Abstract
The medial septum (MS) complex modulates hippocampal function and related behaviors. Septohippocampal projections promote and control different forms of hippocampal synchronization. Specifically, GABAergic and cholinergic projections targeting the hippocampal formation from the MS provide bursting discharges to promote theta rhythm, or tonic activity to promote gamma oscillations. In turn, the MS is targeted by ascending projections from the hypothalamus and brainstem. One of these projections arises from the nucleus incertus in the pontine tegmentum, which contains GABA neurons that co-express the neuropeptide relaxin-3 (Rln3). Both stimulation of the nucleus incertus and septal infusion of Rln3 receptor agonist peptides promotes hippocampal theta rhythm. The Gi/o-protein-coupled receptor, relaxin-family peptide receptor 3 (RXFP3), is the cognate receptor for Rln3 and identification of the transmitter phenotype of neurons expressing RXFP3 in the septohippocampal system can provide further insights into the role of Rln3 transmission in the promotion of septohippocampal theta rhythm. Therefore, we used RNAscope multiplex in situ hybridization to characterize the septal neurons expressing Rxfp3 mRNA in the rat. Our results demonstrate that Rxfp3 mRNA is abundantly expressed in vesicular GABA transporter (vGAT) mRNA- and parvalbumin (PV) mRNA-positive GABA neurons in MS, whereas ChAT mRNA-positive acetylcholine neurons lack Rxfp3 mRNA. Approximately 75% of Rxfp3 mRNA-positive neurons expressed vGAT mRNA (and 22% were PV mRNA-positive), while the remaining 25% expressed Rxfp3 mRNA only, consistent with a potential glutamatergic phenotype. Similar proportions were observed in the posterior septum. The occurrence of RXFP3 in PV-positive GABAergic neurons gives support to a role for the Rln3-RXFP3 system in septohippocampal theta rhythm.
Keywords: arousal, ChAT, emotion, GABA, hippocampus, nucleus incertus, relaxin-3, theta rhythm
Introduction
Intrinsic neural circuits within, and projections from, the MS subserve various roles of this important brain area in different functions ranging from arousal, attention and spatial working memory (Givens and Olton, 1990; Sweeney et al., 1992; Osborne, 1994). Much research on the MS has centered on characterizing its projections to the hippocampus [see (Zaborszky et al., 2012, 2014) for review], in addition to descending projections from the MS to the hypothalamus, raphe nuclei and the NI (Borhegyi and Freund, 1998; Leranth et al., 1999; Sánchez-Pérez et al., 2015). Modulation of septal function has been traditionally viewed to derive strongly from ascending projections from the posterior hypothalamus and brainstem, including the raphe nuclei, which have been described as modulators of hippocampal theta rhythm via activation of the septohippocampal projection system (Vertes and Kocsis, 1997; Vertes, 2005). In addition, descending projections from the somatostatin-positive GABA projection neurons of the hippocampus provide a descending feedback regulation of the MS (Toth et al., 1993; Gulyas et al., 2003; Yuan et al., 2017).
However, the less well-studied projection from the NI in the pontine tegmentum also strongly modulates the MS (Goto et al., 2001; Olucha-Bordonau et al., 2003, 2012). Specifically, NI projections to the MS are associated with modulation of hippocampal theta rhythm. Electrical stimulation of the NI increased theta rhythm band power of the CA1 hippocampal field potential and NI lesions attenuated the increased hippocampal theta rhythm power induced by stimulation of the nucleus reticularis pontis oralis (RPO) in urethane-anesthetized rats (Nuñez et al., 2006).
A major population of GABA neurons in the NI co-express the neuropeptide, Rln3 (Ma et al., 2007) and NI projections and Rln3-positive fibers are in close contact with cholinergic and GABAergic neurons in the MS (Olucha-Bordonau et al., 2012). Moreover, infusion of a Rln3 analog into the MS increased hippocampal theta rhythm, whereas infusion of a Rln3 receptor antagonist impaired the theta rhythm produced by novel environment exploration or RPO stimulation (Ma et al., 2009). Different approaches in recent years have confirmed and extended these observations regarding the role of the NI and its associated peptide Rln3 in subcortical modulation of hippocampal theta rhythm, with an observed synchrony between the firing of NI neurons and different phases of hippocampal theta rhythm (Ma et al., 2013; Martínez-Bellver et al., 2015, 2017).
The cognate receptor for Rln3 is the Gi/o-protein-coupled receptor, RXFP3. In in vitro studies in Chinese hamster ovary cells transfected with RXFP3, bath application of Rln3 results in inhibition of cAMP synthesis and increased ERK phosphorylation (Liu et al., 2003; van der Westhuizen et al., 2005, 2007; Bathgate et al., 2013). In agreement with a potential inhibitory effect of neuronal RXFP3 activation, Rln3 and a selective RXFP3 agonist, RXFP3-A2 (Shabanpoor et al., 2012), hyperpolarized RXFP3-expressing magnocellular neurons in the rat paraventricular and supraoptic hypothalamic nuclei (Kania et al., 2017). Furthermore, following intracerebroventricular (icv) infusion of RXFP3-A2, we observed increased phospho-ERK levels in the MS and disruption of spatial working memory in a spatial alternation test (Albert-Gascó et al., 2017), although the precise relationship between these effects is not known.
The MS is composed of a heterogeneous population of neurons and each neuronal type participates in a different way in septo-hippocampal interactions (Sotty et al., 2003). For example, slow firing cholinergic neurons facilitate hippocampal activity (Sotty et al., 2003), while PV GABAergic projection neurons inhibit hippocampal interneurons (Toth et al., 1997). Somatostatin positive neurons are concentrated in the HDB (Köhler and Eriksson, 1984), but to our knowledge, no functional role has been assigned to these neurons. Different types of calcium-binding protein-expressing neurons and neurons expressing choline acetyltransferase (ChAT) are targeted by NI axons/terminals in the rat (Olucha-Bordonau et al., 2012), but it is not clear which of these neurons express RXFP3. Thus, we explored the distribution of Rxfp3 mRNA expression in different neuronal types of the rat septal area using multiplex in situ hybridization and specific probes for Rxfp3, ChAT, vGAT (slc32a1), PV, and somatostatin (SOM) transcripts.
Materials and Methods
Animals
Experiments were conducted with approval from The Florey Institute of Neuroscience and Mental Health Animal Ethics Committee, in compliance with guidelines of the National Health and Medical Research Council of Australia. Adult male Sprague-Dawley rats weighing 300–320 g were maintained on a 12–12 h light-dark cycle with lights on at 0700 h. Rats were provided free access to food and water.
Multiplex in Situ Hybridization (ISH)
The distribution of septal Rxfp3 mRNA-positive neurons and their GABAergic or cholinergic phenotype was assessed using RNAscope multiplex in situ hybridization. RNAscope® is a commercial method provided by Advanced Cell Diagnostics (ACD, Newark, CA, United States), which involves the incubation of post-fixed, fresh-frozen brain sections with up to three custom probes. Standard probes contain 20 ZZ pairs (25 base pairs/Z) which cover a total of ∼1000 base pairs of the target mRNA. In silico verification of the probes is performed and validated to select oligonucleotides with compatible melting temperature for optimal hybridization under RNAscope assay conditions and minimal cross-hybridization to off-target sequences. There is a verification procedure conducted following each major step during the probe design to guarantee accuracy, according to previously described rules (Wang et al., 2012).
Two naïve rats were deeply anesthetized with pentobarbitone (100 mg/kg, i.p.), decapitated, and brains were quickly extracted and rapidly frozen on dry ice. The fresh-frozen brains were embedded in OCT embedding gel (Tissue-Tek® OCT, Optimum Cutting Temperature, Sakura Finetek USA, Inc., Torrance, CA, United States) and stored at -80°C. Before cryo-sectioning, brains were warmed to -20°C for 2 h and then mounted on a cryostat (Cryocut CM 1800, Leica Microsystems, North Ryde, NSW, Australia) using OCT embedding gel. Coronal sections (16 μm) were cut and thaw-mounted on Superfrost-Plus Slides (Fisher Scientific, Hampton, NH, United States, Cat#12-550-15).
Sections were fixed in 4% paraformaldehyde (PFA) for 16 min at 4°C, rinsed in PBS, and dehydrated in increasing ethanol concentrations (50, 70, and 100%). Once dehydrated the sections were stored in 100% ethanol overnight at -20°C. The next day, slides were air-dried and a hydrophobic barrier was drawn around the sections (ImmEdge hydrophobic PAP pen, Vector Laboratories, Burlingame, CA, United States; Cat #310018). Sections were incubated with protease pretreatment-4 (ACD, Cat #322340) for 16 min. After a PBS rinse, sections were incubated for 2 h at 40°C with three different probe combinations targeting (i) Rxfp3 (ACD, #316181), ChAT (ACD, #430111), and vGAT (Slc32a1; ACD, #424541) mRNA; (ii) Rxfp3, PV (pvalb, ACD, #407828) and SOM (Sst, ACD, #412181-C3) mRNA; (iii) Rxfp3, PV, and vGAT mRNA. Sections were processed in two different trials. Following incubation, sections were rinsed with wash buffer (ACD, Cat#310091) and signals were amplified with ACD amplifier reagents according to manufacturer’s protocol. After 2 × rinses with wash buffer, sections were stained with DAPI (ACD, #320851), covered with fluorescent mounting medium (Fluoromount-G, Southern Biotech, Birmingham, AL, United States, Cat# 17985-10), coverslipped, and stored at -20°C.
