunofluorescence staining of the signaling molecules, phosphorylated spleen tyrosine kinase (pSyk), and downstream signaling molecules of phosphorylated tyrosine (pTyr) as reported previously (41–43). The phosphorylation of Syk and total tyrosine of BCR downstream signaling molecules is the direct outcome of BCR aggregation and therefore has been widely used as the downstream marker for B cell activation. Consistent with the effect on VRC01-BCR microclusters, the monomeric and trimeric V61, V51, and V17 were able to trigger the recruitment of pSyk and downstream signaling molecules of pTyr into the immunological synapses demonstrated by the accumulation and significant colocalization with VRC01-BCR in the TIRFM images (Fig. 5D and Fig. S4A). Quantitative measurement of the contact area and total fluorescence intensity of pSyk and pTyr in the immunological synapse showed significantly more potency for the trimeric than the monomeric form, except for V17 and V17tri where the comparable effect on pSyk was found (Fig. 5, E and F, and Fig. S4, B and C). Furthermore, the trimeric forms were at least equivalent to if not more potent than the positive controls such as the monomeric full-length gp120 glycoprotein (HXB2, CNE54, and CNE55). The negative control P01 showed insufficient ability to induce the phosphorylation of Syk and the total tyrosine of BCR downstream signaling molecules (Fig. 5, E and F, and Fig. S4, B and C).
We also examined the ability of V61, V51, and V17 to induce Ca2+ mobilization in the Ramos-VRC01 B cells, an event following the formation of VRC01-BCR microclusters and the recruitment of pSyk and downstream signaling molecules of pTyr into the immunological synapses. Specifically, Ramos-VRC01-Gcamp5 B cells were generated to express the intracellular calcium indicator Gcamp5, stimulated with the monomeric or trimeric V61, V51, and V17, and then monitored calcium dynamics by flow cytometry. As shown in Fig. 5G, whereas the monomeric V61, V51, and V17 failed to induce Ca2+ flux in the Ramos-VRC01-Gcamp5 B cells, the corresponding trimeric forms were quite capable, although V61 and V17 were more robust than V51. Taken together, these results demonstrate that EAD-VRC01 V61, V51, and V17 can activate B cells through the VRC01-BCR and are therefore appropriate for downstream immunological characterization.
EAD-VRC01 induces neutralizing antibody against autologous and heterologous HIV-1
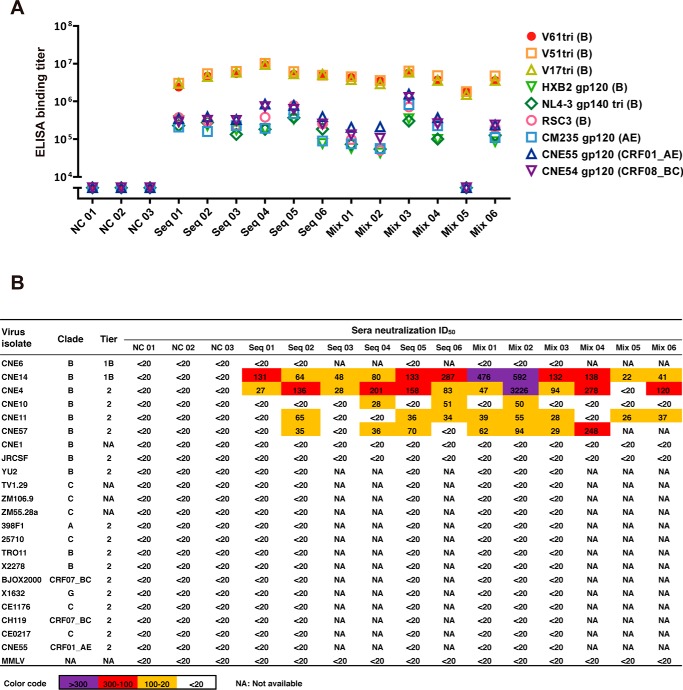
As the trimeric forms of V61, V51, and V17 are superior to the monomeric ones in binding to the soluble VRC01 and activating B cell signaling through the membrane-bound VRC01, we have chosen the trimeric forms to immunize guinea pigs. Two different immunization strategies were explored with the intention to test whether the immunization method itself has significant impact on immunogenicity in inducing neutralizing antibodies. One strategy involved a sequential immunization from the longest to the shortest, starting with the V17tri, followed by V51tri and then V61tri, and with a final boost of V17tri. The other involved immunization with the equal mixture of V61tri, V51tri, and V17tri throughout the course. For each strategy, six animals received four rounds of subcutaneous immunization at weeks 0, 4, 12, and 20, and blood samples were collected at weeks −2 for prebleed and at weeks 14 and 22 after the first immunization (Fig. S5, A and B). Serum-binding activity to a broad array of the envelope recombinant proteins was evaluated using standard ELISA (Fig. 6A). It is clear that both immunization strategies induced high levels of binding antibodies to the autologous trimeric V61, V51, and V17. Binding activity to the heterologous monomeric gp120 (HXB2, CM235, CNE54, and CNE55) and trimeric NL4-3 gp140 as well as RSC3 was also significant, although the actual binding titer was about 1 log less compared with the autologous trimers. The serum sample of Mix 05 in the mixture of the immunized group had equivalent high levels of binding to the autologous trimers but unusually low levels to the heterologous recombinant envelopes for reasons that are currently unknown (Fig. 6A). Serum-neutralizing activity was tested against a panel of autologous and heterologous CNE pseudoviruses originally from China (38) and the global panel and other standard controls from the AIDS Reagent Program, National Institutes of Health (Fig. 6B) (44, 45). It needs to be noted that all of the CNE pseudoviruses as well as those in the global panel were properly tier-phenotyped by Dr. David Montefiori and co-workers at Duke University Medical Center using widely accepted tier phenotype definition (44). Results from multiple independent experiments showed appreciable levels of neutralizing antibodies against the genetically related autologous and heterologous subtype B CNE pseudoviruses, although no detectable neutralizing activity was found against distantly related subtype B or other subtypes or CRFs (Fig. 6B). In general, neutralizing activity in the mixture group was slightly higher than that in the sequentially immunized group, but no significant difference in potency or breadth was noticed. Among all of the animals tested, Mix 02 demonstrated the strongest activity against the autologous (CNE11) as well as the heterologous (CNE4, CNE10, CNE14, and CNE57) strains. However, the prebleed sera did not have any detectable neutralization activity against the same virus panel (Fig. S5C). It is important to note phylogenetic analysis based on the amino acid sequences corresponding to the EAD-VRC01 region revealed that the sensitive viruses were clustered in a single branch, whereas those insensitive ones were rather distant (Fig. S6). These results clearly indicate that the trimeric forms of V61, V51, and V17 were able to induce appreciable levels of neutralizing antibodies in vivo against the autologous and heterologous subtype B CNE strains that are genetically more related than other divergent subtypes or CRFs. The immunization strategy per se does not appear to have significant impact on the potency and breadth of the neutralizing antibodies induced in the experimental animals.
