Abstract
Cardiolipin (CL) is a phospholipid localized in the mitochondria, which is essential for mitochondrial structure and function. Human cardiolipin synthase 1 (CRLS1) is important in regulating phosphatidylglycerol (PG) remodeling and CL biosynthesis. However, the expression and distinct prognostic value of CRLS1 in neoplasms, including non-small cell lung cancer (NSCLC), is not well established. In the present study, the mRNA expression of CRLS1 was investigated using Oncomine analysis and the prognostic value was assessed using the Kaplan-Meier plotter database for patients with NSCLC. The results of the analyses indicated that the expression of CRLS1 in lung cancer was lower, compared with that in normal lung tissues. Notably, a high expression of CRLS1 was found to be associated with improved overall survival (OS) in all patients with NSCLC and lung adenocarcinoma (Ade). However, this was not observed in patients with squamous cell carcinoma (SCC). The results also demonstrated an association between the mRNA expression of CRLS1 and the clinicopathological parameters of patients with NSCLC, including sex, smoking status, tumor grade, clinical stage, lymph node status and chemotherapy. These results indicated that CRLS1 was associated with improved prognosis in patients with NSCLC, particularly at an early stage (T1N1M0). In addition, it was revealed that CRLS1 was co-expressed with well-known genes associated with metabolism using Gene Ontology term enrichment analysis. Kyoto Encyclopedia of Genes and Genomes pathway analysis also showed that tumor-related metabolism and the mitogen-activated protein kinase (MAPK) signaling pathways were enriched with CRLS1-co-expression genes. The results of the present study suggested that CRLS1 may be a novel tumor suppressor involved in regulating lipid and seleno-amino acid metabolism in the tumor microenvironment, and suppressing the MAPK signaling pathway during tumorigenesis and development. Comprehensive evaluation of the expression, prognosis and potential mechanism of CRLS1 is likely to promote an improved understanding of the complexity of the molecular biology of NSCLC.
Keywords: CL, NSCLC, CRLS1, SCC, MAPK
Introduction
The occurrence and development of tumors is modulated by the dual regulation of genetic instability and the tumor microenvironment. Several studies have shown that the tumor microenvironment is important in the development and progression of malignant tumors; however, its mechanisms remain to be fully elucidated. Metabolism is one of the basic biological life activities in the body; changes in the biological properties of normal cells when converted to malignant cells include metabolic reprogramming. In previous years, studies have revealed that the activation of certain oncogenes or tumor suppressor inactivation can regulate metabolic homeostasis, and are involved in promoting or inhibiting cancer progression (1,2). Certain metabolic enzymes can themselves be used as oncogenes or tumor suppressors involved in the process of cancer development.
Cardiolipin (CL) is a major mitochondrial membrane polyglycerophospholipid (3,4). It has been shown to be required for the activation of several pivotal mitochondrial membrane enzymes involved in ATP synthesis and mitochondrial respiratory complexes (5). CL may be involved in the pathway leading to cellular apoptosis (6,7). The final step for the de novo mitochondrial synthesis of CL is based on the substrates phosphatidylglycerol and cytidine diphosphate choline-diacylglycerol. This reaction is catalyzed by the enzyme cardiolipin synthase 1 (CRLS1) (8), which is ubiquitous in eukaryotes, archaebacteria and eubacteria. The human CRLS1 gene has previously been cloned and characterized (9,10).
Although CRLS1 has been demonstrated to be functional in lipid anyagcsere, the exact role in tumorigenesis remains to be fully elucidated, with the exception of investigations in patients with solid and non-solid malignancies, which showed that anti-cardiolipin antibodies were associated with an increased rate of thrombosis (11). The expression, prognostic effects and potential mechanism of CRLS1 in tumors remain to be elucidated. It is known that lung cancer is one of the malignant tumors with high morbidity and mortality rates in the world. Non-small cell lung cancer (NSCLC) accounts for almost 85% of all lung cancer cases and generally presents at an advanced stage at diagnosis with a 5-year survival rate of <5% (12,13). Although standard chemotherapy and molecular targeted therapy have improved overall survival (OS) and quality of life, the survival benefit is limited to those who lack a driver mutation or show resistance to drugs (14). Therefore, it is important to investigate the underlying and intertwined mechanisms of lung tumorigenesis and tumor progression to provide novel ideas and a theoretical basis for the treatment of NSCLC.
