Abstract
The epidermal growth factor receptor (EGFR) is often amplified in glioma, with the most common extracellular domain mutation being EGFR variant III (EGFRvIII). Abnormal EGFRvIII signaling has been shown to be important in driving tumor progression. Centrosomal protein 55 (CEP55), a member of the centrosomal relative proteins family, participates cytokinesis in the cell cycle. It exists in a few normal tissues and various tumor cells. The expression and function of CEP55 in human glioma cells need to investigate. In this study, the expression of CEP55 was detected in 40 cases of glioma tissues and 10 cases of non-tumor brain tissue. The proliferation of glioblastoma U251 cells was analyzed after transfection with EGFRvIII and CEP55 siRNA. We found that the expression of CEP55 was increased significantly in the glioma tissues than in normal brain tissue. The proliferation of U251 cells increased remarkably after transfection with EGFRvIII. Knockdown of CEP55 inhibited proliferation of U251 cells and was able to eliminate the effect of promoting proliferation induced by EGFRvIII in U251 cells. CEP55 played a key role in the proliferation of glioma cells and mediated EGFRvIII-stimulated proliferation in glioma cells. CEP55 might be a novel molecular therapeutic target in patients with gliomas expressing EGFRvIII.
Keywords: centrosomal protein 55, epithelial growth factor receptor vIII, glioma, proliferation
Introduction
Gliomas are the most frequent type of malignant tumor in central nervous system, leading to significant mortality worldwide annually (1,2). It was classified into low-grade gliomas (grade II, e.g., infiltrative astrocytomas and oligodendrogliomas) and high-grade gliomas (grade III and IV, e.g., anaplastic gliomas and glioblastomas) according to the World Health Organization grading system (3). The current therapeutic strategy to glioma consists of surgical resection, radiotherapy, chemotherapy and antiangiogenesis. However, the prognosis of the patients with glioma is still poor in spite of these interventions. Only one third of patients with glioblastoma (GBM) survival for a year and the 5-year survival rate for grade III gliomas is 27% (4). The epidermal growth factor receptor (EGFR) plays a key role in proliferation and pathogenesis of multiple cancer types. Expression of EGFR correlates with poor prognosis and radioresistance in glioma (5).
EGFR amplification and overexpression have been identified in ~50% of GBM patients. EGFR overexpression is often accompanied by the expression of a mutant EGFR named as EGFRvIII (EGFR type III, ΔEGFR), which is expressed in 30% of GBM tumors (6,7). EGFRvIII is generated from a genomic deletion involving exons 2–7 of the EGFR gene, which results in deletion of the extracellular ligand-binding domain. It is unable to bind ligand, yet constitutively activated in a ligand-independent manner leading to overproliferation, survival, resistance to treatment and invasiveness of GBM cells (8,9). Both of the classical subtypes classified by the TCGA working group and the EM subgroup classified by the CGGA working group showed that the characteristics of EGFR activation associated with amplification and mutation displayed the worst prognosis (10–12).
Centrosomal protein 55 (CEP55) is the latest found member in the centrosomal relative protein family, which participates in the regulation of the cell cycle. CEP55 is located in the centrosome in interphase cells and is recruited into the midbody during cytokinesis (13). In addition, CEP55 is required for midbody structure and the completion of cytokinesis at the terminal stage (14). Overexpressing of CEP55 were found in several human tumors and various tumor cell lines (15–17). However, the relationship between EGFRvIII and CEP55 in proliferation regulation of glioma cells and the effect of CEP55 on these malignant features of GBM is poorly understood. Therefore, we detected the expression of CEP55 in human glioma tissues and investigated the proliferation of glioblastoma U251 cells after transfected with EGFRvIII and CEP55 siRNA.
Materials and methods
Tissue samples and cells line
Glioma tissues of 40 patients undergoing resection of lesion were collected from the department of pathology in the Xinqiao hospital from 2010 to 2016. Tumor samples were diagnosed according to the WHO classification of central nervous system tumors. Forty patients with glioma were newly diagnosed and didn't received chemotherapy or radiotherapy. Twenty gliomas were classified as low-grade glioma (WHO I and II) and others were high-grade glioma (WHO III and IV). Ten non-tumor brain tissue were obtained from patients with epilepsy undergoing surgical treatment in our hospital. All patients provided written informed consent according to the research proposals approved by the ethical committee of the Xinqiao Hospital.