Figure 6.
Immunogenicity of EAD-VRC01 trimers in guinea pigs. A, ELISA binding titers of the immunized guinea pig sera against the autologous EAD-VRC01 trimer immunogens (V61tri, V51tri, V17tri, and clade B), HXB2 gp120 (clade B), NL4-3 SOSIP.664 gp140 trimer (clade B), RSC3 (clade B), CM235 gp120 (clade AE), CNE55 gp120 (clade CRF01_AE), and CNE54 gp120 (clade CRF08_BC). The sera tested here were collected 2 weeks (week = 22) after the fourth inoculation. The ELISAs were performed on two independent experiments. B, neutralizing activity of the immune sera against a panel of pseudoviruses as well as 10 strains from the global panel in the TZM-bl assay. Moloney murine leukemia virus (MMLV) was used as a negative control. The sera tested here were collected 2 weeks (week = 22) after the fourth inoculation. The neutralization assays were conducted on at least two independent occasions, and each was in duplicate. The color code was used to define neutralizing ID50 titers, and those <20 were in black, 20–100 in orange, 100–300 in red, and >300 in purple.
Profiling neutralizing sera by competitive ELISA and neutralization depletion
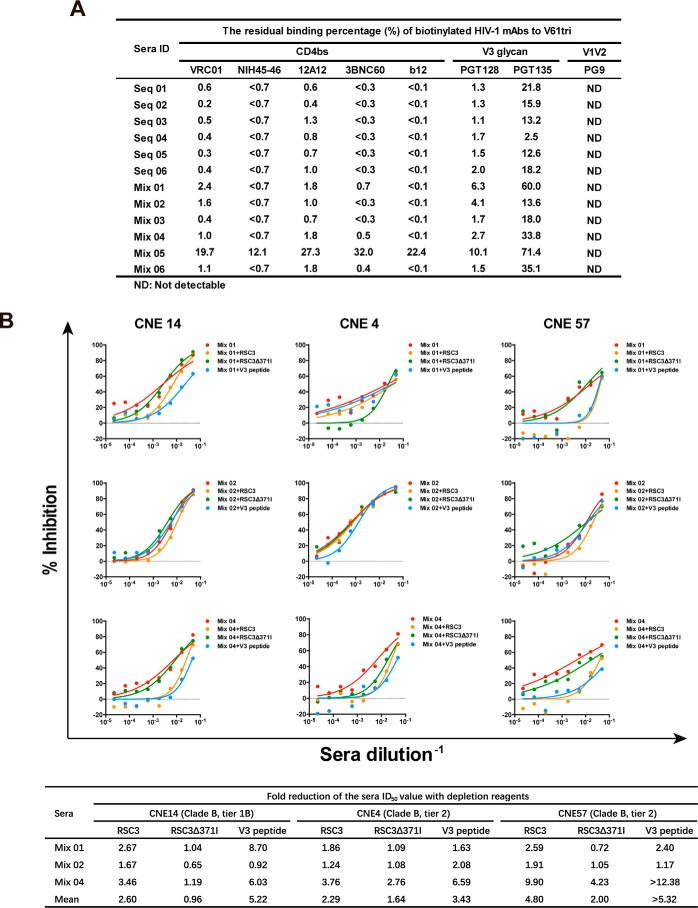
To dissect the target specificity of the neutralizing sera, we first assessed their ability to compete with various mAbs for binding to the immunogens through a previously published ELISA (46). As the immunogens were recognized by CD4bs as well as V3 glycan mAbs, representative mAbs from both classes were biotinylated and tested. Biotinylated PG9 and serum samples from the control group injected with PBS were used as negative controls. Competitions were considered as significant when the binding titer of biotinylated mAbs in the immune sera was reduced to less than half of that in the control sera. Specifically, all serum samples were diluted 1:100 and mixed with a serial dilution of the biotinylated mAbs before being applied to the ELISA plate coated with the immunogen V61tri. The competitive ability of the immune sera was defined as the residual binding percentage of the biotinylated mAbs compared with the controls. As shown in Fig. 7A, all serum samples showed the significant competition with CD4bs mAbs (VRC01, NIH45–46, 12A12, 3BNC60, and b12). In fact, except for the Mix 05, all samples reduced the binding activity of CD4bs mAbs for more than 97%, suggesting antibodies in the immune sera are largely directed to the epitope initially focused in the immunogen design. In addition, all serum samples also demonstrated the significant competition with V3 glycan mAbs, especially a more profound effect on PGT128 than PGT135 (Fig. 7A). The Mix 02 serum with the strongest neutralizing activity (see above) did not seem to have any special competitive edge over other samples. The Mix 05 serum, however, demonstrated the lowest competitive activity consistent with its genuinely low levels of binding as well as neutralizing activity (see above). As expected, no competition effect was detected against the control PG9 mAb. These findings are consistent with the antigenicity of the immunogens analyzed by SPR (Fig. 2) and suggested that some of their antigenic features were translated into the immunogenicity in the experimental animals. It needs to be noted, however, that the competition activity observed here could represent the direct overlaps with the epitope and/or steric interference with the tested mAbs. We need to be cautious when interpreting these results.
Figure 7.
Profiling of the guinea pig immune sera by competitive ELISA and neutralization depletion. A, residual binding percentage (%) of biotinylated HIV-1 mAbs to V61tri measured by competitive ELISA. The binding titers of biotinylated mAbs with known epitopes in the presence of the immune guinea pig sera were evaluated and numerically presented. The sera from the NC group and PG9-biotin mAb were used as negative controls. B, reduction of serum-neutralizing activity in the presence of RSC3, RSC3Δ371I, and the autologous V3 peptide. The three relatively potent neutralizing sera (Mix 01, Mix 02, and Mix 04) were studied for their ability to neutralize the three relatively sensitive pseudoviruses (CNE14, CNE4, and CNE57) in the presence of RSC3, RSC3Δ371I, and the autologous V3 peptide in the TZM-bl assay. The actual neutralization curves (top) and fold reduction in neutralization ID50 (bottom) were showed. The > symbol indicates the complete depletion of neutralizing activity.