Imaging and Quantification of Co-expression of Transcripts
Fluorescence images were taken with an LSM 780 Zeiss Axio Imager 2 confocal laser scanning microscope (Carl Zeiss AG, Jena, Germany). The system is equipped with a stitching stage, and Zen software (Carl Zeiss AG) was used to stitch tiled images taken with a 20 × objective. Quantification of cellular colocalization of transcripts (one section/bregma level, rat and probe combination) was conducted manually using Fiji (Schindelin et al., 2012) [Note: Results consistent with those observed in the sections assessed, were also observed in adjacent brain areas, and in other rat brains, for all probes]. The total number of positive neurons for each region was counted separately, relative to DAPI-stained nuclei, to avoid bias. The percentage co-expression of transcripts was related to the total number of Rxfp3 mRNA-positive neurons in each of the septal areas. Higher-power, (inset) images to illustrate co-localization were taken using a 40 × objective.
Results
In these experiments, we assessed the rostrocaudal distribution of Rxfp3 mRNA-positive neurons in the MS/diagonal band, LS, triangular septal nucleus, and SFi, and determined whether these Rxfp3 mRNA-positive neurons co-expressed ChAT or vGAT mRNA or PV and/or vGAT mRNA (or SOM and/or vGAT mRNA). All these neurotransmitter-related transcripts and their related proteins or peptides have been described as clear markers of the onion-like structure of the septum. According to Wei et al. (2012), the onion-like MS can be described as a five-layer structure with layers determined by their highest density marker (MS-1-MS-3, LSv, and LSi). Layers are distributed from the midline to the LSi with MS-1 on the midline, rich in PV neurons; followed by MS-2, rich in ChAT neurons; followed by MS-3, rich in nNOS; followed by CR (LSV), and CB (LSi). The following results illustrate a high level of co-localization of Rxfp3 and vGAT mRNA in neurons in most septal regions. In contrast, in caudal septal regions and diagonal band, no co-localization of Rxfp3 with vGAT mRNA occurred, suggesting an alternative non-GABAergic phenotype (Table 1).
Table 1.
Semi-quantification of the number of neurons expressing Rxfp3, vGAT, and ChAT mRNA alone and in combination throughout the different regions of the rat septal area.
| Bregma | Area | Rxfp3 | vGAT | PV | SOM | ChAT | Rxfp3/vGAT | Rxfp3/PV/vGAT | Rxfp3/ChAT | Rxfp3/- |
|---|---|---|---|---|---|---|---|---|---|---|
| 1.08 mm | MS | 77 ± 28 | 240 | – | – | 91 | 44 | – | 0 | 5 |
| – | 116 | 0 | – | – | 39 | – | 66 | |||
| VDB | 67 ± 14 | 148 | – | – | 41 | 45 | – | 3 | 5 | |
| – | 106 | 0 | – | – | 37 | – | 35 | |||
| HDB | 62 | – | 16 | 78 | – | – | 1 | – | 53 | |
| 0.6 mm | MS | 107 ± 13 | 500 ± 100 | – | – | 75 | 82 | 7 | 0 | 14 |
| 179 | – | – | 99 | – | – | 12 | ||||
| VDB | 54 ± 12 | 89 | – | – | 9 | 31 | – | 0 | 34 | |
| 237 | 87 | – | – | 23 | 4 | – | 15 | |||
| HDB | 37 ± 6 | 162 | – | – | 26 | 21 | – | 0 | 10 | |
| 234 | 43 | – | – | 24 | 7 | – | 12 | |||
| 0.48 mm | MS | 172 ± 16 | 1500 ± 30 | 126 ± 10 | – | – | 109 ± 8 | 27 ± 8 | – | 22 ± 15 |
| VDB | 50 | 306 ± 20 | 83 ± 15 | – | – | 24 | 14 | – | 12 | |
| HDB | 41.5 ± 13 | 188 ± 16 | 23 ± 5 | – | – | 23 ± 10 | 7 ± 2 | – | 12 ± 5 | |
| 0.24 mm | LSI | 11 | 37 | – | – | 2 | 9 | – | 0 | 2 |
| SFi | 20 | 97 | – | – | 2 | 19 | – | 0 | 1 | |
| MS | 8 | 86 | – | – | 4 | 8 | – | 0 | 0 | |
| -0.24 mm | LSD | 14 | 61 | – | – | 0 | 12 | – | 0 | 2 |
| SFi | 100 | 314 | – | – | 0 | 75 | – | 0 | 25 | |
| TS | 245 | 800 | – | – | 3 | 193 | – | 0 | 52 | |
| LSV | 223 | 307 | – | – | 4 | 142 | – | 1 | 80 | |
| SFO | 30 | 4 | – | 6 | 2 | – | 0 | 28 |
– Mean ± SEM are indicated on cases where more than one subject was analyzed. – indicates not determined probe combinations and split rows indicate each of different trials.
Rxfp3 mRNA-Positive Neurons in MS Co-express vGAT, But Not ChAT mRNA
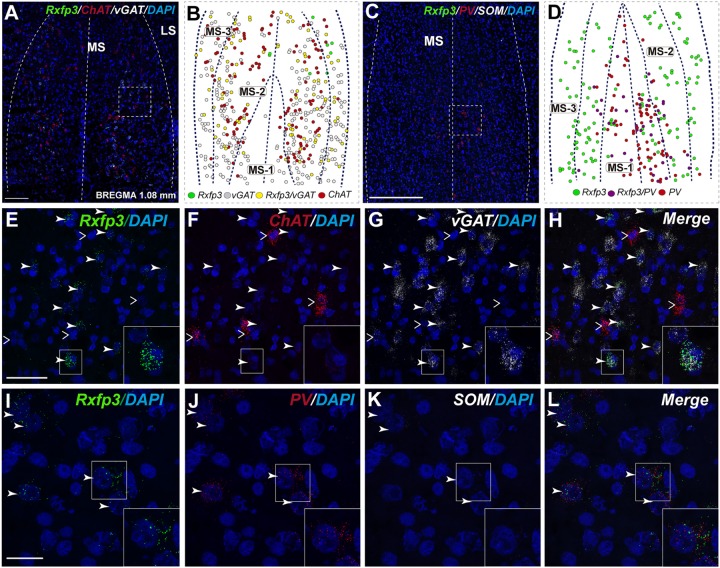
At the most rostral level of the MS (bregma ∼1.08 mm), Rxfp3 mRNA-expressing neurons were mainly located between the MS-1 and MS-3 layers (Figures 1A–D). The majority of Rxfp3 mRNA-positive neurons in these layers co-expressed vGAT mRNA (∼90%; 44/49) of expressing neurons) while only ∼10% (5/49 neurons) of Rxfp3 mRNA-positive neurons lacked vGAT and ChAT mRNA (Figures 1E–H, 2). Given the distribution of these neurons and the co-localization of Rxfp3 mRNA with vGAT and not ChAT mRNA, this labeling is consistent with expression of RXFP3 by GABA neurons (Ma et al., 2009, 2017; Olucha-Bordonau et al., 2012). With a different combination of probes for Rxfp3/PV/SOM mRNA, ∼37% of Rxfp3 mRNA-expressing-neurons in the MS (39/105 neurons), expressed PV mRNA, and were distributed within MS-1, while 63% (66/105) of Rxfp3 mRNA-positive neurons that did not co-express PV mRNA (Figures 1I–L), were located within MS-2 and 3 (Figure 1D).
FIGURE 1.
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), and ChAT mRNA relative to DAPI-stained nuclei, and Rxfp3, SOM, and PV mRNA in the rat MS at bregma +1.08 mm (A,C); and a schematic map illustrating the different neuronal phenotypes based on mRNA co-expression, and their distribution (B,D). Thick dotted lines indicate the midline and the medial and lateral septal border and thin dotted lines, the layers within MS. Higher magnification images illustrating co-localization of Rxfp3 (E), ChAT (F), and vGAT (G) mRNA and merged signals (H). High-magnification images illustrating the co-localization of Rxfp3 (I), PV (J), and SOM (K) mRNA and merged signals (L). Arrowheads indicate neurons double-labeled for Rxfp3 and vGAT mRNA (E,F) and Rxfp3 and PV mRNA (I,J). No colocalization of Rxfp3 and ChAT mRNA was observed (open arrowheads). Insets (lower right) are high magnification images of the boxed area in (E–L), illustrating a neuron double-labeled for Rxfp3 and vGAT mRNA or Rxfp3 and PV mRNA consistent with a GABAergic phenotype. Calibration bar in (A) 250 μm, (C) 250 μm, (E–H) 50 μm, and (I–L) 20 μm.
FIGURE 2.
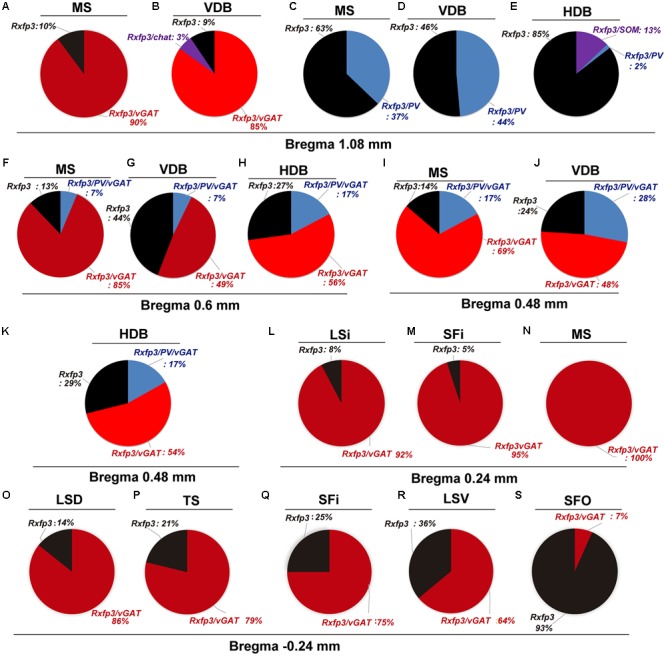
Percentage co-localization of Rxfp3, vGAT, PV, and SOM mRNA throughout the septal complex. Rostral to caudal levels of MS (A,C), VDB (B,D), and HDB (E) at +1.08 mm from bregma; MS (F), VDB (G) and HDB (H) at +0.6 mm from bregma; MS (I), VDB (J) and HDB (K) at +0.48 mm from bregma; LSi (L), SFi (M) and MS (N) at +0.24 mm from bregma; and LSD (O), TS (P), SFi (Q), LSV (R) and SFO (S) at –0.24 mm from bregma.