Human glioblastoma U251 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS), 50 IU/ml benzyl penicillin G potassium, and 100 µg/ml streptomycin sulfate in a humidified incubator at 37°C and 5% CO2. U251 glioma cells were purchased from Riken Cell Bank (Tsukuba, Japan).
TCGA data analysis
Gene expression data from TCGA was analyzed in Firebrowse (http://firebrowse.org/). Input ‘CEP55’ in View Expression Profile box; click ‘Filter on’ and choose ‘GBMLGG’; submit. The Kaplan Meier Plot was analyzed in web (https://xenabrowser.net/heatmap/#). Firstly, choose ‘Visualization’ at the top of the web. Then, choose ‘TCGA lower grade glioma and glioblastoma (GBMLGG)’ in ‘Cohort’; choose ‘gene expression RNAseq (polyA+ Illumina HiSeq)’ in ‘samples in’; click ‘+Date’, ‘gene expression RNAseq’, ‘gene expression RNAseq (polyA+ Illumina HiSeq)’ in turn; click ‘next’; input CEP55 in Genes box; click ‘Done’. Finally, click ‘Column menu (Inverted triangle symbol)’ and choose ‘Kaplan Meier plot’.
Western-blot analysis
For western blot analyses, protein was harvested from cells plated to 70–80% confluence. Cell lysates were prepared using a RIPA buffer (25 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate; pH 7.6). Protease inhibitors were added prior to use. The protein extracts were loaded, size-fractionated by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). After blocking, the membranes were incubated with the specific first antibodies (Abcam, Cambridge, UK) in dilution buffer at 4°C overnight. The blots were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000, anti-rabbit/anti-mouse; Boster Biological Technology Co., Ltd., Wuhan, China) at room temperature for 2 h and then chemiluminescence signals were detected using a UVI gel imaging system camera.
Quantitative real-time PCR
Total RNA was extracted from frozen tissues and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions and was reverse transcribed into cDNA using PrimeScript RT reagent (Takara Bio Inc., Shiga, Japan). The reaction system (20 µl) contained the corresponding cDNA. The CEP55 primers used in quantitative real-time PCR were as follows: forward primer of 5′-TTGGAACAACAGATGCAGGC-3′ and reverse primer of 5′-GAGTGCAGCAGTGGGACTTT-3′. GAPDH, used as an internal control, was amplified with forward primer 5′-TGGACTCCACGACGTACTCAG-3′ and reverse primer 5′-CGGGAAGCTTGTCATCAATGGAA-3′. All procedures were performed in triplicate.
RNAi and transfection
To further identify the role of CEP55 in tumor and the relationship of CEP55 and EGFRvIII, the CEP55 knockdown lentiviral vector (siCEP55) was constructed (Shanghai GeneChem Co., Ltd., Shanghai, China). A GFP lentiviral vector was used as negative control (NC). The day before transfection, cells were seeded in 24-well plates at a density of 50,000 cells/well. The lentiviruses were transfected according to the manufacturer's instruction with MOI=10, and stably transfection cells were selected by puromycin (6 µg/ml). All lentiviral vectors expressed GFP and Puromycin, which enabled us to select stably transfection cells. The siRNA sequences are as following: 5′-GTGGGAAAGGAAAGCTGAC-3′. The high expression of EGFRvIII lentiviral vector was also constructed (Shanghai GeneChem Co., Ltd.). The sequences resulted from NCBI. The following are the NCBI reference sequence: NM_001346941.1. A RFP lentiviral vector was used as negative control.
Proliferation assay of U251 cells
The Cell Counting Kit-8 (CCK-8) assay
U251 cells (3,000 cells/well) were seeded into 96-well plates in 100 µl complete medium. The CCK-8 (Dojindo Laboratories, Kumamoto, Japan) was used to measure cell viability according to the manufacturer's instructions. The plates were incubated for 7 days. The number of viable cells was assessed by measurement of the spectrophotometric absorbance at 450 nm with a microplate reader iMARK (Bio-Rad Laboratories, Inc.).
EdU assay
Cells that had undergone different interventions were seeded in 96-well plates at a density of 3×103 cells/well. EdU assay was done following manufacturer's instructions after culturing for 48 h Cell. The proliferation was assessed by calculating the percentage of EdU-positive cells in all cells.