To further profile the neutralizing sera, we also performed ELISA analysis against a 15-mer peptide library derived from the autologous CNE11 EAD-VRC01 envelope sequence. All serum samples were 1:1000 diluted before applied to the ELISA plate and their binding activity was measured as the absorbance (A) at 450 nm. Fig. S7 shows the binding profiles of all serum samples across the entire HIV-1 CNE11 EAD-VRC01 sequence. Each individual profile is presented together with the average values from the sequential and mixed immunization group. For comparative purposes, regions that have important structural and functional relevance are also indicated along the top of each profile. It is clear that binding profiles of all serum samples were confined to the region starting from the upstream of loop D all the way to the downstream of the V5 loop, matching the sequences of EAD-VRC01 immunogens (Fig. 1C and Fig. S7). However, the binding activity was not evenly distributed but was largely focused on six stretches of amino acid residues in the upstream of loop D, the V3 loop, the CD4-binding loop, the V4, the β20/β21, and the V5 regions. In fact, these residues overlap somewhat with those in the CD4bs and V3 region recognized by VRC01 class and V3 glycan mAbs previously identified through the crystal structure analysis (40, 47, 48). Furthermore, no significant difference was found in the average profiles between the sequential and mixed immunization groups demonstrated by the almost overlapping orange dashed and green dotted lines in Fig. S7. Taken together, these results suggest that the targeted specificity of the neutralizing sera is confined to the intended regions of our initial design and overlaps with the CD4bs and V3 epitope recognized by VRC01 class and V3 glycan mAbs, respectively.
To further map the neutralizing sera, we conducted neutralization depletion experiments in the presence of recombinant RSC3, RSC3Δ371I, and the autologous V3 peptide. We hypothesized that if the sera contained the CD4bs- and V3-directed neutralizing antibodies, such mixing would result in a spontaneous absorption and therefore a reduction of the neutralizing activity of immune sera. To this end, the three relatively potent neutralizing sera (Mix 01, Mix 02, and Mix 04) were studied for their ability to neutralize the three sensitive pseudoviruses (CNE14, CNE4, and CNE57) in the presence or absence of RSC3, RSC3Δ371I, and the autologous V3 peptide in the TZM-bl assay. The fold of reduction in sera ID50 was then compared and recorded. Throughout the entire analysis, RSC3 was on average more than twice as efficient as RSC3Δ371I in reducing the serum-neutralizing activity, although apparent differences existed for different samples and different viruses (Fig. 7B). For instance, the serum sample Mix 04 was the most sensitive and Mix 02 was the least sensitive to RSC3 depletion reflected by the actual fold of reduction in serum ID50. Among the tested viruses, however, CNE14 (tier 1B) was the most responsive to RSC3 depletion (RSC3/RSC3Δ371I = 2.60/0.96 = 2.71) followed by CNE57 (tier 2) (4.80/2.00 = 2.40) and then by CNE4 (tier 2) (2.29/1.64 = 1.39). Similarly, the V3 peptide was also able to reduce neutralizing activity of all serum samples, although Mix 04 was the most responsive and Mix 02 was the least responsive. These results clearly indicate that the immune sera from Mix 01, Mix 02, and Mix 04 contained CD4bs- and V3-directed neutralizing antibodies that can be absorbed and reduced by RSC3 and the V3 peptide, respectively. However, as sequence and structure differences exist between the EAD-VRC01 immunogens and RSC3 and the V3 peptide, the observed reduction in neutralizing activity and its extrapolated target specificity could only be interpreted with caution. More definitive answer for target specificity would have to rely on isolation and characterization of a sizable number of monoclonal antibodies from the immune animals.
Discussion
Identification of vulnerable sites on the HIV-1 envelope glycoprotein by bnAbs has provided precise targets for HIV-1 vaccine design and development. Among many other approaches being explored, the epitope-focused strategy aims to target the vulnerable sites while eliminating the surrounding antigenic regions to minimize unnecessary or deleterious immune responses in vivo. However, as many of the epitopes are conformational in nature, carving them out from the entire envelope glycoprotein while maintaining and stabilizing their native structures remains a technical challenge. In this study, we demonstrated a successful isolation of the epitope-focused antigenic domain recognized by VRC01 (EAD-VRC01) through FACS sorting of the combinatorial antigen library displayed on the surface of yeast. Sequences and structural analysis showed that EAD-VRC01 had a significant structural overlap with the outer domain of gp120, covering the upstream of loop D all the way to V3, V4, and the downstream of V5 in sequence (33, 40). Biochemical characterizations of the three representatives EAD-VRC01 (V61, V51, and V17) revealed their strong binding to VRC01 class as well as to V3 glycan mAbs and therefore contained both CD4bs and V3 glycan-vulnerable sites. V61, V51, and V17 were also able to attenuate the neutralizing activity of VRC01 Fab in a pseudovirus-based assay and to trigger B cell activation through the formation of microclusters of the membrane-bound VRC01-BCRs and downstream signalings, the critical steps for B cells to differentiate into antibody-producing plasma cells. In all these settings, the trimeric forms of V61, V51, and V17 were invariably more potent than the monomeric ones. In particular, the sequential or mixed immunization of guinea pigs with the trimers was able to induce appreciable levels of neutralizing antibodies against genetically related autologous and heterologous subtype B CNE strains from China, although no detectable neutralizing activity was found against distantly related subtype B or other subtypes or CRFs. Systematic profiling of the immune sera by competitive ELISA, autologous peptide library, and neutralization depletion identified appreciable levels of CD4bs- and V3-directed neutralizing antibodies that can be absorbed by RSC3 and the V3 peptide, respectively. Taken together, these results demonstrated that the rational design of epitope-focused vaccine is not only theoretically viable but also technically feasible. The EADs identified here represent a promising collection of possible targets in the rational design of HIV-1 vaccines and lay the foundation for harnessing the specific antigenicity of CD4bs for protective immunogenicity in vivo.
Designing immunogens targeting the various sites of vulnerability and dissecting the antibody response in animal models will provide a critical guidance for the selection and optimization of vaccine candidates (20, 32–34). Studies on EAD-VRC01 here represent one of the first examples that a portion instead of a complete envelope immunogen can induce appreciable levels of neutralizing antibody in guinea pigs. We speculate that such immunogenicity could result from the structural and biochemical properties of EAD-VRC01 where the CD4bs might have become more stabilized and exposed through the trimeric formation thereby to improve the accessibility and affinity for VRC01 class and the related antibodies. In other words, our selection criteria for EAD-VRC01, such as the structural mimicry and stability, the antigenic similarity and multiplicity, as well as the ability to trigger B cell activation in vitro, have served as a meaningful gatekeeper for the immunogen selection and animal immunization. In fact, these criteria are somewhat similar to those for evaluating the eOD-GT8 immunogen except it is precisely engineered to trigger the germ line version of VRC01 in the transgenic animals (14). It is certain that future optimization would be required for EAD-VRC01 to obtain additional desired features such as improved binding to the intermediate or germ line ancestors of the mature VRC01 to trigger and enhance the development process of VRC01-like antibodies in vivo. EAD-VRC01 without the V3 region would also be a viable alternative to see whether deletion of the V3 region could further drive the immune response toward the CD4bs. In addition, detailed analyses of the neutralizing antibody responses in the immunized guinea pigs, including isolating monoclonal antibodies and next-generation sequencing, will certainly provide a more concrete reference for antibody specificity and the subsequent rounds of optimization. Furthermore, as the different animal models contain the distinct antibody repertoire, it is imperative to test the immunogenicity of EAD-VRC01 in those closest to humans to provide the most relevant and precise reference for downstream human use.