In the mid-anterior dorsal part of the MS (bregma ∼0.6 mm), Rxfp3 mRNA-expressing neurons were present mainly in the MS-1 layer, characterized as containing PV neurons, and in more lateral layers containing lower PV neuron densities (Kiss et al., 1990; Wei et al., 2012). The highest number of Rxfp3 mRNA-expressing neurons was located between MS-2 and MS-3. In the ventral part of this mid-MS level, Rxfp3 mRNA-positive neurons were limited to the MS-2 (Figures 3A–D). At this level, the majority of Rxfp3 mRNA-expressing neurons co-expressed vGAT mRNA (∼87% (82/94) of labeled neurons), while only ∼13% (13/94 neurons) of labeled cells expressed Rxfp3 mRNA in the absence of vGAT and ChAT mRNA (Figures 3E–H). In sections labeled with the Rxfp3/PV/vGAT probe combination, some Rxfp3 mRNA-expressing-neurons co-expressed PV mRNA (∼6%; 7/111 neurons), but most co-expressed vGAT mRNA, distributed within MS-2 (82% (91/111) of labeled neurons). Only 12% (14/111 neurons) of Rxfp3 mRNA-expressing neurons did not co-express either transcript (Figures 2, 3I–L).
FIGURE 3.
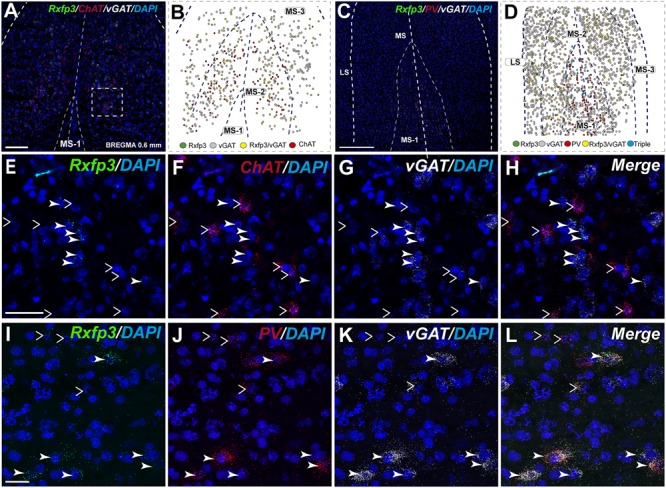
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), ChAT, and PV mRNA relative to DAPI-stained nuclei in the rat MS at bregma +0.60 mm (A,C), and a schematic map illustrating the different neuronal phenotypes based on mRNA co-expression, and their distribution (B,D). Thick dotted lines indicate the midline and the medial and lateral septal border and thin dotted lines, the layers within MS. High-magnification images illustrating colocalization of Rxfp3 (E), ChAT (F), and vGAT (G) mRNA, and merged signals (H). High-magnification images illustrating co-localization of Rxfp3 (I), PV (J), and vGAT (K) mRNA and merged signals (L). Arrowheads indicate neurons double-labeled for Rxfp3 and vGAT mRNA (E–H) and Rxfp3, vGAT and PV mRNA (I–L). No colocalization of Rxfp3 and ChAT mRNA was observed (open arrowheads). Calibration bar in (A) 250 μm, (C) 250 μm, (E–H) 50 μm, and (I–L).
In the mid-posterior part of the septal area (bregma ∼0.48 mm), the Rxfp3/PV/SOM probe combination revealed that Rxfp3 mRNA-expressing neurons were distributed across MS-1 to MS-3. Rxfp3 mRNA-positive neurons that co-expressed PV mRNA (17%; 27/157 neurons) were mostly located near the midline (Figures 4A–D). In the MS-2 and MS-3 layers, 69% (109/157 neurons) of Rxfp3 mRNA-positive neurons co-expressed vGAT mRNA and 14% (22/172 neurons) did not co-express either transcript (Figures 2,‘4E–L).
FIGURE 4.
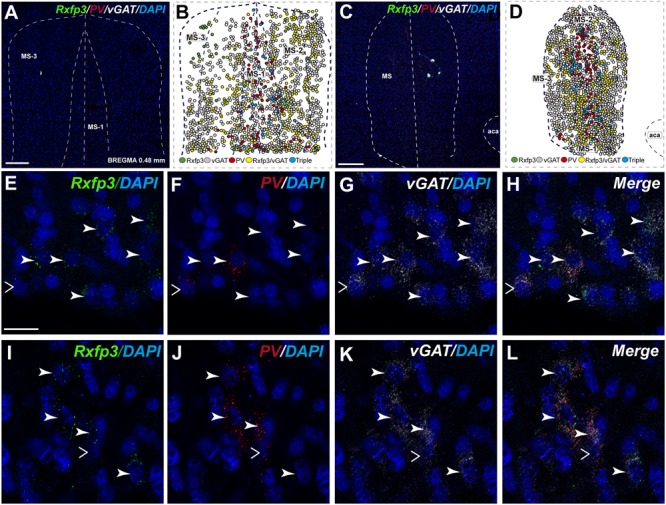
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), ChAT, and PV mRNA relative to DAPI-stained nuclei in the rat MS at bregma +0.48 mm (A,C), and a schematic map illustrating the different neuronal phenotypes based on mRNA co-expression, and their distribution (B,D). Thick dotted lines indicate the midline and the medial and lateral septal border and thin dotted lines the layers within MS. High-magnification images illustrating colocalization of Rxfp3 (E,I), PV (F,J), and vGAT (G,K) mRNA and merged signals (H,L). Arrowheads indicate neurons double-labeled for Rxfp3 and vGAT mRNA (E–H) and Rxfp3, vGAT, and PV mRNA (I–L). No colocalization of Rxfp3 and PV mRNA was indicated with open arrowheads. Calibration bar in (A) 200, (C) 500, and (E–L) 50 μm.
In the posterior septum (bregma ∼0.24 mm), Rxfp3 mRNA-positive neurons were present in the MS and were more dense in the SFi and the LSI (Figures 5A,B). In the MS, Rxfp3 mRNA-positive neurons co-expressed vGAT mRNA in ∼100% of cases (8/8 neurons), but did not express ChAT mRNA (Figures 2, 5C–F). In the SFi ∼95% (19/20 neurons) of Rxfp3 mRNA-positive neurons co-expressed vGAT mRNA, and ∼5% (1/20 neurons) of Rxfp3 mRNA-positive neurons lacked vGAT and ChAT mRNAs (Figures 2, 5G–J). Finally, in the LSI ∼92% (24/26 neurons) of detected neurons co-expressed Rxfp3 and vGAT mRNAs whereas only ∼8% (2/26 neurons) of Rxfp3 mRNA-positive neurons lacked vGAT and ChAT mRNA (Figures 2, 5K–N).
FIGURE 5.
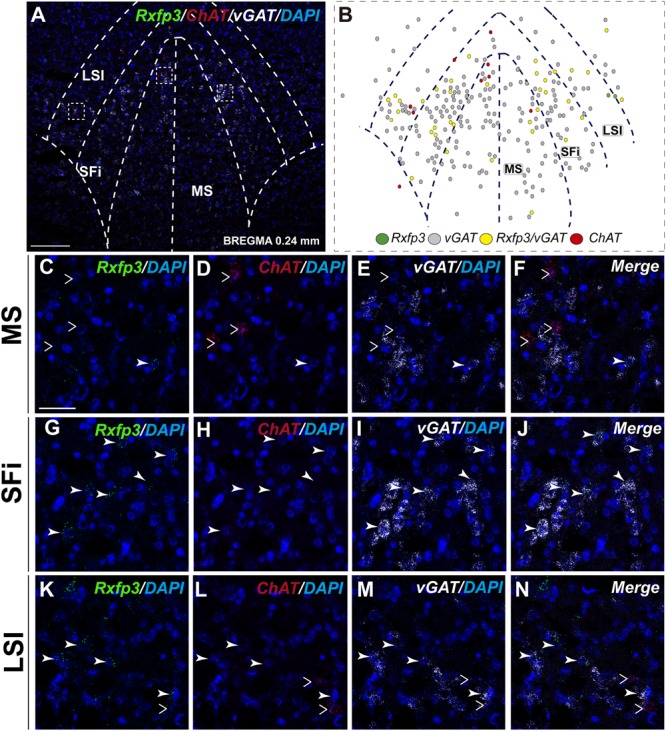
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), and ChAT mRNA, relative to DAPI-stained nuclei in the rat MS, SFi, and LSI at bregma +0.24 mm (A) and a schematic map illustrating different neuronal phenotypes based on mRNA co-expression, and their distribution (B). Dotted lines indicate the midline and the medial and lateral septal and septofimbrial borders. High-magnification images illustrate co-localization of Rxfp3 (C,G,K), ChAT (D,H,L), vGAT (E,I,M) mRNA and merged signals (F,J,N) in the MS, SFi, and LSI, respectively. No co-localization of Rxfp3 and ChAT mRNA was observed (open arrowheads). Calibration bar in (A) 125 μm and (C–N) 50 μm.