Statistical analysis
Statistical analysis for TCGA was described above. SPSS software program (version 18.0; SPSS Inc., Chicago, IL, USA) and Graphpad Prism 5 software program (Graphpad Software, Inc., San Diego, CA, USA) were used for statistical analyses. All results were presented as mean ± standard deviation. The horizontal bars in histograms represented mean values. The one-way ANOVA was used to analyze the differences between groups for in vivo or in vitro studies. If the variance was homogeneous, Tukey's test was used for the post-hoc test; If not, Dunnett's test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of CEP55 increased in human glioma cells and related to survival of patients with glioma
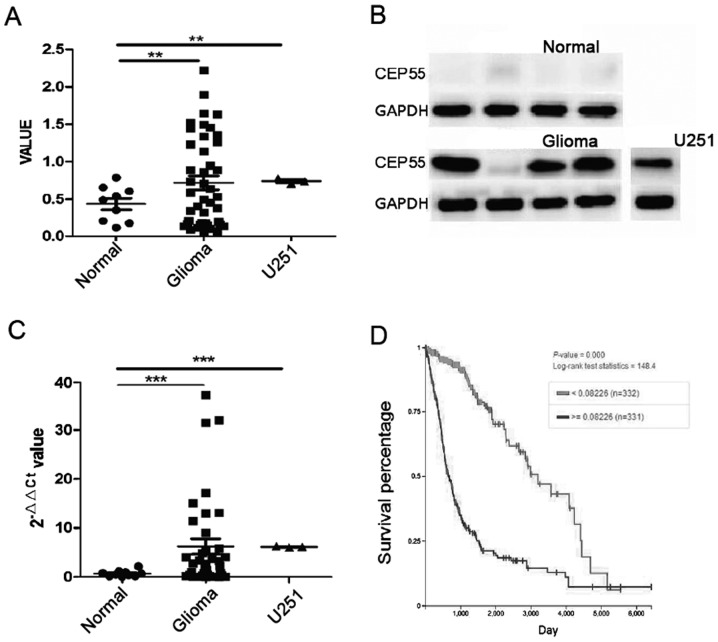
CEP55 played an important role in proliferation of cells and overexpressed in several human tumors. The mRNA and protein of CEP55 were detected by qPCR and western blot analysis in 40 cases of glioma tissues and 10 cases of normal brain tissues. The mRNA of CEP55 in glioma tissues increased significantly than it in normal brain tissues (P<0.01) (Fig. 1A). The result of western blot analysis showed that CEP55 proteins increased in 40 cases of glioma tissues accompany to improving of CEP55 mRNA. The level of CEP55 proteins in glioma was 7 times higher than it in normal brain tissues (P<0.001) (Fig. 1B and C). There was overexpression of CEP55 in GBM U251 cells too, compared with normal brain tissues (Fig. 1A-C). Our results of qPCR and western blot analysis confirmed that expression of CEP55 in glioma cells rocket up markedly than it in brain tissues.
Figure 1.
Expression of CEP55 in human glioma and GBM U251 cells. (A) mRNA of CEP55 in human glioma tissues and normal brain tissues and U251 cells. (B) Representative results of CEP55 protein expression measured by western blot analysis. (C) Statistical analysis of CEP55 protein expression by western blot analysis. (D) The patient survival database from TCGA. **P<0.05, ***P<0.001. CEP55, centrosomal protein 55; TCGA, The Cancer Genome Atlas; GBM, glioblastoma.
To discover the value of CEP55 overexpression in glioma cells, we investigated the relationship of CEP55 expression and survival time in 663 cases of patients with glioma in The Cancer Genome Atlas (TCGA) database. The patients with glioma were classified to overexpression of CEP55 group (n=332) and low expression of CEP55 group (n=331) according to the threshold value (>=0.00226 and <0.00226) of CEP55 mRNA. The median overall survival (OS) of patients were 21.5 months in overexpression group and 107.1 months in low expression group. The result showed that expression of CEP55 was significantly associated to survival of patient with glioma and overexpression of CEP55 mean shorter survival time to patient with glioma (P<0.001) (Fig. 1D).