It needs to be stressed that our combinatorial antigen library approach offers several advantages pertaining to the epitope-focused vaccine strategies. The sheer size and diversity of the library undoubtedly improve the likelihood of success in identifying those antigenic domains containing the vulnerable sites that may otherwise be impossible by manual design on a one by one basis. More importantly, the vulnerable sites in the antigenic domains maintain their native conformation and closely resemble those in the full-length envelope expressed in mammalian cells with the post-translational modifications such as glycosylation and disulfide isomerization (49). This is largely due to the fact that yeast is a eukaryotic species, and the selection of antigenic domains is conducted in solution by FACS, which likely reflects the intrinsic interaction between antigen and antibody within the host. Our ability to identify EAD-VRC01 and EAD-PG09 that contain the complex conformational epitopes as well as the sugar moiety is a clear testimony for these advantages. Furthermore, random mutagenesis could also be applied to the selected EAD to generate the novel library for further selection of EADs with the additional desired attributes such as improved binding to the mature mAb or to its intermediate and germ line ancestors. As all of these procedures are conducted through the yeast-surface display system, and there is no need to produce each individual EAD in either prokaryotic or eukaryotic system and hence to greatly improve the chance and efficiency in identifying EADs with the desired properties. The versatility with high throughput capacity has therefore made the combinatorial library approach potentially the method of choice and serve as the critical starting point toward the successful epitope-focused vaccine strategies.
In summary, our results demonstrated that the rational design of epitope-focused vaccine is not only theoretically viable but also technically feasible. The EADs identified here represent a promising collection of possible targets in the rational design of HIV-1 vaccines and lay the foundation for harnessing the specific antigenicity of CD4bs for protective immunogenicity in vivo. The yeast surface display system will offer a powerful tool for the epitope-focused vaccine strategies targeting other major vulnerable sites on the envelope glycoprotein of HIV-1.
Experimental procedures
Ethics statement
The guinea pig experimental protocol for immunization and blood collection was approved by the Institutional Animal Care and Use Committee (IACUC) at Tsinghua University. The actual experiments were designed and performed in strict accordance with the government rules and regulations for animal welfare and the principles of the 3Rs (Replacement, Refinement, and Reduction).
Construction of the HIV-1 gp160 combinatorial antigen library
The combinatorial antigen fragment library of HIV-1 gp160 was constructed on the surface of yeast Saccharomyces cerevisiae as described previously (35, 36). In brief, the full-length envelope gene of CNE11 derived from a subtype B HIV-1 chronically infected patient in China (38) was amplified by PCR, purified by electrophoresis (QIAquick DNA purification kit, Qiagen), and then digested into about 50-bp fragments by DNase I. Next, the digested fragments were reassembled into larger fragments about 100–800 bp in length through controlled rounds of PCR amplification, which were then A-tailed (DNA A-tailing kit, Takara) and ligated to the modified pCTCON2-T vector. The ligation products were transformed into the Escherichia coli competent cells, amplified, extracted, and then further transformed into the competent S. cerevisiae EBY100 by electroporation. Transformed yeast cells were partially spread on SDCAA Amp plates and incubated overnight at 30 °C to estimate the number and sequences of recombinant colonies for quality control purpose. Conditions and procedures for yeast growing and induction of the surface antigen expression in solution have been previously described (50).
Identification of the epitope-focused antigenic domain by FACS and sequence analysis
Immunofluorescence staining and sorting of the yeast clones expressing the appropriate antigenic domains were performed as described previously (35, 36). The EAD was defined as the antigenic regions closest to the mAb epitope previously identified by crystallography while excluding the surrounding irrelevant regions. Specifically, the induced yeast cells (106–107) of the combinatorial antigen fragment library were stained with VRC01 or other HIV-1 mAbs (10 μg/ml), and the positive clones were sorted by FACS using Aria II (BD Biosciences). The sorted yeast clone mixture was spread onto the SDCAA Amp plates before single colonies were picked, grown, and further confirmed for binding with mAbs. To ensure accurate determination and representation of the EAD, multiple yeast clones were analyzed in parallel for their reactivity with the selected HIV-1 mAb. Those with detectable and desired reactivity to the mAb were further grown in SDCAA before plasmids were extracted (yeast plasmid kit, Omega Bio-tek) for sequencing and sequence analysis (Sequencher 5.0, Gene Codes Corp.). The EAD selected by VRC01 (EAD-VRC01) was also modeled onto the HIV-1 gp120 monomer to determine its spatial location using the program PyMOL (DeLano Scientific).
Production of recombinant EAD-VRC01, Fab, and their complex
The EAD-VRC01 was further expressed by Bac-to-Bac Baculovirus Expression System (Invitrogen) in the monomeric and trimeric forms, according to the manufacturer's instructions. All EAD-VRC01 sequences were fused with a gp67 signal peptide at the N terminus and a His6 tag at the C terminus. For expression of the trimeric EAD-VRC01, a foldon domain (GYIPEAPRDGQAYVRKDGEWVLLSTFL) of bacteriophage T4 fibritin was added upstream of the His6 tag at the C terminus. Recombinant EAD-VRC01 expressed in the supernatants of Sf9 cells were concentrated and purified by the nickel-nitrilotriacetic acid metal-affinity chromatography (Qiagen) and size-exclusion chromatography using a Superdex 200 column (GE Healthcare).
Antigen-binding fragments (Fab) of mAbs were produced by the protease digestion of full-length antibodies using human rhinovirus 3C or Lys-C (Sigma) protease. The complex of the trimeric EAD-VRC01 and VRC01 Fab was generated by the on-column cleavage strategy. Additional details regarding the generation of Fab and the complex can be found in the supporting Methods.