Diagonal Band Neurons Co-express Rxfp3 and vGAT (slc32a1) mRNA
In the anterior (VDB; bregma ∼1.08 mm), Rxfp3 mRNA-positive neurons were evenly distributed laterally at a similar distance from the midline (Figures 6A–D′). Rxfp3 mRNA-positive neurons were present in the vicinity of ChAT mRNA-expressing neurons, but Rxfp3 and ChAT mRNA were sparsely co-expressed in ∼6% (3/53 neurons) of total Rxfp3 mRNA-positive cells, while ∼85% (45/53 neurons) co-expressed Rxfp3 and vGAT mRNA, and Rxfp3 transcripts were present in the absence of the other markers in only ∼9% (5/53 neurons) of identified neurons (Figures 2, 6E–H). In sections incubated in a different combination of probes, Rxfp3 mRNA-expressing neurons in the VDB co-localized with PV mRNA (44%; 35/80 neurons) and did not co-localize with any marker in 46% (37/80) of neurons (Figures 2, 6I–L) In contrast, in the HDB, some Rxfp3 mRNA-expressing neurons co-expressed SOM (13%; 8/62) and PV (2% 1/62) mRNA, but most did not co-express either of these transcripts (85%; 53/62 neurons) (Figures 2, 7A,B,E–H).
FIGURE 6.
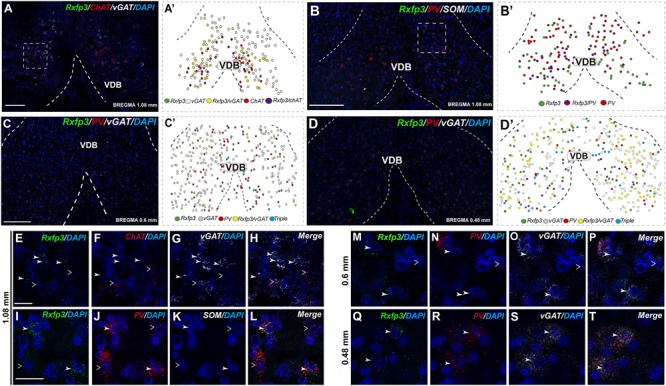
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), ChAT, and PV mRNAs relative to DAPI-stained nuclei in the vertical limb of the diagonal band at bregma +1.08 (A,B), +0.6 (C), and 0.48 mm (D) and schematic map illustrating different colocalization neuronal phenotypes and their distribution (A′–D′). Dotted lines indicate the midline and the VDB border. Higher magnification images illustrate colocalization of Rxfp3 (E), ChAT (F), vGAT mRNA (G), and merged signals (H). High-magnification images illustrate co-localization of Rxfp3 (I,M,Q), PV (J,N,R), SOM (K) vGAT (O,S) mRNA and merged signals (L,P,T) at different bregma levels. Arrowheads indicate neurons double-labeled for Rxfp3 and vGAT (E–H) or Rxfp3, vGAT, and PV (I–T) mRNA and open arrowheads indicate ChAT or PV mRNA-positive neurons that do not express Rxfp3 mRNA. Calibration bar in (A) 100 μm, (B–D) 125 μm, (E–H) 50 μm, and (I–T) 20 μm.
FIGURE 7.
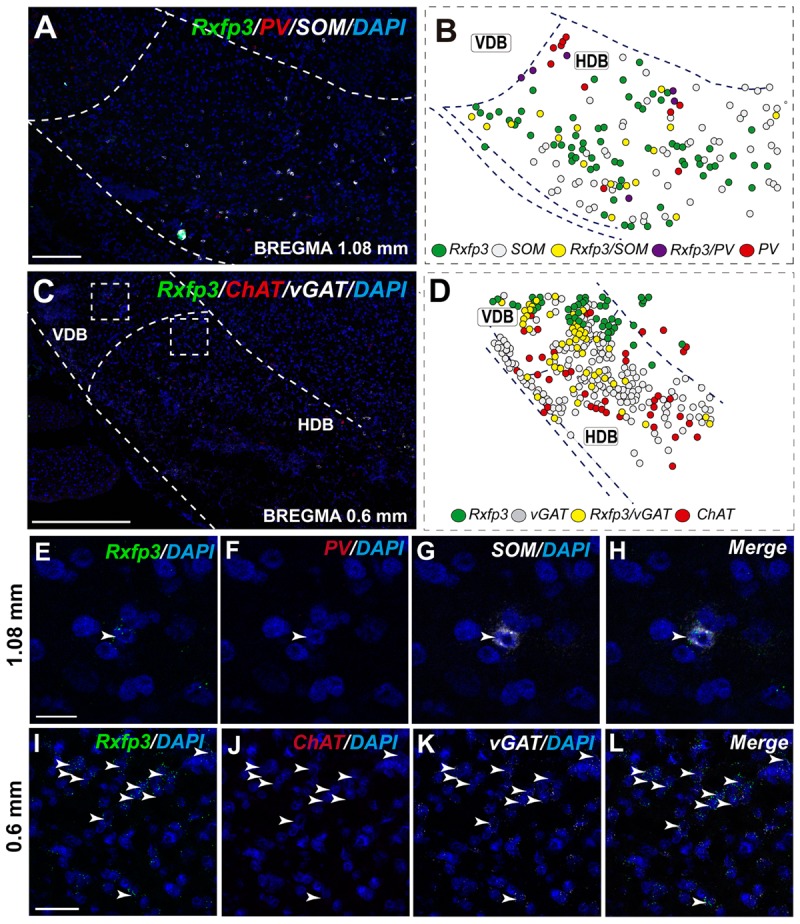
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), ChAT, and PV mRNAs relative to DAPI-stained nuclei in the HDB at bregma +1.08 mm (A) +0.60 mm (C), and schematic map illustrating different colocalization neuronal phenotypes and their distribution (B,D). Dotted lines indicate the vertical and horizontal limbs of the diagonal band border. Higher magnification images illustrate colocalization of Rxfp3 (E), PV (F), SOM mRNA (G), and merged signals (H); Rxfp3 (I), ChAT (J), vGAT mRNA (K), and merged signals (L) in the HDB, respectively. Arrowheads indicate neurons double-labeled for Rxfp3, SOM, and PV (E–H) or Rxfp3 and vGAT (I–L). No colocalization of Rxfp3 and ChAT mRNAs was observed. Calibration bar in (A) 125 μm, (C) 200 μm, (E–H) 20 μm, and (I–L) 50 μm.
At more posterior levels (bregma ∼0.6 mm), Rxfp3 mRNA-positive neurons were present in the VDB and the HDB (Figures 6C,C′, 7C,D). In the VDB, Rxfp3 mRNA-expressing neurons were present in two clusters. From the total amount of Rxfp3 mRNA positive neurons ∼48% (31/65) of them co-expressed vGAT mRNA. A second cluster/population of Rxfp3 mRNA-positive neurons, ∼52%; 34/65 neurons, did not co-express vGAT or ChAT mRNA. In sections labeled for Rxfp3/PV/vGAT mRNA some Rxfp3 mRNA-expressing neurons expressed PV mRNA (7%; 4/56), while 49% (27/56) expressed vGAT mRNA and 44% (25/56) did not express either of the other transcripts (Figures 2, 6M–P).
The number of Rxfp3 mRNA-positive neurons in the HDB was lower than in the VDB (Figures 7C,D). In contrast to the VDB, in the HDB the majority (∼68%; 21/31 neurons) of Rxfp3 mRNA-positive neurons expressed vGAT mRNA, while the remaining were vGAT mRNA and ChAT mRNA negative (∼32%; 10/31 neurons) (Figures 2, 7I–L). Rxfp3 mRNA colocalized with vGAT mRNA (56%; 23/41 neurons), and with PV/vGAT mRNA (17%; 7/41 neurons) and was also expressed in the absence of either transcript (27%; 11/41) (Figures 2, 8A,B,E–H).
FIGURE 8.
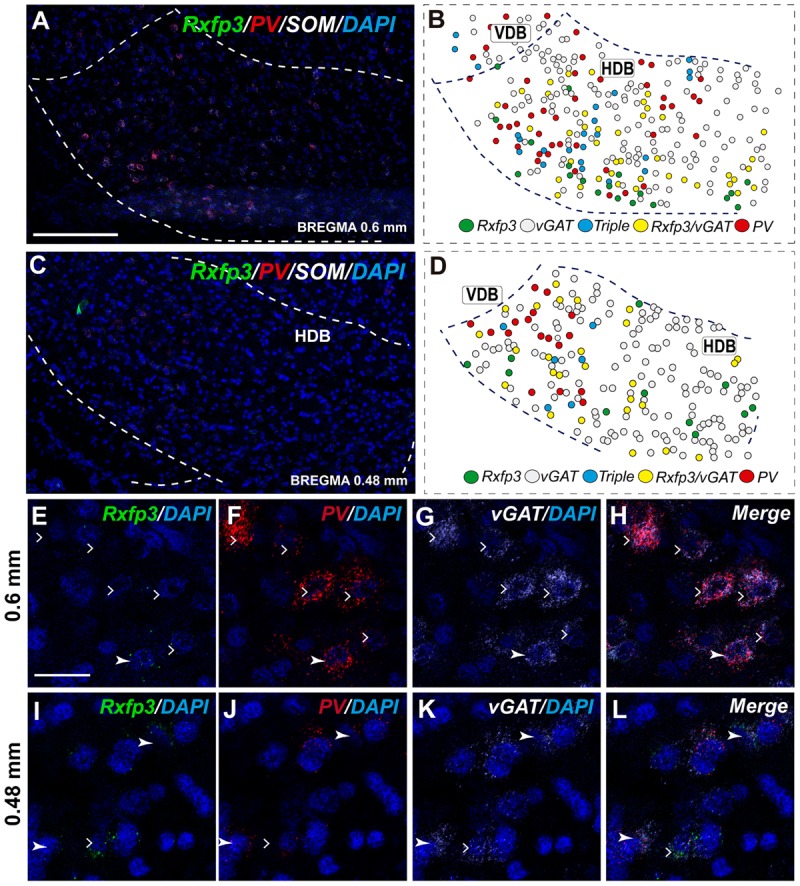
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), ChAT, and PV mRNAs relative to DAPI-stained nuclei in the HDB at bregma +0.6 mm (A) +0.48 mm (C) and schematic map illustrating different colocalization neuronal phenotypes and their distribution (B,D). Dotted lines indicate the vertical and horizontal limbs of the diagonal band border. Higher magnification images illustrate colocalization of Rxfp3 (E), PV (F), vGAT mRNA (G), and merged signals (H); Rxfp3 (I), PV (J), vGAT mRNA (K), and merged signals (L) in the HDB, respectively. Arrowheads indicate neurons double-labeled for Rxfp3, vGAT, and PV (E–H) or Rxfp3 and vGAT (I–L). Lack of colocalization of Rxfp3 and PV mRNA indicated with open arrowheads. Calibration bar in (A,C) 125 μm, (E–L) 20 μm.