Proliferation of U251 cells was suppressed by CEP55 RNAi
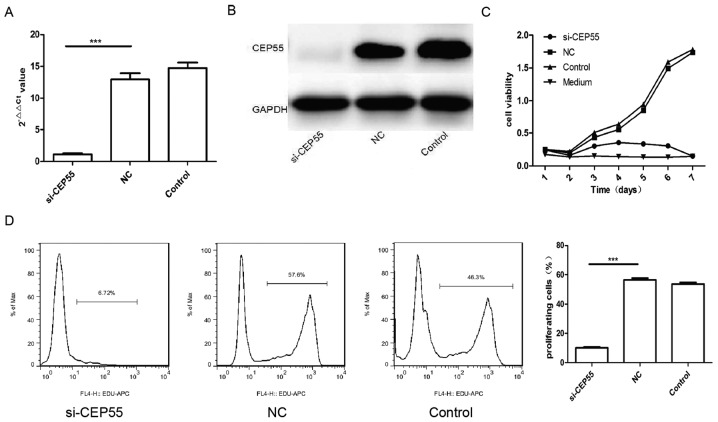
CEP55 acted as a member of the centrosomal participated in mitosis of cell. Then, we supposed that expression of CEP55 was related to proliferation of glioma cells. To survey the effect of CEP55 on proliferation in glioma cells, the expression of CEP55 was knockdown with RNAi mediated by lentiviral vector in GBM U251 cells. The mRNA and protein of CEP55 in U251 cells decreased remarkably assayed by qPCR and western blot analysis after the cells infected by CEP55 siRNA lentiviral (Fig. 2A and B). The proliferation of three groups of U251 cells (control, NC and si-CEP55) were analyzed with CCK-8 kit and EdU. The result of CCK-8 showed that the proliferation of U251 cells with CEP55 knockdown dropped significantly than the cells without CEP55 intervened. The proliferation curve of U251 cells with CEP55 RNAi became flat similar to the medium (Fig. 2C). There were 57.6 and 46.3% cells being reduplication phase separately in NC group and control group of U251 cells labeled with EdU. But only 6.72% cells were being proliferation phase in si-CEP55 group of U251 cells detected with flow cytometry. The rate of U251 cells being proliferation in si-CEP55 group was lower markedly than it in NC group and control group (Fig. 2D). The result demonstrated that CEP55 RNAi was able to induce the inhibition of proliferation in U251 cells.
Figure 2.
CEP55 RNAi induced suppression of proliferation in U251 cells. (A and B) Knockdown effect of CEP55 in U251 cells measured by (A) quantitative polymerase chain reaction and (B) western blot analysis. (C) Proliferation of human glioma cell line U251 treated with or without CEP55 siRNA measured by CCK-8. (D) EdU staining results of human glioma cell line U251 treated with or without CEP55 siRNA measured by flow cytometry. ***P<0.001. si, small interfering; CEP55, centrosomal protein 55.
EGFRvIII promoted the proliferation and expression of CEP55 in U251 cells
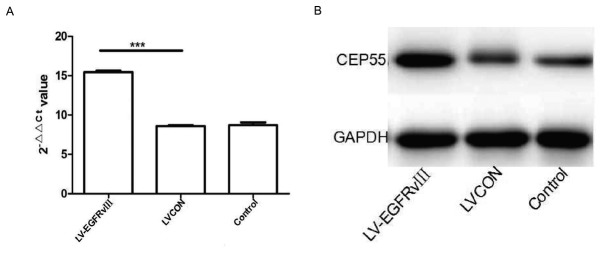
CEP55 is a conserved gene to preventing genetic damage and its protein acts as a regulator of cell cycle progression ensuring proper abscission, the final stage of cytokinesis (18). Therefore, the expression of CEP55 might be regulated by other driver gene. The EGFRvIII is one frequent driver gene of GBM cells and plays an important role of proliferation in GBM cells. Then, EGFRvIII gene were transfected into U251 cells by lentiviral vector and the expression of EGFRvIII in U251 cells were detected with qPCR and Western-blot. The results of qPCR and western blot analysis verified that the mRNA and proteins of EGFRvIII increased significantly in U251 cells infected by EGFRvIII gene lentiviral vector compared with the cells infected by control lentiviral vector group (LVCON) and control group (Fig. 3A and B). CCK-8 kit was used to analyze the proliferation of U251 cells after transduced with EGFRvIII gene. The results of curve showed that proliferation of U251 transduced with EGFRvIII gene increased considerably compared to the cells of LVCON group and control group (P<0.001) (Fig. 3C). Expression of EGFRvIII was able to promote the proliferation of U251 cells.