Surface plasmon resonance
The binding kinetics of the EAD-VRC01 with various mAbs were determined by SPR on a Biacore T200 (GE Healthcare). All experiments were performed at 25 °C with HBS-EP buffer (10 mm Hepes, 150 mm NaCl, 3 mm EDTA, and 0.005% Surfactant P20, pH 7.4). The monomeric or trimeric EAD-VRC01 was immobilized onto CM5 sensor chips at 200–300 response units by an amine coupling kit (GE Healthcare). The Fab of HIV-1 mAbs or a control mAb 3C11 was injected through flow cells at the concentration ranging from 1 to 500 nm followed by a series of 2-fold increase in concentration. The flow rate was 30 μl/min for 3 min for association and for 5–30 min for dissociation. Regenerations were conducted with 10 mm glycine HCl, pH 2.0, at a flow rate of 50 μl/min for 1 min. Biacore T200 Evaluation Software (GE Healthcare) was performed to subtract blank references. The sensorgrams were fitted globally in a 1:1 Langmuir model, and several KD values (denoted by asterisk symbols in Fig. 2A) were determined by the steady-state affinity model as the interactions were too fast to provide kinetic information.
Neutralization and neutralization depletion experiments
Neutralizing activities of guinea pig immune sera and mAbs were evaluated using a single round infection of TZM-bl target cells by pseudoviruses as previously described (51). The pseudoviruses were selected in such a way that included the autologous and heterologous CNE viruses initially isolated from the Chinese patients (38) and the subtype B and C and the global panel from the AIDS Reagent Program, National Institutes of Health (44, 45). It needs to be noted that all of the CNE viruses were properly tier-phenotyped by Dr. David Montefiori and co-workers at Duke University Medical Center using the same protocol for widely accepted standard strains, including those in the global panel (44). Moloney murine leukemia virus envelope was used as a negative control. Neutralization depletion assay was conducted for VRC01 Fab in the presence of the EAD-VRC01 to verify whether the EAD-VRC01 was able to bind and attenuate the neutralizing activity. Neutralizing depletion assay was also performed for the immune sera of guinea pigs in the presence of RSC3, RSC3Δ371I, and autologous V3 peptide to study whether neutralizing activity could be competed out by these specific domains of gp120. Specifically, the competitor proteins or peptides (30 μg/ml) were incubated with a serial dilution of mAbs or sera at 37 °C for 1 h before adding to 100 TCID50 of pseudoviruses. V3 peptide (KSIHLGQGRAWYTTG) derived from CNE11 was designed according to the format (catalog no. 1830) listed in the AIDS Reagent Program, National Institutes of Health, and synthesized by ChinaPeptides (Shanghai, China). The depletion effect of the competitor proteins or peptides was assessed by the changes in half-maximal inhibitory concentrations (IC50) or half-maximal inhibitory dilutions (ID50) compared with the mock controls.
Electron microscopy and image reconstruction
Three microliters of the purified V51tri (0.01 mg/ml) or V51tri–VRC01 Fab complex (0.01 mg/ml) were applied to a pre-glow discharged carbon coated EM grid and then stained with 1.5% uranyl acetate. Micrographs of the V51tri (101 micrographs) and the V51tri–VRC01 Fab complex (115 micrographs) were collected on a 120-kV FEI Tecnai Spirit microscope using a charge-coupled device camera at a nominal magnification of 49,000 (the calibrated pixel size is 2.27 Å). The structure model of the V51tri was generated using the crystal structures of the BG505 SOSIP.664 gp140 trimer (PDB code 4NCO). Because the V3 loop was truncated in the gp120-VRC01 Fab crystal structure (PDB code 3NGB), the V51–VRC01 Fab complex model was generated as a chimera with the V51 fragment from the crystal structure of the BG505 SOSIP.664 gp140 trimer (PDB code 4NCO) and the VRC01 Fab from the crystal structure of the gp120–VRC01 Fab complex (PDB code 3NGB). Model Fitting was performed by manual adjustment and the “fit in map” function in the UCSF Chimera software (52). Map comparison was accomplished with EMAN2 e2pdb2mrc.py package (53) and the fit in map function in Chimera. Additional details of data processing and model fitting can be found in the supporting Methods.
Triggering VRC01-BCR signaling by EAD-VRC01
The Ramos B cell line was first constructed to express VRC01-BCR on the cell membrane by lentiviral infection, according to the supporting Methods. The EAD-VRC01 triggering VRC01-BCR signaling was recorded by an Olympus IX-81 TIRFM, following our well-established protocols (41, 42, 54, 55). Additional details related to the entire procedure for molecule staining and TIRFM imaging could be found in the supporting Methods.
Calcium mobilization analysis
Ca2+ mobilization in Ramos-VRC01 B cells was measured using intracellular calcium indicator Gcamp5 after EAD-VRC01 stimulation. Briefly, Ramos-VRC01-Gcamp5 cells were washed with Hanks' balanced salt solution (HBSS) once and then incubated at 37 °C for 3 min in HBSS buffer (with 1.26 mm Ca2+). After the basal level of Ca2+ concentration was monitored for 30 s, cells were directly stimulated with 100 nm EAD-VRC01 monomer, trimer, or control antigens. The fluorescence intensity of Gcamp5 was measured on BD Accuri C6 and analyzed in FlowJo software (Tree Star). The Gcamp5 mFI was normalized against the average basal level.
Animal immunization and sample collection
Approximately 8–10-week-old Dunkin Hartley female guinea pigs (Beijing Vital River Laboratory) were immunized subcutaneously with the trimeric EAD-VRC01 either sequentially or together as illustrated in Fig. S5. Specifically, the sequential immunization group (n = 6) was immunized sequentially with 300 μg (in 300 μl of PBS) each of EAD-VRC01 V17tri, V51tri, and then V61tri. The mix immunization group (n = 6) was immunized with a 300-μg (in 300 μl PBS) mixture of the three EAD trimers and each contributed 100 μg. The first immunization was conducted at week 0 with Freund's complete adjuvant (300 μl), and the following boosts were at weeks 4, 12, and 20 with Freund's incomplete adjuvant (300 μl) (Sigma). The negative control group (n = 3) was injected with PBS and then adjuvant. Blood samples were collected from the anesthetized animals at week −2 for prebleed and at weeks 14 and 22 after the first immunization, and no sign of distress was noticed during the experiment.
ELISA, competitive ELISA, and sera profiling
ELISA was used to define the binding activities of mAbs or sera against various antigens as described previously (35, 56). The end-point titers of the sera were determined at the dilution where A450 was 3-fold higher than the negative control animal sera. To study the antibody specificity of immunized animals, competitive ELISA was conducted against various biotinylated mAbs with known epitopes, following the previous protocol (46). The sera profiling of the immunized guinea pigs was performed by ELISA against the autologous 15-mer peptides library, overlapping by 11 residues, derived from the CNE11 envelope sequence (clade B). These peptides (5 μg/ml) were coated onto 96-well plates followed by incubation with 1:1000 dilution of the sera as described previously (57). Additional details for the profiling experiment could be found in the supporting Methods.