The present study focused on the association between metabolism-related genes and CRLS1 in addition to NSCLC, and comprehensively analyzed the mRNA expression of CRLS1 in tumors and patient prognosis. Furthermore, the mechanism in tumor development was examined using web-based and soft-based microarray databases. Together, the results suggested that CRLS1 is a promising candidate gene for prognostic and diagnostic approaches in NSCLC, and warrants further investigation.
Materials and methods
Oncomine analysis
Oncomine, a cancer microarray database and integrated data-mining platform, was used to facilitate the identification from genome-wide expression analyses, and the transcriptome data in major types of cancer were compared with respective normal tissues (15,16). The metabolism-related gene expression of CRLS1 (Affymetrix ID, 241741_at) was analyzed using Oncomine™ research premium edition (http://oncomine.org). The mRNA levels in NSCLC vs. normal patient datasets were compared. P<0.0001, fold-change=2, and top 10% gene rank were adjusted as the thresholds. Subsequently, the log2 median centered intensity, and the 10 and 90th percentile data from Oncomine concerning CRLS1 were plotted using GraphPad Prism software (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA).
Analysis of survival prognosis using the Kaplan-Meier plotter
The correlation between survival prognosis and the mRNA expression of CRLS1 in NSCLC was evaluated using the Kaplan-Meier plotter (www.kmplot.com) (17), an online database containing gene expression data and clinical data for lung cancer, breast cancer, stomach cancer and ovarian cancer. The patient samples were divided into two cohorts according to the median expression of the gene (high vs. low expression); this limited the cohorts in accordance with the clinical characteristics of patients, including sex, pathological type, clinical stage and treatment. OS was analyzed in patients with NSCLC using a Kaplan-Meier survival plot and CRLS1 was uploaded into the database to obtain the Kaplan-Meier survival plots. Subsequently, the plot data were exported as text and the Kaplan-Meier survival curve was plotted using GraphPad Prism software.
Potential mechanism in NSCLC meta-analysis of CRLS1
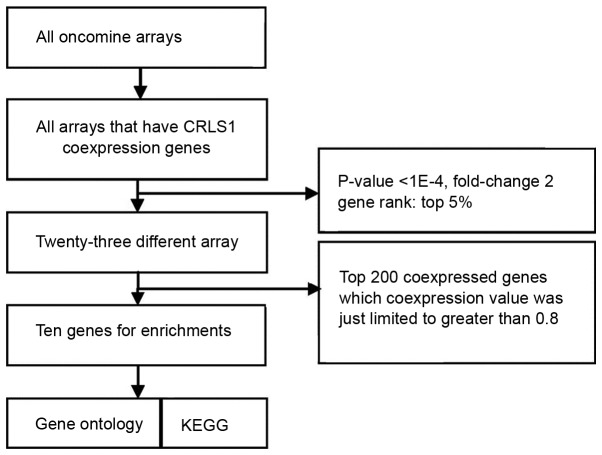
To understand the function of CRLS1, co-expression analysis was performed using the Oncomine database (http://oncomine.org). The threshold was adjusted to P<0.0001, fold-change=2, and gene rank of top 5%. A total of 23 different arrays fulfilled these criteria and the top 200 co-expressed genes, where the co-expression value was limited to >0.8, were extracted and filtered to obtain one representative gene per study following the removal of duplicates. These filtered gene lists were then compared with searches for repeatedly co-expressed genes over multiple studies, with the two studies (>30% of five studies) frequency cut-off. Through this, a meta-analysis list for CRLS1 was generated, and enriched Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed for the terms of the gene lists produced via co-expression data analysis on account of the web-based Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov) (18,19). The Benjamini and Hochberg false discovery rate correction was used to correct the results for multiple testing.