In the mid-posterior part of the septal area (bregma ∼0.48 mm) sections labeled with Rxfp3/PV/vGAT probes displayed Rxfp3 mRNA expressing neurons in the VDB (Figures 6D,D′) that co-expressed PV/vGAT mRNA (28%; 14/50 neurons), and vGAT mRNA (48%; 24/50 neurons) (Figures 2, 6Q–T), but some Rxfp3 mRNA-positive neurons did not express either transcript (24%; 12/50 neurons). Likewise, analysis of the HDB, revealed that the majority of Rxfp3 mRNA-positive neurons expressed vGAT mRNA (54%; 23/42 neurons) and a small proportion expressed PV/vGAT mRNA (17%; 7/42 neurons) or neither of the other transcripts (29%; 12/42 neurons) (Figures 2, 8I–L).
Triangular Septal Area, and Septofimbrial and Dorsolateral Septal Area Contain Heterogeneous Populations of Rxfp3 mRNA-Positive Neurons
In the most caudal region of the septum analyzed (bregma -0.24 mm), the distribution and phenotype of Rxfp3 mRNA-positive neurons varied within the different nuclei. In the LSD, Rxfp3 mRNA-positive neurons were widely and evenly distributed (Figures 9A,B) and were mainly vGAT mRNA-positive (∼86%; 12/14 neurons), with a small number of neurons located near the SFi that were vGAT mRNA negative (∼14%; 2/14 neurons; Figures 2, 9K–N).
FIGURE 9.
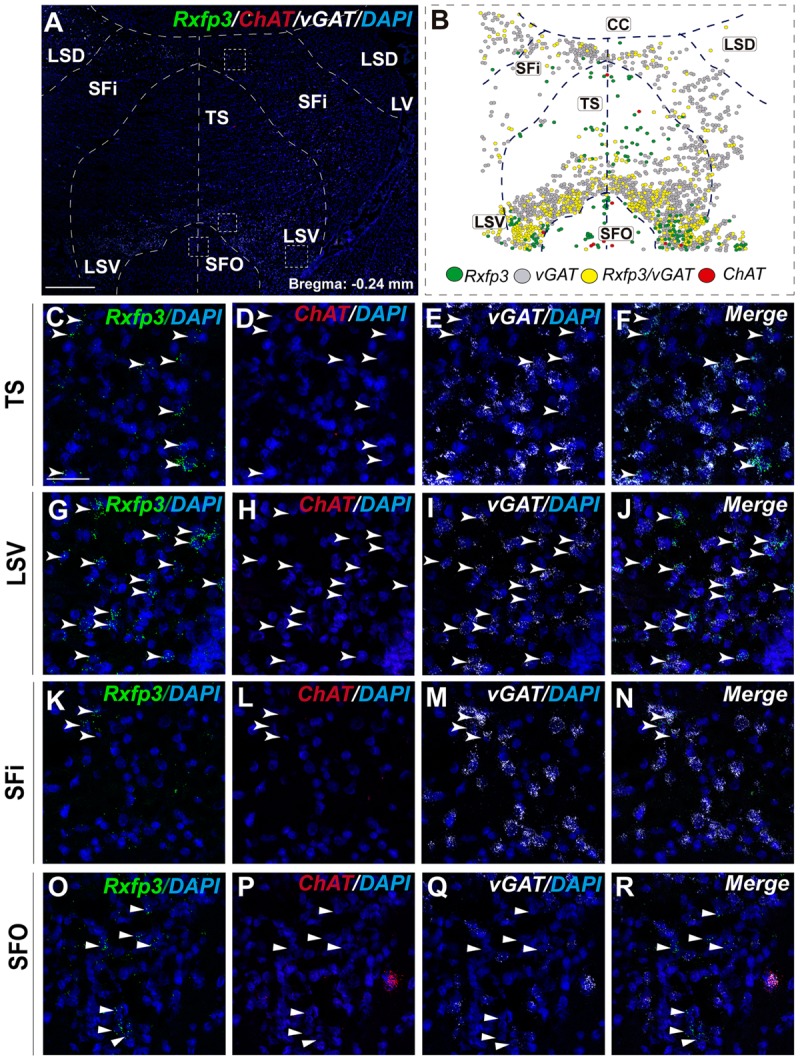
Distribution of neurons expressing Rxfp3, vGAT (slc32a1), and ChAT mRNA, relative to DAPI-stained nuclei in LSD, SFi, TS, LSV, and SFO at bregma –0.24 mm (A), and a schematic map illustrating the different neuronal phenotypes and their distribution (B). Dotted lines indicate the midline and borders between the different regions. High-magnification images illustrate the co-localization of Rxfp3 (C,G,K,O), ChAT (D,H,L,P), vGAT (E,I,M,Q) mRNA and merged signals (F,J,N,R) in the TS, LSV, SFi, and SFO, respectively. Arrowheads indicate neurons double-labeled for Rxfp3 and vGAT mRNA. No co-localization of Rxfp3 and ChAT mRNA was observed (open arrowheads). Calibration bar in (A) 500 μm and (C–R) 50 μm.
Rxfp3 mRNA-positive neurons in the SFi were mainly distributed in the most dorsal part of the nucleus near the corpus callosum (cc). In the ventral SFi, Rxfp3 mRNA-positive neurons were lower in number (Figures 9A,B). Throughout the dorsal and ventral SFi, the majority of Rxfp3 mRNA-positive neurons co-expressed vGAT mRNA (∼75%; 75/100 neurons; Figures 2, 9L–O).
The triangular septal area (TS) contained three Rxfp3 mRNA-positive neuron populations based on their differential phenotype and distribution. Dispersed Rxfp3 mRNA-positive neurons were present in the most dorsal portion near the midline, while in the most ventral TS, a large, densely packed population of Rxfp3 mRNA-positive neurons were distributed alongside the border with the SFO (Figures 9A,B). The ventral TS area was rich in vGAT mRNA-expressing neurons, while the dorsal TS was not. Rxfp3 mRNA-positive neurons in the dorsal TS were generally vGAT mRNA-negative, while in the ventral TS, Rxfp3 mRNA-positive neurons were generally vGAT mRNA-positive. In the lateral part of the ventral TS, there was a population of Rxfp3 mRNA-positive neurons which were vGAT mRNA-negative. Overall, ∼79% of Rxfp3 mRNA-positive neurons in TS co-expressed vGAT mRNA (193/245 neurons), while the remainder were negative (∼21%; 52/245 neurons; Figures 2, 9C–K).
Similar to ventral TS, the LSV contained a large population of vGAT mRNA-positive neurons and most were Rxfp3/vGAT mRNA-positive (64%; 142/223 neurons). In addition to these GABAergic neurons, this area also contained a large non-GABAergic population (36%; 80/223 neurons; Figures 2, 9G,H). Finally, we noted that within the SFO, a vast majority of Rxfp3 mRNA-positive neurons were vGAT mRNA-negative (93%; 28/30 cells; Figures 2, 9O–R).
Discussion
In this study we have employed RNAscope multiplex in situ hybridization (Wang et al., 2014a,b; Li and Kim, 2015) to characterize the neurochemical phenotype of Rxfp3 mRNA-positive neurons in the rat septal area. The highly specific nature of the method means that these data represent a more accurate estimation of the distribution of RXFP3 than studies using putative antisera against the receptor protein (Meadows and Byrnes, 2014), although this powerful approach does not provide information about the subcellular location of RXFP3, which might be available with alternative protein detection methods.
In the septal area, neurons expressing Rxfp3 transcripts were concentrated in the MS complex, including the diagonal band nuclei, and in the posterior septum, including the SFi and TS nuclei. Some Rxfp3 mRNA-positive neurons were also detected in LS divisions. Our findings are consistent with previous studies of the presence and distribution of Rxfp3 mRNA in the rat septal area detected using radioactive oligonucleotide probes. Specifically, MS and HDB displayed moderate to high levels of Rxfp3 mRNA, while in LSI and VDB, expression was moderate (Sutton et al., 2004; Ma et al., 2007). These findings are consistent with concurrent studies of these and other transcripts in rat hippocampus (Ma and Gundlach, unpublished data). Therefore, in light of the strong innervation of the rat septal region by Rln3-positive nerve fibers, the presence of septal RXFP3 binding sites, and functional studies (Ma et al., 2007, 2009), we conclude that the detection of Rxfp3 mRNA reflects the expression of functional RXFP3 protein by these neurons.