Sequence and statistical analysis
The sequences corresponding to EAD-VRC01 were downloaded from the GenBankTM and aligned using the ClustalW program. Phylogenetic analysis was performed by the neighbor-joining method, and the reliability of the branching orders was detected by bootstrap analysis of 1000 replicates. In the pseudovirus-based neutralization and neutralization depletion experiments, IC50 and ID50 values were calculated by the dose-response inhibition model in GraphPad Prism 5.0 (GraphPad Software). In the BCR clustering and downstream signaling molecule imaging analysis, the contact area and total fluorescent intensity among the study groups were analyzed using a two-tailed unpaired t test (*** means p < 0.001). In the ELISA and competitive ELISA, data were also analyzed in GraphPad Prism 5.0.
Author contributions
H. W., Y. X., W. L., and L. Z. designed the research. H. W., X. C., D. W., C. Y., Q. W., J. X., and X. S. performed the experimental work. Specifically, H. W., C. Y., Q. W., J. X., and X. S. performed the EAD selection and expression, SPR, neutralization assay, ELISA, and the animal experiments. X. C. performed the B cell trigger assay. D. W. conducted the EM analysis. H. W., X. C., D. W., Y. X., W. L., and L. Z. analyzed the data and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. K. Dane Wittrup and Annie Gai at Massachusetts Institute of Technology for providing the yeast surface display vector pCTCON2; Drs. John Mascola at Vaccine Research Center, National Institutes of Health, and Michel Nussenzweig at Rockefeller University for providing HIV-1 bnAbs; Drs. Peter Kwong and Tongqing Zhou at National Institutes of Health for providing RSC3 and RSC3Δ371I proteins; Drs. David Montefiori and Hongmei Gao at Duke University Medical Center for tier phenotying of the CNE pseudovirues; Dr. Xinquan Wang and student Dr. Xi Liu at Tsinghua University for providing the Sf9 cell line, pFastBacTM dual vector, and technical suggestions; Dr. Susan K. Pierce at National Institutes of Health for providing the Human Ramos B cell line; Dr. Chenqi Xu at Chinese Academy of Science for providing the Gcamp5 gene; Dr. Yi Shi at Chinese Academy of Science for providing the NL4-3 SOSIP.664 gp140 trimer protein, and Kelly Arledge previously of our laboratory for providing valuable suggestions and input. National Institutes of Health Contract HHSN27201100016C from NIAID was awarded to Duke University Medical Center.
This work was supported by National Natural Science Foundation Award 81530065, Grand Challenge China Grant 81661128042, National Science and Technology Major Projects 012ZX10001-006, -004, and -009, Ministry of Science and Technology of China Grant 2014CB542500-03, and Grand Challenges Explorations of The Bill and Melinda Gates Foundation Award OPP1021992-004. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7, supporting Methods, and supporting Refs. 1–3.
- bnAbs
- broadly neutralizing antibody
- Env
- envelope glycoprotein
- SPR
- surface plasmon resonance
- EAD
- epitope-focused antigenic domain
- CD4bs
- CD4-binding site
- CRF
- circulating recombinant form
- MPER
- membrane proximal external region
- aa
- amino acid
- PDB
- Protein Data Bank
- TIRFM
- total internal reflection fluorescence microscopy
- HBSS
- Hanks' balanced salt solution
- BCR
- B cell receptor
- CNE
- China Envelope.
References
- 1. Mascola J. R., and Montefiori D. C. (2010) The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28, 413–444 10.1146/annurev-immunol-030409-101256 [DOI] [PubMed] [Google Scholar]
- 2. Fauci A. S., and Marston H. D. (2014) Ending AIDS–is an HIV vaccine necessary? N. Engl. J. Med. 370, 495–498 10.1056/NEJMp1313771 [DOI] [PubMed] [Google Scholar]
- 3. Haynes B. F., and Bradley T. (2015) Broadly neutralizing antibodies and the development of vaccines. JAMA 313, 2419–2420 10.1001/jama.2015.2427 [DOI] [PubMed] [Google Scholar]
- 4. Burton D. R., and Mascola J. R. (2015) Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576 10.1038/ni.3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burton D. R., and Hangartner L. (2016) Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 34, 635–659 10.1146/annurev-immunol-041015-055515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. West A. P. Jr., Scharf L., Scheid J. F., Klein F., Bjorkman P. J., and Nussenzweig M. C. (2014) Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156, 633–648 10.1016/j.cell.2014.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwong P. D., and Mascola J. R. (2012) Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425 10.1016/j.immuni.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Letvin N. L., and Walker B. D. (2003) Immunopathogenesis and immunotherapy in AIDS virus infections. Nat. Med. 9, 861–866 10.1038/nm0703-861 [DOI] [PubMed] [Google Scholar]
- 9. Overbaugh J., and Bangham C. R. (2001) Selection forces and constraints on retroviral sequence variation. Science 292, 1106–1109 10.1126/science.1059128 [DOI] [PubMed] [Google Scholar]
- 10. Ross A. L., Bråve A., Scarlatti G., Manrique A., and Buonaguro L. (2010) Progress towards development of an HIV vaccine: Report of the AIDS Vaccine 2009 Conference. Lancet Infect. Dis. 10, 305–316 10.1016/S1473-3099(10)70069-4 [DOI] [PubMed] [Google Scholar]
- 11. Wei X., Decker J. M., Wang S., Hui H., Kappes J. C., Wu X., Salazar-Gonzalez J. F., Salazar M. G., Kilby J. M., Saag M. S., Komarova N. L., Nowak M. A., Hahn B. H., Kwong P. D., and Shaw G. M. (2003) Antibody neutralization and escape by HIV-1. Nature 422, 307–312 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- 12. Kwong P. D., Doyle M. L., Casper D. J., Cicala C., Leavitt S. A., Majeed S., Steenbeke T. D., Venturi M., Chaiken I., Fung M., Katinger H., Parren P. W., Robinson J., Van Ryk D., Wang L., et al. (2002) HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 10.1038/nature01188 [DOI] [PubMed] [Google Scholar]