Results
Gene expression of CRLS1 in lung cancer
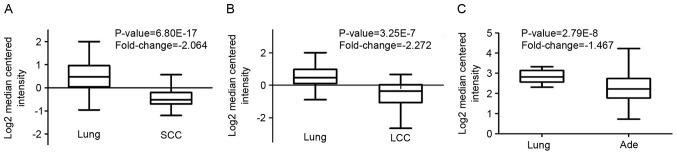
Through Oncomine analysis, the present study investigated the mRNA levels of CRLS1 in human cancer cases. There were more datasets showing a downregulation of CRLS1 than an upregulation of CRLS1 in the various types of tumors, compared with the normal tissues (Table I). This was similar in lung cancer, vs. normal tissues, for which three datasets showed a downregulation of CRLS1 and none showed an upregulation of CRLS1 under the threshold of P=0.05, fold-change=1.5, and gene rank of 10%. The mRNA levels of CRLS1 in squamous cell carcinoma (SCC) and large cell lung cancer (LCC) were lower, compared with those in normal tissues (fold-change, −2.046 and −2.272, respectively; Fig. 1A and B). However, compared with the normal tissues, no changes in the mRNA levels of CRLS1 were found in adenocarcinoma (Ade), with a fold change of −1.467 (Fig. 1C). Taken together, these data confirmed that CTLS1 was downregulated in lung tumors, compared to normal tissues (Table II).
Table I.
Number of datasets correlated with upregulation and downregulation of the CRLS1 gene in cancer vs. normal tissues, determined via oncomine analysis at different P-values.
| Datasets for CRLS1 | ||
|---|---|---|
| P-value | Upregulated (n) | Downregulated (n) |
| 0.05 | 8 | 10 |
| 0.01 | 8 | 10 |
| 1E-03 | 8 | 8 |
| 1E-04 | 5 | 8 |
| 1E-05 | 4 | 8 |
| 1E-06 | 3 | 7 |
| 1E-07 | 2 | 6 |
| 1E-08 | 1 | 6 |
Fold-change=2; gene rank=top 10%. CRLS1, cardiolipin synthase 1.
Figure 1.
CRLS1 gene analysis in lung SCC, Ade and LCC using the Oncomine database. Results were derived from gene expression data in Oncomine comparing expression levels of CRLS1 family genes in normal and cancerous tissue of the lung, plotted using GraphPad Prism software. The y-axis represents the median intensity, 10 and 90th percentile data. (A) Comparison of CRLS1 mRNA expression in SCC. (B) Comparison of CRLS1 mRNA expression in LCC. (C) Comparison of CRLS1 mRNA expression in Ade. SCC, squamous cell carcinoma; Ade, adenocarcinoma; LCC, large cell carcinoma; CRLS1, cardiolipin synthase 1.
Table II.
Gene expression of CRLS1 is elevated in lung cancer, determined using the Oncomine database.
| Sample size (n) | ||||||
|---|---|---|---|---|---|---|
| Comparison | P-value | Fold-change | Dataset | Normal | Cancer | Total |
| Lung Ade vs. normal | 2.79E-8 | −1.567 | Okayama | 20 | 226 | 246 |
| Squamous cell carcinoma vs. normal | 6.80E-17 | −10.564 | Hou | 65 | 27 | 92 |
| Large cell carcinoma vs. normal | 3.25E-7 | −6.603 | Hou | 65 | 19 | 84 |
CRLS1, cardiolipin synthase 1; Ade, adenocarcinoma.