In the MS, VBD, and HDB, the vast majority of Rxfp3 mRNA-positive neurons co-expressed vGAT mRNA. Furthermore, a population of these presumed GABAergic Rxfp3 mRNA-positive neurons are PV mRNA-positive (Wei et al., 2012). Septal PV/GABA neurons are the main source of the GABAergic projections to the hippocampus and specifically target hippocampal interneurons (Freund and Antal, 1988; Freund and Gulyas, 1997). A number of studies have demonstrated that PV/GABA neuron activity is crucial for hippocampal theta rhythm (Borhegyi et al., 2004; Bassant et al., 2005; Simon et al., 2006). The modulation of the GABAergic inter-neuronal inhibition of hippocampal pyramidal neurons has been reported to be a source for hippocampal theta rhythm synchronization (Toth et al., 1997). In addition, septal PV/GABA neurons expressing cyclic nucleotide activated, non-selective cation channels play a role in driving hippocampal theta rhythm (Varga et al., 2008; Hangya et al., 2009). Notably, RXFP3 activation results in inhibition of cellular cAMP synthesis in cell-based assays in vitro (Liu et al., 2003; van der Westhuizen et al., 2007, 2010), consistent with a similar interaction in vivo (see further discussion below).
In contrast to the strong association with GABAergic neurons, only a small number of cholinergic (ChAT mRNA-positive) neurons co-expressed Rxfp3 mRNA. However, anterograde neural tract-tracing and immunohistochemical studies suggest that cholinergic (ChAT-positive) septal neurons receive a robust innervation from the Rln3 rich NI (Olucha-Bordonau et al., 2012). Thus, the influence of NI neurons on the septal cholinergic system might be mediated by NI neurons that contain GABA only or other peptides, such as cholecystokinin, which is expressed in the NI (Kubota et al., 1983; Olucha-Bordonau et al., 2003) (Ma and Gundlach, unpublished data).
The discovery that Rxfp3 mRNA is absent from MS cholinergic neurons provides new insights into the nature of the coordinated neural actions that result in the generation and modulation of hippocampal theta rhythm, and since RXFP3 activation often produces neuronal inhibition in vitro (Blasiak et al., 2013; Kania et al., 2017), it is possible that pERK activation in MS cholinergic neurons occurs via RXFP3-mediated inhibition of non-PV, GABAergic interneurons (Leranth and Frotscher, 1989). In this regard, optogenetic activation of cholinergic septohippocampal neurons suppressed ripple sharp waves and enhance theta rhythm oscillations (Vandecasteele et al., 2014) and local circuit inhibitory actions on cholinergic neurons are a primary process in the generation of septal rhythmicity (Leão et al., 2015). Furthermore, icv infusion of an RXFP3 agonist (RXFP3-A2; Shabanpoor et al., 2012) resulted in increased phosphorylation of ERK in the MS, mainly in ChAT-immunoreactive neurons (Albert-Gascó et al., 2017). Given the observed absence of Rxfp3- and ChAT mRNA-positive neurons in the MS in the present study, and the observation that RXFP3 activation routinely induces neuronal inhibition (Kania et al., 2017), there is a possibility that the pERK activation within the cholinergic neurons occurs via a reduction in local circuit inhibition within the MS.
In addition, ∼25% of the Rxfp3 mRNA-positive neurons in the MS were non-GABAergic, non-cholinergic in nature. Although further studies are required to better identify the phenotype of these neurons, it is presumed that some or many are glutamatergic neurons, since they constitute ∼25% of the total MS neuron population (Colom et al., 2005; Gritti et al., 2006). Glutamatergic neurons provide both local and septohippocampal projections (Manseau et al., 2005; Henderson et al., 2010; Huh et al., 2010) and interestingly, optogenetic activation of MS glutamatergic neurons produces strong theta rhythm synchronization, mainly mediated by local septal circuits (Robinson et al., 2016).
Considerable data suggest a strong link between RXFP3 activation in the MS and modulation of hippocampal theta rhythm. Hippocampal theta rhythm has been traditionally associated with arousal mechanisms which are directly involved in attentional mechanisms of memory (Vinogradova, 1995). The NI, along with other brainstem areas, the hypothalamus and the basal forebrain, promote arousal and fast electroencephalographic (EEG) rhythms (Brown and McKenna, 2015; Korotkova et al., 2018). Moreover, stimulation of the NI promotes arousal and is associated with cortical EEG desynchronization, increased locomotor activity, and head-scanning vigilance behavior during fear recall (Ma et al., 2017). In addition, ipsilateral NI stimulation induces locomotion and rotation at latencies consistent with a role in the modulation of premotor areas like the basal forebrain (Farooq et al., 2016). Furthermore, Rln3 and Rxfp3 gene knockout mice display reduced voluntary running wheel activity during the dark, active phase (Smith et al., 2012; Hosken et al., 2015) providing further evidence for a likely role for this signaling system in sustained arousal and related locomotor and exploratory activity.
Indeed, the MS controls exploratory behavior (Köhler and Srebro, 1980; Poucet, 1989; Mamad et al., 2015; Gangadharan et al., 2016). Different forms of memory, including spatial working memory and object recognition can be affected by manipulations of the MS (Givens and Olton, 1994; Fitz et al., 2008; Roland et al., 2014; Okada et al., 2015; Gangadharan et al., 2016). Interestingly, interference with global or septal Rln3/RXFP3 signaling in the rat results in disruption of spatial working memory in the spontaneous alternation test (Ma et al., 2009; Albert-Gascó et al., 2017).
Conclusion
The strong expression of Rxfp3 mRNA by GABAergic neurons in the rat MS and adjacent nuclei, is consistent with the central role of these neurons in the control of hippocampal theta rhythm by actions on local septal circuits. In turn, these actions may indirectly influence septal cholinergic neurons/circuits and hippocampal interneurons via septohippocampal projections. Notably, independent studies have revealed a strong Rln3 innervation of the hippocampus and identified Rxfp3 mRNA expression by hippocampal GABA neurons in the rat (Ma and Gundlach, unpublished data), consistent with direct actions of Rln3/RXFP3 signaling on these circuits. Therefore, further studies of the neurotransmitter and neurochemical phenotype of septal and hippocampal neurons that express Rxfp3 mRNA and their precise functional roles are warranted in both normal adult rats and mice, and in models of neuropathology and cognitive and psychiatric disorders.
Author Contributions
HA-G and SM designed and performed the experiments, prepared the figures and drafted the manuscript. FR-B and AMS-P contributed to some experiments, figure preparation and corrections of the draft manuscript. ALG and FEO-B planned the research, supervised the experiments, edited the figures, and prepared the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the following grants: Universitat Jaume I FPI-UJI Predoctoral Research Scholarship PREDOC/2014/35 (HA-G); E-2016-43 Research Travel Grant (HA-G); NHMRC (Australia) Project Grant 1067522 (ALG); Dorothy Levien Foundation Research Grant (ALG); and Universitat Jaume I Research Grant UJI-B2016-40 (FEO-B).
Abbreviations
- ChAT
choline acetyl transferase
- HDB
horizontal diagonal band
- LS
lateral septum
- LSD
lateral septum dorsal
- LSI
lateral septum intermediate
- LSV
lateral septum ventral
- LV
lateral ventricle
- MS
medial septum
- NI
nucleus incertus
- nNOS
neuronal nitric oxide synthase
- PV
parvalbumin
- Rln3
relaxin-3
- Rxfp3
relaxin-family peptide receptor 3
- SFi
septofimbrial nucleus
- SFO
subfornical organ
- TS
triangular septum
- VDB
vertical diagonal band
- vGAT (slc32a1)
vesicular GABA transporter
References
- Albert-Gascó H., García-Avilés Á, Moustafa S., Sánchez-Sarasua S., Gundlach A. L., Olucha-Bordonau F. E., et al. (2017). Central relaxin-3 receptor (RXFP3) activation increases ERK phosphorylation in septal cholinergic neurons and impairs spatial working memory. Brain Struct. Funct. 222 449–463. 10.1007/s00429-016-1227-8 [DOI] [PubMed] [Google Scholar]
- Bassant M.-H. H., Simon A., Poindessous-Jazat F., Csaba Z., Epelbaum J., Dournaud P. (2005). Medial septal GABAergic neurons express the somatostatin sst2A receptor: functional consequences on unit firing and hippocampal theta. J. Neurosci. 25 2032–2041. 10.1523/JNEUROSCI.4619-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathgate R. A., Halls M. L., van der Westhuizen E. T., Callander G. E., Kocan M., Summers R. J. (2013). Relaxin family peptides and their receptors. Physiol. Rev. 93 405–480. 10.1152/physrev.00001.2012 [DOI] [PubMed] [Google Scholar]
- Blasiak A., Blasiak T., Lewandowski M. H., Hossain M. A., Wade J. D., Gundlach A. L. (2013). Relaxin-3 innervation of the intergeniculate leaflet of the rat thalamus - neuronal tract-tracing and in vitro electrophysiological studies. Eur. J. Neurosci. 37 1284–1294. 10.1111/ejn.12155 [DOI] [PubMed] [Google Scholar]
- Borhegyi Z., Freund T. F. (1998). Dual projection from the medial septum to the supramammillary nucleus in the rat. Brain Res. Bull. 46 453–459. 10.1016/S0361-9230(98)00038-0 [DOI] [PubMed] [Google Scholar]