- 13. Burton D. R., Ahmed R., Barouch D. H., Butera S. T., Crotty S., Godzik A., Kaufmann D. E., McElrath M. J., Nussenzweig M. C., Pulendran B., Scanlan C. N., Schief W. R., Silvestri G., Streeck H., Walker B. D., et al. (2012) A blueprint for HIV vaccine discovery. Cell Host Microbe 12, 396–407 10.1016/j.chom.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jardine J. G., Kulp D. W., Havenar-Daughton C., Sarkar A., Briney B., Sok D., Sesterhenn F., Ereño-Orbea J., Kalyuzhniy O., Deresa I., Hu X., Spencer S., Jones M., Georgeson E., Adachi Y., et al. (2016) HIV-1 broadly neutralizing antibody precursor B cells revealed by germ line-targeting immunogen. Science 351, 1458–1463 10.1126/science.aad9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jardine J., Julien J. P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P. S., MacPherson S., Jones M., Nieusma T., Mathison J., Baker D., Ward A. B., Burton D. R., et al. (2013) Rational HIV immunogen design to target specific germ line B cell receptors. Science 340, 711–716 10.1126/science.1234150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanders R. W., Derking R., Cupo A., Julien J. P., Yasmeen A., de Val N., Kim H. J., Blattner C., de la Peña A. T., Korzun J., Golabek M., de Los Reyes K., Ketas T. J., van Gils M. J., King C. R., et al. (2013) A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 10.1371/journal.ppat.1003618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Georgiev I. S., Joyce M. G., Yang Y., Sastry M., Zhang B., Baxa U., Chen R. E., Druz A., Lees C. R., Narpala S., Schön A., Van Galen J., Chuang G. Y., Gorman J., Harned A., et al. (2015) Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J. Virol. 89, 5318–5329 10.1128/JVI.03451-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma S. K., de Val N., Bale S., Guenaga J., Tran K., Feng Y., Dubrovskaya V., Ward A. B., and Wyatt R. T. (2015) Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 11, 539–550 10.1016/j.celrep.2015.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pritchard L. K., Vasiljevic S., Ozorowski G., Seabright G. E., Cupo A., Ringe R., Kim H. J., Sanders R. W., Doores K. J., Burton D. R., Wilson I. A., Ward A. B., Moore J. P., and Crispin M. (2015) Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep. 11, 1604–1613 10.1016/j.celrep.2015.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanders R. W., van Gils M. J., Derking R., Sok D., Ketas T. J., Burger J. A., Ozorowski G., Cupo A., Simonich C., Goo L., Arendt H., Kim H. J., Lee J. H., Pugach P., Williams M., et al. (2015) HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223 10.1126/science.aac4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Taeye S. W., Ozorowski G., Torrents de la Peña A., Guttman M., Julien J. P., van den Kerkhof T. L., Burger J. A., Pritchard L. K., Pugach P., Yasmeen A., Crampton J., Hu J., Bontjer I., Torres J. L., Arendt H., et al. (2015) Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163, 1702–1715 10.1016/j.cell.2015.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jardine J. G., Ota T., Sok D., Pauthner M., Kulp D. W., Kalyuzhniy O., Skog P. D., Thinnes T. C., Bhullar D., Briney B., Menis S., Jones M., Kubitz M., Spencer S., Adachi Y., et al. (2015) HIV-1 vaccines. Priming a broadly neutralizing antibody response to HIV-1 using a germ line-targeting immunogen. Science 349, 156–161 10.1126/science.aac5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Escolano A., Steichen J. M., Dosenovic P., Kulp D. W., Golijanin J., Sok D., Freund N. T., Gitlin A. D., Oliveira T., Araki T., Lowe S., Chen S. T., Heinemann J., Yao K. H., Georgeson E., et al. (2016) Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell 166, 1445–1458 10.1016/j.cell.2016.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Briney B., Sok D., Jardine J. G., Kulp D. W., Skog P., Menis S., Jacak R., Kalyuzhniy O., de Val N., Sesterhenn F., Le K. M., Ramos A., Jones M., Saye-Francisco K. L., Blane T. R., et al. (2016) Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 166, 1459–1470 10.1016/j.cell.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian M., Cheng C., Chen X., Duan H., Cheng H. L., Dao M., Sheng Z., Kimble M., Wang L., Lin S., Schmidt S. D., Du Z., Joyce M. G., Chen Y., DeKosky B. J., et al. (2016) Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell 166, 1471–1484 10.1016/j.cell.2016.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sok D., Briney B., Jardine J. G., Kulp D. W., Menis S., Pauthner M., Wood A., Lee E. C., Le K. M., Jones M., Ramos A., Kalyuzhniy O., Adachi Y., Kubitz M., MacPherson S., et al. (2016) Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560 10.1126/science.aah3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steichen J. M., Kulp D. W., Tokatlian T., Escolano A., Dosenovic P., Stanfield R. L., McCoy L. E., Ozorowski G., Hu X., Kalyuzhniy O., Briney B., Schiffner T., Garces F., Freund N. T., Gitlin A. D., et al. (2016) HIV vaccine design to target germ line precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496 10.1016/j.immuni.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dosenovic P., von Boehmer L., Escolano A., Jardine J., Freund N. T., Gitlin A. D., McGuire A. T., Kulp D. W., Oliveira T., Scharf L., Pietzsch J., Gray M. D., Cupo A., van Gils M. J., Yao K. H., et al. (2015) Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515 10.1016/j.cell.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy L. E., van Gils M. J., Ozorowski G., Messmer T., Briney B., Voss J. E., Kulp D. W., Macauley M. S., Sok D., Pauthner M., Menis S., Cottrell C. A., Torres J. L., Hsueh J., Schief W. R., et al. (2016) Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 16, 2327–2338 10.1016/j.celrep.2016.07.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng Y., Tran K., Bale S., Kumar S., Guenaga J., Wilson R., de Val N., Arendt H., DeStefano J., Ward A. B., and Wyatt R. T. (2016) Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous Tier 2 neutralizing antibodies. PLoS Pathog. 12, e1005767 10.1371/journal.ppat.1005767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burton D. R. (2002) Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2, 706–713 10.1038/nri891 [DOI] [PubMed] [Google Scholar]