Survival prognosis analysis
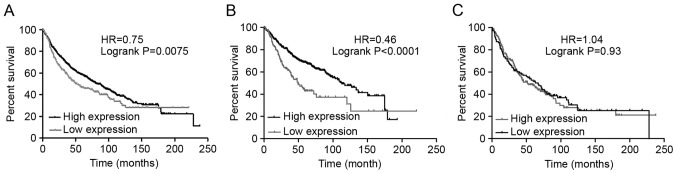
The association between the mRNA expression of CRLS1 and clinical outcomes was examined using the Kaplan-Meier plotter (www.kmplot.com). Survival curves were plotted for all patients with NSCLC (Fig. 2A), Ade (Fig. 2B), and SCC (Fig. 2C). Consequently, a high mRNA expression of CRLS1 was found to be associated with significantly improved OS for all patients with NSCLC [HR=0.75 (0.61–0.93); P=0.0075]. In addition, a high mRNA expression of CRLS1 was found to be associated with significantly improved OS in Ade [HR=0.46 (0.33–0.64); P<0.0001]. However, no statistically significant difference was found in the expression levels in patients with SCC [HR=1.04 (0.76–1.42); P=0.83].
Figure 2.
Prognostic values associated with the expression of CRLS1. Data were analyzed using the Kaplan-Meier Plotter. (A) Survival curves were plotted for all patients with NSCLC (n=1,165). (B) Survival curves were plotted for patients with Ade (n=673). (C) Survival curves were plotted for patients with SCC (n=271). CRLS1, cardiolipin synthase 1; SCC, squamous cell carcinoma; Ade, adenocarcinoma; LCC, large cell carcinoma.
The association between CRLS1 and the clinical characteristics for patients with NSCLC, including sex, smoking status, tumor grade, clinical stage, lymph node status, chemotherapy and radiotherapy, was also investigated. CRLS1 was associated with significantly improved OS in stage I tumors and N0 lymph node status in patients with NSCLC. No significant difference was found in stages II–IV tumors or N1 and N2 lymph node status. CRLS1 was not significantly associated with sex or smoking status in patients with NSCLC. No association between CRLS1 and chemotherapy was observed, as the sample size was too small. The results of the associations are shown in Table III.
Table III.
Correlation between CRLS1 and tumor clinical characteristics of patients with non-small cell lung cancer.
| Characteristic | Cases low (n) | Cases high (n) | HR (95% CI) | P-value |
|---|---|---|---|---|
| Stage | ||||
| I | 224 | 225 | 0.36 (0.26–0.51) | 2.7E-09 |
| II | 80 | 81 | 0.69 (0.43–1.09) | 0.11 |
| III | 22 | 22 | 0.54 (0.27–1.09) | 0.08 |
| Topography | ||||
| I | 112 | 112 | 0.63 (0.42–0.94) | 0.023 |
| II | 95 | 95 | 0.81 (0.55–1.17) | 0.26 |
| III | 14 | 15 | 1.06 (0.47–2.38) | 0.89 |
| Lymph node status | ||||
| N0 | 162 | 162 | 0.72 (0.52–0.98) | 0.037 |
| N1 | 51 | 51 | 0.81 (0.49–1.34) | 0.42 |
| N2 | 16 | 16 | 0.56 (0.27–1.2) | 0.13 |
| Metastasis | ||||
| M0 | 231 | 231 | 0.69 (0.54–0.89) | 0.0037 |
| M1 | – | – | – | – |
| Smoking status | ||||
| Never Smoked | 151 | 149 | 0.51 (0.33–0.78) | 0.0018 |
| Smoked | 70 | 71 | 0.33 (0.13–0.83) | 0.014 |
| Sex | ||||
| Male | 330 | 329 | 0.54 (0.44–0.67) | 8.7E-09 |
| Female | 189 | 186 | 0.45 (0.31–0.65) | 8.6E-06 |
| Chemotherapy | ||||
| Yes | 17 | 17 | 0.41 (0.12–1.34) | 0.13 |
| No | 10 | 10 | 3.94 (0.46–33.95) | 0.18 |
CRLS1, cardiolipin synthase 1.