- Borhegyi Z., Varga V., Szilágyi N., Fabo D., Freund T. F. (2004). Phase segregation of medial septal GABAergic neurons during hippocampal theta activity. J. Neurosci. 24 8470–8479. 10.1523/jneuroscience.1413-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. E., McKenna J. T. (2015). Turning a negative into a positive: ascending gabaergic control of cortical activation and arousal. Front. Neurol. 6:135. 10.3389/fneur.2015.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom L. V., Castaneda M. T., Reyna T., Hernandez S., Garrido-Sanabria E. (2005). Characterization of medial septal glutamatergic neurons and their projection to the hippocampus. Synapse 58 151–164. 10.1002/syn.20184 [DOI] [PubMed] [Google Scholar]
- Farooq U., Kumar J. R., Rajkumar R., Dawe G. S. (2016). Electrical microstimulation of the nucleus incertus induces forward locomotion and rotation in rats. Physiol. Behav. 160 50–58. 10.1016/j.physbeh.2016.03.033 [DOI] [PubMed] [Google Scholar]
- Fitz N. F., Gibbs R. B., Johnson D. A. (2008). Selective lesion of septal cholinergic neurons in rats impairs acquisition of a delayed matching to position T-maze task by delaying the shift from a response to a place strategy. Brain Res. Bull. 77 356–360. 10.1016/j.brainresbull.2008.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund T. F., Antal M. (1988). GABA-containing neurons in the septum control inhibitory interneurons in the hippocampus. Nature 336 170–173. 10.1038/336170a0 [DOI] [PubMed] [Google Scholar]
- Freund T. F., Gulyas A. I. (1997). Inhibitory control of GABAergic interneurons in the hippocampus. Can. J. Physiol. Pharmacol. 75 479–487. 10.3389/fnins.2012.00165 [DOI] [PubMed] [Google Scholar]
- Gangadharan G., Shin J., Kim S.-W., Kim A., Paydar A., Kim D.-S., et al. (2016). Medial septal GABAergic projection neurons promote object exploration behavior and type 2 theta rhythm. Proc. Natl. Acad. Sci. U.S.A. 113 6550–6555. 10.1073/pnas.1605019113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens B., Olton D. S. (1994). Local modulation of basal forebrain: effects on working and reference memory. J. Neurosci. 14 3578–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens B. S., Olton D. S. (1990). Cholinergic and GABAergic modulation of medial septal area: effect on working memory. Behav. Neurosci. 104 849–855. 10.1037/0735-7044.104.6.849 [DOI] [PubMed] [Google Scholar]
- Goto M., Swanson L. W., Canteras N. S. (2001). Connections of the nucleus incertus. J. Comp. Neurol. 438 86–122. 10.1002/cne.1303 [DOI] [PubMed] [Google Scholar]
- Gritti I., Henny P., Galloni F., Mainville L., Mariotti M., Jones B. E. (2006). Stereological estimates of the basal forebrain cell population in the rat, including neurons containing choline acetyltransferase, glutamic acid decarboxylase or phosphate-activated glutaminase and colocalizing vesicular glutamate transporters. Neuroscience 143 1051–1064. 10.1016/j.neuroscience.2006.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyas A. I., Hajos N., Katona I., Freund T. F. (2003). Interneurons are the local targets of hippocampal inhibitory cells which project to the medial septum. Eur. J. Neurosci. 17 1861–1872. 10.1046/j.1460-9568.2003.02630.x [DOI] [PubMed] [Google Scholar]
- Hangya B., Borhegyi Z., Szilagyi N., Freund T. F., Varga V. (2009). GABAergic neurons of the medial septum lead the hippocampal network during theta activity. J. Neurosci. 29 8094–8102. 10.1523/jneurosci.5665-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson Z., Lu C., Janzsó G., Matto N., McKinley C. (2010). Distribution and role of Kv3. 1b in neurons in the medial septum diagonal band complex. Neuroscience 166 952–969. 10.1016/j.neuroscience.2010.01.020 [DOI] [PubMed] [Google Scholar]
- Hosken I. T., Sutton S. W., Smith C. M., Gundlach A. L. (2015). Relaxin-3 receptor (Rxfp3) gene knockout mice display reduced running wheel activity: implications for role of relaxin-3/RXFP3 signalling in sustained arousal. Behav. Brain Res. 278 167–175. 10.1016/j.bbr.2014.09.028 [DOI] [PubMed] [Google Scholar]
- Huh C. Y. L., Goutagny R., Williams S. (2010). Glutamatergic neurons of the mouse medial septum and diagonal band of Broca synaptically drive hippocampal pyramidal cells: relevance for hippocampal theta rhythm. J. Neurosci. 30 15951–15961. 10.1523/jneurosci.3663-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A., Gugula A., Grabowiecka A., de Ávila C., Blasiak T., Rajfur Z., et al. (2017). Inhibition of oxytocin and vasopressin neuron activity in rat hypothalamic paraventricular nucleus by relaxin-3-RXFP3 signalling. J. Physiol. 595 3425–3447. 10.1113/JP273787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J., Patel A. J., Baimbridge K. G., Freund T. F. (1990). Topographical localization of neurons containing parvalbumin and choline acetyltransferase in the medial septum-diagonal band region of the rat. Neuroscience 36 61–72. 10.1016/0306-4522(90)90351-4 [DOI] [PubMed] [Google Scholar]
- Köhler C., Srebro B. (1980). Effects of lateral and medial septal lesions on exploratory behavior in the albino rat. Brain Res. 182 423–440. 10.1016/0006-8993(80)91199-3 [DOI] [PubMed] [Google Scholar]
- Köhler C., Eriksson L. G. (1984). An immunohistochemical study of somatostatin and neurotensin positive neurons in the septal nuclei of the rat brain. Anat. Embryol. (Berl) 170 1–10. 10.1007/BF00319452 [DOI] [PubMed] [Google Scholar]
- Korotkova T., Ponomarenko A., Monaghan C. K., Poulter S. L., Cacucci F., Wills T., et al. (2018). Reconciling the different faces of hippocampal theta: the role of theta oscillations in cognitive, emotional and innate behaviors. Neurosci. Biobehav. Rev. 85 65–80. 10.1016/j.neubiorev.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Kubota Y., Inagaki S., Shiosaka S., Cho H. J., Tateishi K., Hashimura E., et al. (1983). The distribution of cholecystokinin octapeptide-like structures in the lower brain stem of the rat: an immunohistochemical analysis. Neuroscience 9 587–604. 10.1016/0306-4522(83)90176-8 [DOI] [PubMed] [Google Scholar]
- Leão R. N., Targino Z. H., Colom L. V., Fisahn A. (2015). Interconnection and synchronization of neuronal populations in the mouse medial septum/diagonal band of Broca. J. Neurophysiol. 113 971–980. 10.1152/jn.00367.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leranth C., Carpi D., Buzsaki G., Kiss J. (1999). The entorhino-septo-supramammillary nucleus connection in the rat: morphological basis of a feedback mechanism regulating hippocampal theta rhythm. Neuroscience 88 701–718. 10.1016/S0306-4522(98)00245-0 [DOI] [PubMed] [Google Scholar]
- Leranth C., Frotscher M. (1989). Organization of the septal region in the rat brain: cholinergic-GABAergic interconnections and the termination of hippocampo-septal fibers. J. Comp. Neurol. 289 304–314. 10.1002/cne.902890210 [DOI] [PubMed] [Google Scholar]
- Li Y., Kim J. (2015). Neuronal expression of CB2 cannabinoid receptor mRNAs in the mouse hippocampus. Neuroscience 311 253–267. 10.1016/j.neuroscience.2015.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Eriste E., Sutton S., Chen J., Roland B., Kuei C., et al. (2003). Identification of Relaxin-3/INSL7 as an endogenous ligand for the orphan G-protein-coupled receptor GPCR135. J. Biol. Chem. 278 50754–50764. 10.1074/jbc.M308995200 [DOI] [PubMed] [Google Scholar]
- Ma S., Allocca G., Ong-Pålsson E. K. E., Singleton C. E., Hawkes D., McDougall S. J., et al. (2017). Nucleus incertus promotes cortical desynchronization and behavioral arousal. Brain Struct. Funct. 222 515–537. 10.1007/s00429-016-1230-0 [DOI] [PubMed] [Google Scholar]
- Ma S., Bonaventure P., Ferraro T., Shen P.-J., Burazin T. C. D., Bathgate R. A. D., et al. (2007). Relaxin-3 in GABA projection neurons of nucleus incertus suggests widespread influence on forebrain circuits via G-protein-coupled receptor-135 in the rat. Neuroscience 144 165–190. 10.1016/j.neuroscience.2006.08.072 [DOI] [PubMed] [Google Scholar]
- Ma S., Olucha-Bordonau F. E., Hossain M. A., Lin F., Kuei C., Liu C., et al. (2009). Modulation of hippocampal theta oscillations and spatial memory by relaxin-3 neurons of the nucleus incertus. Learn. Mem. 16 730–742. 10.1101/lm.1438109 [DOI] [PubMed] [Google Scholar]
- Ma S. K., Blasiak A., Olucha-Bordonau F. E., Verberne A. J. M., Gundlach A. L. (2013). Heterogeneous responses of nucleus incertus neurons to corticotrophin-releasing factor and coherent activity with hippocampal theta rhythm in the rat. J. Physiol. 591 3981–4001. 10.1113/jphysiol.2013.254300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamad O., McNamara H. M., Reilly R. B., Tsanov M. (2015). Medial septum regulates the hippocampal spatial representation. Front. Behav. Neurosci. 9:166. 10.3389/fnbeh.2015.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau F., Danik M., Williams S. (2005). A functional glutamatergic neurone network in the medial septum and diagonal band area. J. Physiol. 566 865–884. 10.1113/jphysiol.2005.089664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Bellver S., Cervera-Ferri A., Luque-García A., Martínez-Ricós J., Valverde-Navarro A., Bataller M., et al. (2017). Causal relationships between neurons of the nucleus incertus and the hippocampal theta activity in the rat. J. Physiol. 595 1775–1792. 10.1113/JP272841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Bellver S., Cervera-Ferri A., Martínez-Ricós J., Ruiz-Torner A., Luque-García A., Blasco-Serra A., et al. (2015). Regular theta-firing neurons in the nucleus incertus during sustained hippocampal activation. Eur. J. Neurosci. 41 1049–1067. 10.1111/ejn.12934 [DOI] [PubMed] [Google Scholar]