- 32. Correia B. E., Bates J. T., Loomis R. J., Baneyx G., Carrico C., Jardine J. G., Rupert P., Correnti C., Kalyuzhniy O., Vittal V., Connell M. J., Stevens E., Schroeter A., Chen M., Macpherson S., et al. (2014) Proof of principle for epitope-focused vaccine design. Nature 507, 201–206 10.1038/nature12966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joyce M. G., Kanekiyo M., Xu L., Biertümpfel C., Boyington J. C., Moquin S., Shi W., Wu X., Yang Y., Yang Z. Y., Zhang B., Zheng A., Zhou T., Zhu J., Mascola J. R., et al. (2013) Outer domain of HIV-1 gp120: antigenic optimization, structural malleability, and crystal structure with antibody VRC-PG04. J. Virol. 87, 2294–2306 10.1128/JVI.02717-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang X., Totrov M., Li W., Sampson J. M., Williams C., Lu H., Wu X., Lu S., Wang S., Zolla-Pazner S., and Kong X. P. (2016) Rationally designed immunogens targeting HIV-1 gp120 V1V2 induce distinct conformation-specific antibody responses in rabbits. J. Virol. 90, 11007–11019 10.1128/JVI.01409-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin S., Ji Y., Wang Q., Wang H., Shi X., Han X., Zhou T., Shang H., and Zhang L. (2016) Spatiotemporal hierarchy in antibody recognition against transmitted HIV-1 envelope glycoprotein during natural infection. Retrovirology 13, 12 10.1186/s12977-016-0243-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zuo T., Shi X., Liu Z., Guo L., Zhao Q., Guan T., Pan X., Jia N., Cao W., Zhou B., Goldin M., and Zhang L. (2011) Comprehensive analysis of pathogen-specific antibody response in vivo based on an antigen library displayed on surface of yeast. J. Biol. Chem. 286, 33511–33519 10.1074/jbc.M111.270553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zuo T., Sun J., Wang G., Jiang L., Zuo Y., Li D., Shi X., Liu X., Fan S., Ren H., Hu H., Sun L., Zhou B., Liang M., Zhou P., Wang X., and Zhang L. (2015) Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection. Nat. Commun. 6, 8855 10.1038/ncomms9855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shang H., Han X., Shi X., Zuo T., Goldin M., Chen D., Han B., Sun W., Wu H., Wang X., and Zhang L. (2011) Genetic and neutralization sensitivity of diverse HIV-1 env clones from chronically infected patients in China. J. Biol. Chem. 286, 14531–14541 10.1074/jbc.M111.224527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Julien J. P., Cupo A., Sok D., Stanfield R. L., Lyumkis D., Deller M. C., Klasse P. J., Burton D. R., Sanders R. W., Moore J. P., Ward A. B., and Wilson I. A. (2013) Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 10.1126/science.1245625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou T., Georgiev I., Wu X., Yang Z. Y., Dai K., Finzi A., Kwon Y. D., Scheid J. F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M. C., Sodroski J., Shapiro L., et al. (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 10.1126/science.1192819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu W., Meckel T., Tolar P., Sohn H. W., and Pierce S. K. (2010) Intrinsic properties of immunoglobulin IgG1 isotype-switched B cell receptors promote microclustering and the initiation of signaling. Immunity 32, 778–789 10.1016/j.immuni.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu W., Meckel T., Tolar P., Sohn H. W., and Pierce S. K. (2010) Antigen affinity discrimination is an intrinsic function of the B cell receptor. J. Exp. Med. 207, 1095–1111 10.1084/jem.20092123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen X., Pan W., Sui Y., Li H., Shi X., Guo X., Qi H., Xu C., and Liu W. (2015) Acidic phospholipids govern the enhanced activation of IgG-B cell receptor. Nat. Commun. 6, 8552 10.1038/ncomms9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seaman M. S., Janes H., Hawkins N., Grandpre L. E., Devoy C., Giri A., Coffey R. T., Harris L., Wood B., Daniels M. G., Bhattacharya T., Lapedes A., Polonis V. R., McCutchan F. E., Gilbert P. B., et al. (2010) Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84, 1439–1452 10.1128/JVI.02108-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Georgiev I. S., Doria-Rose N. A., Zhou T., Kwon Y. D., Staupe R. P., Moquin S., Chuang G. Y., Louder M. K., Schmidt S. D., Altae-Tran H. R., Bailer R. T., McKee K., Nason M., O'Dell S., Ofek G., et al. (2013) Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756 10.1126/science.1233989 [DOI] [PubMed] [Google Scholar]
- 46. Crooks E. T., Tong T., Chakrabarti B., Narayan K., Georgiev I. S., Menis S., Huang X., Kulp D., Osawa K., Muranaka J., Stewart-Jones G., Destefano J., O'Dell S., LaBranche C., Robinson J. E., et al. (2015) Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4-binding site. PLoS Pathog. 11, e1004932 10.1371/journal.ppat.1004932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu X., Yang Z. Y., Li Y., Hogerkorp C. M., Schief W. R., Seaman M. S., Zhou T., Schmidt S. D., Wu L., Xu L., Longo N. S., McKee K., O'Dell S., Louder M. K., Wycuff D. L., et al. (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou T., Lynch R. M., Chen L., Acharya P., Wu X., Doria-Rose N. A., Joyce M. G., Lingwood D., Soto C., Bailer R. T., Ernandes M. J., Kong R., Longo N. S., Louder M. K., McKee K., et al. (2015) Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161, 1280–1292 10.1016/j.cell.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu C., and Ng D. T. (2015) Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 16, 742–752 10.1038/nrm4073 [DOI] [PubMed] [Google Scholar]
- 50. Chao G., Lau W. L., Hackel B. J., Sazinsky S. L., Lippow S. M., and Wittrup K. D. (2006) Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768 10.1038/nprot.2006.94 [DOI] [PubMed] [Google Scholar]
- 51. Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., et al. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 10.1128/JVI.79.16.10108-10125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 53. Ludtke S. J. (2010) 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol. Biol. 673, 157–173 10.1007/978-1-60761-842-3_9 [DOI] [PubMed] [Google Scholar]
- 54. Liu W., Won Sohn H., Tolar P., Meckel T., and Pierce S. K. (2010) Antigen-induced oligomerization of the B cell receptor is an early target of Fc gamma RIIB inhibition. J. Immunol. 184, 1977–1989 10.4049/jimmunol.0902334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu W., Chen E., Zhao X. W., Wan Z. P., Gao Y. R., Davey A., Huang E., Zhang L., Crocetti J., Sandoval G., Joyce M. G., Miceli C., Lukszo J., Aravind L., Swat W., et al. (2012) The scaffolding protein synapse-associated protein 97 is required for enhanced signaling through isotype-switched IgG memory B cell receptors. Sci. Signal. 5, ra54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Q., Gui M., Niu X., He S., Wang R., Feng Y., Kroeker A., Zuo Y., Wang H., Wang Y., Li J., Li C., Shi Y., Shi X., Gao G. F., Xiang Y., Qiu X., Chen L., and Zhang L. (2016) Potent neutralizing monoclonal antibodies against Ebola virus infection. Sci. Rep. 6, 25856 10.1038/srep25856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang Z., Li J., Liu Q., Yuan T., Zhang Y., Chen L. Q., Lou Q., Sun Z., Ying H., Xu J., Dimitrov D. S., and Zhang M. Y. (2015) Identification of non-HIV immunogens that bind to germ line b12 predecessors and prime for elicitation of cross-clade neutralizing HIV-1 antibodies. PLoS ONE 10, e0126428 10.1371/journal.pone.0126428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.