CRLS1 is co-expressed with genes involved in tumor metabolism and the mitogen-activated protein kinase (MAPK) signaling pathway
Using the Oncomine cancer microarray database, a search was performed of CRLS1 with co-expressed genes. The retrieval flow of the meta-analysis and the selected multi-array studies for CRLS1 are shown in Fig. 3. Following meta-analysis, 10 genes were found as being co-expressed in two or more studies, comprising G protein subunit γ10, membrane associated ring-CH-type finger 7 (MARCH7), MARCH5, DAZ interacting zinc finger protein 1 (DZIP1), selenophosphate synthetase 2 (SEPHS2), Obg-like ATPase 1 (OLA1), SEPHS1, dual-specificity phosphatase 7 (DUSP7), protein phosphatase, Mg2+/Mn2+-dependent 1A (PPM1A) and protein tyrosine phosphatase receptor type R (PPTRR) in five datasets that met the criteria (Jia Liver, Hoek Melanoma 2, Tomida Lung, Kreike Breast, and Fandy Leukemia). Web-based DAVID was used to perform GO term enrichment analysis to obtain the specific functions of the above-mentioned co-expressed genes, which provided a list of gene functions in a gene set. Analysis of the 10 CRLS1-co-expressed genes with the DAVID functional annotation tool (GOTERM_BP/MF_FAT) resulted in eight annotation clusters matching the statistical criteria (P<0.05; count ≥2; fold enrichment >2; Table IV). Subsequently, the above-mentioned DAVID functional annotation tool was used for the identification of putative KEGG pathways associated with the CRLS1-co-expressed genes. Consequently, one pathway associated with seleno-amino acid metabolism and another associated with the MAPK signaling pathway in cancer. These two signaling pathways were significantly enriched with CRLS1-co-expressed genes (P<0.05 and fold enrichment >2; Table V).
Figure 3.
Retrieval flow of the meta-analysis and the selected multi-arrays for CRLS1. CRLS1, cardiolipin synthase 1; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table IV.
Functional enrichment of CRLS1-co-expressed genes.
| Term | Count | % | P-value | Fold-change |
|---|---|---|---|---|
| GO:0004756-selenide, water dikinase activity | 2 | 20 | 1.4E-3 | 1,298.3 |
| GO:0016781-phosphotransferase activity | 2 | 20 | 1.4E-3 | 1,298.3 |
| GO:0006470-protein amino acid dephosphorylation | 3 | 30 | 2.6E-3 | 33.9 |
| GO:0016311-dephosphorylation | 3 | 30 | 3.4E-3 | 29.3 |
| GO:0004721-phosphoprotein phosphatase activity | 3 | 30 | 5.5E-3 | 23.6 |
| GO:0016791-phosphatase activity | 3 | 30 | 1.2E-2 | 15.6 |
| GO:0008430-selenium binding | 2 | 20 | 2.2E-2 | 81.1 |
| GO:0004722-protein serine/threonine phosphatase activity | 2 | 20 | 3.1E-2 | 57.7 |
CRLS1, cardiolipin synthase 1; GO, Gene Ontology.
Table V.
Pathway-based enrichment of CRLS1-co-expressed genes.
| Term | Count | % | P-value | Fold-change |
|---|---|---|---|---|
| Hsa04010:MAPK signaling pathway | 3 | 30 | 2.5E-2 | 9.5 |
| Hsa00450:Selenoamino acid metabolism | 2 | 20 | 3.8E-2 | 65.2 |
CRLS1, cardiolipin synthase 1.
CRLS1 is differentially expressed in various types of cancer
The expression of CRLS1 in various types of cancer was investigated using publicly available gene expression data from Oncomine, the results of which are shown in Table VI.
Table VI.