- Meadows K. L., Byrnes E. M. (2014). Sex- and age-specific differences in relaxin family peptide receptor expression within the hippocampus and amygdala in rats. Neuroscience 284 337–348. 10.1016/j.neuroscience.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Nuñez A., Cervera-Ferri A., Olucha-Bordonau F., Ruiz-Torner A., Teruel V. (2006). Nucleus incertus contribution to hippocampal theta rhythm generation. Eur. J. Neurosci. 23 2731–2738. 10.1111/j.1460-9568.2006.04797.x [DOI] [PubMed] [Google Scholar]
- Okada K., Nishizawa K., Kobayashi T., Sakata S., Kobayashi K. (2015). Distinct roles of basal forebrain cholinergic neurons in spatial and object recognition memory. Sci. Rep. 5:13158. 10.1038/srep13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olucha-Bordonau F. E., Otero-Garcia M., Sanchez-Perez A. M., Nunez A., Ma S., Gundlach A. L. (2012). Distribution and targets of the relaxin-3 innervation of the septal area in the rat. J. Comp. Neurol. 520 1903–1939. 10.1002/cne.23018 [DOI] [PubMed] [Google Scholar]
- Olucha-Bordonau F. E., Teruel V., Barcia-González J., Ruiz-Torner A., Valverde-Navarro A. A., Martínez-Soriano F. (2003). Cytoarchitecture and efferent projections of the nucleus incertus of the rat. J. Comp. Neurol. 464 62–97. 10.1002/cne.10774 [DOI] [PubMed] [Google Scholar]
- Osborne P. G. (1994). A GABAergic mechanism in the medial septum influences cortical arousal and locomotor activity but not a previously learned spatial discrimination task. Neurosci. Lett. 173 63–66. 10.1016/0304-3940(94)90150-3 [DOI] [PubMed] [Google Scholar]
- Poucet B. (1989). Object exploration, habituation, and response to a spatial change in rats following septal or medial frontal cortical damage. Behav. Neurosci. 103 1009–1016. 10.1037/0735-7044.103.5.1009 [DOI] [PubMed] [Google Scholar]
- Robinson J., Manseau F., Ducharme G., Amilhon B., Vigneault E., El Mestikawy S., et al. (2016). Optogenetic activation of septal glutamatergic neurons drive hippocampal theta rhythms. J. Neurosci. 36 3016–3023. 10.1523/jneurosci.2141-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland J. J., Stewart A. L., Janke K. L., Gielow M. R., Kostek J. A., Savage L. M., et al. (2014). Medial septum-diagonal band of Broca (MSDB) GABAergic regulation of hippocampal acetylcholine efflux is dependent on cognitive demands. J. Neurosci. 34 506–514. 10.1523/JNEUROSCI.2352-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pérez A. M., Arnal-Vicente I., Santos F. N., Pereira C. W., El Mlili N., Sanjuan J., et al. (2015). Septal projections to the nucleus incertus: bidirectional pathways for modulation of hippocampal function. J. Comp. Neurol. 523 565–588. 10.1002/cne.23687 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabanpoor F., Akhter Hossain M., Ryan P. J., Belgi A., Layfield S., Kocan M., et al. (2012). Minimization of human relaxin-3 leading to high-affinity analogues with increased selectivity for relaxin-family peptide 3 receptor (RXFP3) over RXFP1. J. Med. Chem. 55 1671–1681. 10.1021/jm201505p [DOI] [PubMed] [Google Scholar]
- Simon A. P., Poindessous-Jazat F., Dutar P., Epelbaum J., Bassant M. H. (2006). Firing properties of anatomically identified neurons in the medial septum of anesthetized and unanesthetized restrained rats. J. Neurosci. 26 9038–9046. 10.1523/JNEUROSCI.1401-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. M., Hosken I. T., Sutton S. W., Lawrence A. J., Gundlach A. L. (2012). Relaxin-3 null mutation mice display a circadian hypoactivity phenotype. Genes Brain Behav. 11 94–104. 10.1111/j.1601-183X.2011.00730.x [DOI] [PubMed] [Google Scholar]
- Sotty F., Danik M., Manseau F., Laplante F., Quirion R., Williams S. (2003). Distinct electrophysiological properties of glutamatergic, cholinergic and GABAergic rat septohippocampal neurons: novel implications for hippocampal rhythmicity. J. Physiol. 551 927–943. 10.1113/jphysiol.2003.046847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton S. W., Bonaventure P., Kuei C., Roland B., Chen J., Nepomuceno D., et al. (2004). Distribution of G-protein-coupled receptor (GPCR)135 binding sites and receptor mRNA in the rat brain suggests a role for relaxin-3 in neuroendocrine and sensory processing. Neuroendocrinology 80 298–307. 10.1159/000083656 [DOI] [PubMed] [Google Scholar]
- Sweeney J. E., Lamour Y., Bassant M. H. (1992). Arousal-dependent properties of medial septal neurons in the unanesthetized rat. Neuroscience 48 353–362. 10.1016/0306-4522(92)90495-N [DOI] [PubMed] [Google Scholar]
- Toth K., Borhegyi Z., Freund T. F. (1993). Postsynaptic targets of GABAergic hippocampal neurons in the medial septum-diagonal band of Broca complex. J. Neurosci. 13 3712–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth K., Freund T. F., Miles R. (1997). Disinhibition of rat hippocampal pyramidal cells by GABAergic afferents from the septum. J. Physiol. 500(Pt 2) 463–474. 10.1113/jphysiol.1997.sp022033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Westhuizen E. T., Christopoulos A., Sexton P. M., Wade J. D., Summers R. J. (2010). H2 relaxin is a biased ligand relative to H3 relaxin at the relaxin family peptide receptor 3 (RXFP3). Mol. Pharmacol. 77 759–772. 10.1124/mol.109.061432 [DOI] [PubMed] [Google Scholar]
- van der Westhuizen E. T., Sexton P. M., Bathgate R. A., Summers R. J. (2005). Responses of GPCR135 to human gene 3 (H3) relaxin in CHO-K1 cells determined by microphysiometry. Ann. N. Y. Acad. Sci. 1041 332–337. 10.1196/annals.1282.053 [DOI] [PubMed] [Google Scholar]
- van der Westhuizen E. T., Werry T. D., Sexton P. M., Summers R. J. (2007). The relaxin family peptide receptor 3 activates extracellular signal-regulated kinase 1/2 through a protein kinase C-dependent mechanism. Mol. Pharmacol. 71 1618–1629. 10.1124/mol.106.032763 [DOI] [PubMed] [Google Scholar]
- Vandecasteele M., Varga V., Berenyi A., Papp E., Bartho P., Venance L., et al. (2014). Optogenetic activation of septal cholinergic neurons suppresses sharp wave ripples and enhances theta oscillations in the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 111 13535–13540. 10.1073/pnas.1411233111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Hangya B., Kranitz K., Ludanyi A., Zemankovics R., Katona I., et al. (2008). The presence of pacemaker HCN channels identifies theta rhythmic GABAergic neurons in the medial septum. J. Physiol. 586 3893–3915. 10.1113/jphysiol.2008.155242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertes R. P. (2005). Hippocampal theta rhythm: a tag for short-term memory. Hippocampus 15 923–935. 10.1002/hipo.20118 [DOI] [PubMed] [Google Scholar]
- Vertes R. P., Kocsis B. (1997). Brainstem-diencephalo-septohippocampal systems controlling the theta rhythm of the hippocampus. Neuroscience 81 893–926. 10.1016/S0306-4522(97)00239-X [DOI] [PubMed] [Google Scholar]
- Vinogradova O. S. (1995). Expression, control, and probable functional significance of the neuronal theta-rhythm. Prog. Neurobiol. 45 523–583. 10.1016/0301-0082(94)00051-I [DOI] [PubMed] [Google Scholar]
- Wang F., Flanagan J., Su N., Wang L.-C., Bui S., Nielson A., et al. (2012). RNAscope. J. Mol. Diagnostics 14 22–29. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Su N., Wang L.-C., Wu X., Bui S., Nielsen A., et al. (2014a). Dual-color ultrasensitive bright-field RNA in situ hybridization with RNAscope. Methods Mol. Biol. 1211 139–149. 10.1007/978-1-4939-1459-3_12 [DOI] [PubMed] [Google Scholar]
- Wang H., Su N., Wang L.-C., Wu X., Bui S., Nielsen A., et al. (2014b). Quantitative ultrasensitive bright-field RNA in situ hybridization with RNAscope. Methods Mol. Biol. 1211 201–212. 10.1007/978-1-4939-1459-3_16 [DOI] [PubMed] [Google Scholar]
- Wei B., Huang Z., He S., Sun C., You Y., Liu F., et al. (2012). The onion skin-like organization of the septum arises from multiple embryonic origins to form multiple adult neuronal fates. Neuroscience 222 110–123. 10.1016/j.neuroscience.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Yuan M., Meyer T., Benkowitz C., Savanthrapadian S., Ansel-Bollepalli L., Foggetti A., et al. (2017). Somatostatin-positive interneurons in the dentate gyrus of mice provide local- and long-range septal synaptic inhibition. Elife 6:e21105. 10.7554/eLife.21105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborszky L., Duque A., Gielow M., Gombkoto P., Nadasdy Z., Somogyi J. (2014). “Organization of the basal forebrain cholinergic projection system: specific or diffuse?” in The Rat Nervous System 4th Edn ed. Paxinos G. (Amsterdam: Elsevier; ) 491–507. [Google Scholar]
- Zaborszky L., van den Pol A. N., Gyengesi E. (2012). “The basal forebrain cholinergic projection system in mice,” in The Mouse Nervous System eds Watson C., Paxinos G., Puelles L. (Amsterdam: Elsevier; ) 684–718. [Google Scholar]