Figure 3.
Expression of EGFRvIII promoted the proliferation of U251 cells. Transfection effect of EGFRvIII measured by (A) quantitative polymerase chain reaction and (B) western blot analysis. (C) Proliferation of U251 cells with or without EGFRvIII transfection detected by CCK-8 kit. ***P<0.001. EGFRvIII, epithelial growth factor receptor variant III.
Meanwhile, the qPCR and western blot analysis were performed to measuring the mRNA and protein of CEP55 in U251 cells transduced with EGFRvIII gene. The level of CEP55 mRNA was higher in U251 cells of EGFRvIII group than in cells of LVCON group and control group. After EGFRvIII gene transduced and expressed in U251 cells, the protein of CEP55 increased drastically compared to the cells of control group (Fig. 4A and B). The results of qPCR and western blot analysis revealed that expression of EGFRvIII promoted the proliferation and expression of CEP55 in GBM U251 cells.
Figure 4.
Overexpression of CEP55 induced by expression of EGFRvIII. (A) RNA expression level of CEP55 in U251 after EGFRvIII transfection. (B) Protein level of CEP55 in U251 after EGFRvIII transfection. ***P<0.001. CEP55, centrosomal protein 55; EGFR, epithelial growth factor receptor; LV, lentivirus.
CEP55 RNAi inhibited the proliferation induced by EGFRvIII in U251 cells
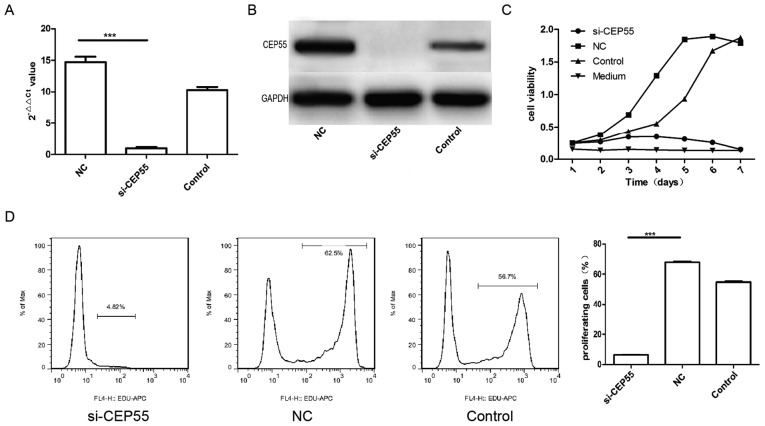
To detect the relationship of the proliferation promoted by EGFRvIII and expression of CEP55 in U251 cells, the expression of CEP55 was suppressed by RNAi in U251 cells transduced with EGFRvIII. The mRNA and protein of CEP55 assayed by qPCR and western blot analysis were reduced significantly in U251 cells with EGFRvIII overexpressing after it transduced with CEP55 siRNA (Fig. 5A and B). The curve of growth was analyzed with CCK-8 in U251 cells with EGFRvIII overexpressing and CEP55 siRNA transduced. The results indicated that the proliferation of U251 cells declined markedly after the expression of CEP55 was inhibited by RNAi (Fig. 5C). The proliferation of U251 cells with EGFRvIII overexpressing and CEP55 siRNA transduced was also measured by flow cytometry stained with EDU. The rate of proliferation in U251 cells expressing EGFRvIII declined to 4.82% after the expression of CEP55 was suppressed with CEP55 siRNA, and the rates of proliferation in U251 cells of NC group and control group were 62.5 and 56.7% respective (Fig. 5D). The result implied that the proliferation of U251 cells induced by EGFRvIII was inhibited by CEP55 RNAi and EGFRvIII promoted proliferation in U251 cells through regulating CEP55 overexpression.
Figure 5.
Proliferation promoted by EGFRvIII is inhibited by CEP55 RNA interference in U251 cells. (A and B) Knockdown effect of CEP55 in U251 cells with overexpressed EGFRvIII measured by qPCR and WB. (C) Proliferation of cells with or without CEP55 knockingdown by siRNA after overexpressed EGFRvIII detected by CCK-8. (D) Proliferation of cells with or without CEP55 knocking down by siRNA after overexpressed EGFRvIII measured by flowcytometry. ***P<0.001. CEP55, centrosomal protein 55; EGFR, epithelial growth factor receptor; si, small interfering; NC, negative control; APC, allophytocyanin.