Differential expression of CRLS1 in cancer tissue, compared with their normal tissue via the Oncomine cancer microarray database.
| Type of cancer | Over expressed | Under expressed | Dataset count |
|---|---|---|---|
| Brain tumor | + | + | 4/1 |
| Leucocythemia | + | 5 | |
| Lung cancer | + | 2 | |
| Pancreatic carcinoma | + | 1 | |
| Lymphoma | + | 1 | |
| Tongue cancer | + | 1 | |
| Mouth floor carcinoma | + | 1 |
CRLS1, cardiolipin synthase 1.
Discussion
Warburg first suggested that the prime cause of cancer was impaired energy metabolism, which involved irreversible injury to cellular respiration (20,21). The majority of normal mammalian cells achieve a constant delta G' of ATP hydrolysis of ~257 kJ/mol as energy required for normal life activities through respiration (22,23). By contrast, tumor cells maintain this level through a combination of respiration and glycolysis (24,25). Elevated glycolysis is a metabolic characteristic of almost all types of tumor, which can be observed through the use of labeled glucose analogs (26,27). Structural and biochemical abnormalities that have occurred in tumor cell mitochondria can compromise oxidative respiratory function. Several studies have assessed the lipid composition of tumor mitochondria (28,29), however, few have evaluated the content and the composition of CL in tumor mitochondria, which is a complex mitochondrial-specific phospholipid regulating numerous enzyme activities, particularly those associated with oxidative phosphorylation and coupled respiration (30,31). Kiebish et al investigated the content and composition of CL for the first time in purified mitochondria from mouse brain tumors. The results showed that the composition and content of CL in mouse brain tumor mitochondria differed distinctly from that derived from syngeneic host normal brain tissue. The mouse brain tumors contained CL abnormalities, which were unique to each tumor type. In addition, these CL abnormalities were associated with significant hypofunction in the electron transport chain (ETC). Taken together, the results suggested that abnormal CL underlies irreversible respiratory injury in tumors, which further supports the importance of CL in maintaining the structural integrity of the inner mitochondrial membrane and lined mitochondrial lipid defects to the Warburg theory of tumorigenesis (30).
CRLS1, also known as C20 or f155, is the human candidate gene for CL synthase, is localized in the mitochondria and is expressed at high levels in tissues, including the heart, skeletal muscle, lung and liver, belonging to the CDP-alcohol phosphatidyltransferase class-I family. It predominantly catalyzes the reversible phosphatidy l group transfer from one phosphatidylglycerol molecule to another to form CL and glycerol. As a result of these findings, the present study hypothesized that the reduction of CRLS1 caused by various factors contributes either directly or indirectly to abnormalities in CL synthesis or remodeling, and leads to damage to mitochondrial intima integrity and hypofunction in ETC activities. This suggests a reliance on glycolysis for cell survival, eventually resulting in the occurrence and development of tumors. In addition to the enzyme-mediated inhibition of CL synthesis, inherited mutations, a variety of epigenetic causes and environmental insults, including necrosis, hypoxia, ischemia, dietary imbalances and reactive oxygen species, can also produce mitochondrial defects to cause CL abnormalities (31,32), contributing to tumor initiation and progression. However, enzyme-mediated CL synthesis may be the most direct element and was investigated as one of the primary aims of the present study. In addition, based-wed DAVID GO term enrichment analysis showed that CRLS1-co-expressed genes were involved in phosphoprotein phosphatase activity, protein amino acid dephosphorylation, protein serine/threonine phosphatase activity and phosphate metabolic process. According to the co-expression analysis theory, which is based on hierarchical clustering in which groups of genes may have similar functions if they have similar expression patterns under a given pathological condition, it is possible to examine whether CRLS1 possesses putative and analogical functional activities with its co-expressed genes. These functional abnormalities caused by abnormalities in lipid metabolism may also be complementary contributors in tumorigenesis.