Discussion
CEP55 was known as a centrosome and midbody associated protein which might cooperate with members of endosomal sorting complex required for transport machinery to allow abscission (19,20). Soon after, overexpression of CEP55 was found in lung adenocarcinoma, gastric carcinoma and hepatocellular carcinoma, where it promote cell proliferation and invasion (17,21,22). It was among the top 70 most highly overexpressed genes in an analysis across 12 cancer types including lung, lymphoma, medulloblastoma and breast, and was identified in prognostic signatures for these cancers (18,23). In this study, we found that CEP55 expression increased significantly in glioma and GBM U251 cells than in normal brain tissue. The level of CEP55 expression was significantly associated to survival of patient with glioma through analyzing CEP55 mRNA and survival in 663 patients from TCGA database. These results implied that CEP55 may play an important role on proliferation or invasion of glioma cells. To observe the effect of CEP55 overexpression on glioma cells, expression of CEP55 were inhibited by RNAi in GBM U251 cells in this study. The proliferation of U251 cells was markedly suppressed after CEP55 knockdown. This result was confirmed in Wang et al study, which provided evidence of inhibiting cell proliferation and inducing cell apoptosis by knockdown of CEP55 in glioma cells (24).
CEP55 expresses in various cancers and is barely detectable in normal tissues except for testis and thymus (24). Its overexpression and activation was regulated via aberrantly upregulated PI3K/AKT pathways and Forkhead box protein M1 in lung cancer, hepatocellular carcinoma and head-neck squamous cell carcinoma (17,21,25). In glioma, the EGFR and its mutations contributed to tumorigenesis and biology via various ways. EGFR amplification and mutation were highly frequent genetic change in gliomas, which resulted in expression of EGFRvIII and overexpression of EGFR. Several studies demonstrated that they were associated with proliferation, progression, and chemo-radiotherapy resistance of gliomas (5–9). The PI3K/AKT pathway activated by EGFR and EGFRvIII was an important molecular mechanism which gave rise to promoting proliferation and resistance of drug and radiation (9,26). Both EGFR/EGFRvIII and CEP55 were associated with proliferation of glioma cells and activation of PI3K/AKT pathway. What is the relationship between CEP55 and EGFR, especially EGFRvIII which is a ligand-independent constitutively active variant of EGFR, in regulation of glioma cells proliferation? To investigate the regulating mechanism of proliferation by EGFRvIII and CEP55 in glioma cells, U251 cells were firstly transfected with EGFRvIII gene in this study. Expression of EGFRvIII promoted significantly the proliferation of U251 cells transfected with EGFRvIII gene. Meanwhile, the CEP55 expression increased remarkably in U251 cells expressing EGFRvIII compared with cells of control group. This result suggested that CEP55 participated the regulating mechanism of proliferation induced by EGFRvIII in U251 cells.
EGFRvIII, a mutant of EGFR, was able to over-activate various downstream signaling pathways, including the PI3K/AKT and RAS-RAF-ERK and regulated many biological outputs that are beneficial to glioma cell proliferation including their initiation and progression of cell cycle (27). CEP55 participated directly abscission in mitosis of cell. To verify CEP55 mediating the proliferation of U251 cells promoted by EGFRvIII, the expression of CEP55 was inhibited by CEP55 siRNA in U251 cell with EGFRvIII transfecting. The proliferation of U251 cells promoted by EGFRvIII declined significantly after CEP55 was knockdown. These results demonstrated that CEP55 participated the proliferation regulated by EGFRvIII and knockdown CEP55 was able to eliminate the effect of promoting proliferation induced by EGFRvIII in U251 cells. Certainly, CEP55 is a mitotic phosphoprotein, acting as an effector of proliferation pathways, which required tightly precise regulation in cytokinesis. The effect of deregulated CEP55 expression in the glioma cells on upstream factor of regulating proliferation such as EGFR remain unresolved. The molecular mechanism and pathways of EGFRvIII upregulating CEP55 expression in detail need further research in our future study.
In conclusion, in this study, we found that CEP55 overexpression played a key role in proliferation of glioma cells and mediated EGFRvIII stimulating proliferation in glioma cells. Knockdown of CEP55 inhibited proliferation of U251 cells induced by EGFRvIII and CEP55 may be a novel molecular therapeutic target to patients with gliomas expressing EGFRvIII. The mechanism underlying proliferation regulated by EGFRvIII-CEP55 in glioma cells need further investigation.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (no. 81372408).
References
- 1.Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Network TC. Corrigendum: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2013;494:506. doi: 10.1038/nature11903. [DOI] [PubMed] [Google Scholar]
- 6.Viana-Pereira M, Lopes JM, Little S, Milanezi F, Basto D, Pardal F, Jones C, Reis RM. Analysis of EGFR overexpression, EGFR gene amplification and the EGFRvIII mutation in Portuguese high-grade gliomas. Anticancer Res. 2008;28:913–920. [PubMed] [Google Scholar]
- 7.Johnson H, Del Rosario AM, Bryson BD, Schroeder MA, Sarkaria JN, White FM. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol Cell Proteomics. 2012;11:1724–1740. doi: 10.1074/mcp.M112.019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S, Hatanpaa KJ, Mickey B, Madden C, Maher E, et al. EGFRvIII and DNA double-strand break repair: A molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 11.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Zhang W, Chen D, Lv Y, Zheng J, Lilljebjörn H, Ran L, Bao Z, Soneson C, Sjögren HO, et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA; Proc Natl Acad Sci USA; 2014; pp. 3538–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, Gabrielli BG, Robinson PJ, Nigg EA, Ono Y, Khanna KK. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9:477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhao WM, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17:3881–3896. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doxsey S. Duplicating dangerously: Linking centrosome duplication and aneuploidy. Mol Cell. 2002;10:439–440. doi: 10.1016/S1097-2765(02)00654-8. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Garay I, Rustom A, Gerdes HH, Kutsche K. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics. 2006;87:243–253. doi: 10.1016/j.ygeno.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Chen CH, Lu PJ, Chen YC, Fu SL, Wu KJ, Tsou AP, Lee YC, Lin TC, Hsu SL, Lin WJ, et al. FLJ10540-elicited cell transformation is through the activation of PI3-kinase/AKT pathway. Oncogene. 2007;26:4272–4283. doi: 10.1038/sj.onc.1210207. [DOI] [PubMed] [Google Scholar]
- 18.Jeffery J, Sinha D, Srihari S, Kalimutho M, Khanna KK. Beyond cytokinesis: The emerging roles of CEP55 in tumorigenesis. Oncogene. 2016;35:683–690. doi: 10.1038/onc.2015.128. [DOI] [PubMed] [Google Scholar]
- 19.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 20.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CH, Lai JM, Chou TY, Chen CY, Su LJ, Lee YC, Cheng TS, Hong YR, Chou CK, Whang-Peng J, et al. VEGFA upregulates FLJ10540 and modulates migration and invasion of lung cancer via PI3K/AKT pathway. PLoS One. 2009;4:e5052. doi: 10.1371/journal.pone.0005052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao J, Zhi X, Tian Y, Li Z, Zhu Y, Wang W, Xie K, Tang J, Zhang X, Wang L, Xu Z. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumor Biol. 2014;35:4389–4399. doi: 10.1007/s13277-013-1578-1. [DOI] [PubMed] [Google Scholar]
- 23.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Liu M, Wang H, Yu S, Jiang Z, Sun J, Han K, Shen J, Zhu M, Lin Z, et al. Centrosomal protein of 55 regulates glucose metabolism, proliferation and apoptosis of glioma cells via the Akt/mTOR signaling pathway. J Cancer. 2016;7:1431–1440. doi: 10.7150/jca.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M, Sugden D, Thurlow JK, Cheong SC, Teo SH, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS One. 2009;4:e4849. doi: 10.1371/journal.pone.0004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, Bi Y, Kong J, Zhang J, Wang B, Li K, Tian M, Pan X, Shi B, Gu J, et al. Combination of an anti-EGFRvIII antibody CH12 with Rapamycin synergistically inhibits the growth of EGFRvIII+PTEN-glioblastoma in vivo. Oncotarget. 2016;7:24752–24765. doi: 10.18632/oncotarget.8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9:E52. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]