In previous years, studies have found that oncogene activation or tumor suppressor gene inactivation can regulate metabolic homeostasis in order to promote or inhibit cancer. Furthermore, certain metabolic enzymes can be used as oncogenes or tumor suppressors involved in the process of cancer progression. Members of the MAPK family, including the extracellular signal-regulated protein kinase (ERK) pathway, c-Jun N-terminal kinase (JNK) pathway, p38 pathway and ERK5 pathway, have been reported to be important in cell proliferation and apoptosis (33). From the extracellular stimulus to the cells to the corresponding biological effects, the MAPK signal transduction pathway involves a signaling cascade of multistage protein kinases, including three key kinases, MAPK, MAPKK and MAPKKK (34). The analysis of KEGG pathways via DAVID showed that CRLS1 may be involved in the MAPK signaling pathway as it is co-expressed with PPM1A, protein tyrosine phosphatase receptor type R protein (PTPRR) and DUSP7. PPM1A is a phosphatase belonging to the PP2C family, which dephosphorylates and inactivates a broad range of substrates, including MAPKs, p38 and JNK, and is involved in tumor suppression (35,36), Su et al (37) characterized the tumor suppressor function of PTPRR, particularly its role in metastasis via the MAPK signaling pathway, in a cervical cancer model. Taken together, it was suggested that PTPRR affects malignant phenotypes by dephosphorylating substrates, including ERK5 and the p38 pathway, which has also been observed in other species (38). However, the extent of kinases targeted by PTPRR in NSCLC has not been investigated. DUSPs show marked substrate preferences for specific MAPKs, which have been reported to limit the duration of MAPK phosphorylation and reduce microglial activation (39). Sharing a substrate preference for ERK1/2 may be categorized as inducible or constitutive according to their expression. Inducible DUSPs dephosphorylate JNK, p38 and ERK, whereas constitutive DUSPs are specific for ERK (40,41). Several studies of DUSPs in tumors have focused on constitutive and inducible DUSP7, which downregulates p38 and ERK in major inflammatory pathways (42,43). To date, few studies have reported on the roles of DUSP6 and other DUSPs in microglia.
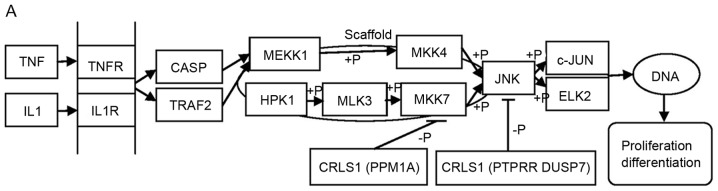
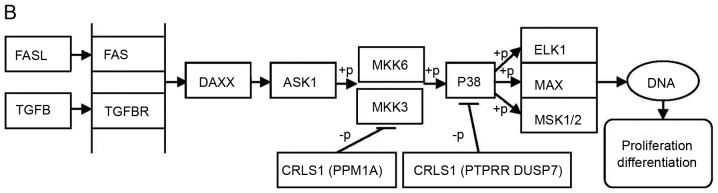
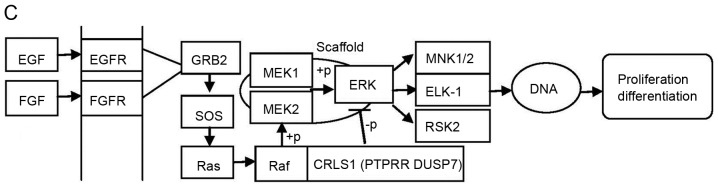
The MAPK pathway consists of a three-tiered kinase cascade and the sequential phosphorylation of each kinase causes the activation of effector kinases JNK, p38, and ERK1/2. Three classes of protein phosphatases modulate the activities of upstream kinases: Type-2 Ser/Thr phosphatases (PP2A) including PPM1A, PTPs including PTPRR, and dual-specificity (Thr/Tyr) phosphatases (DUSPs), including DUSP7 (42). Using the DAVID functional annotation chart, several subclasses of MAPK signaling pathways may reveal the role of CRLS1 in tumorigenesis based on the co-expression analysis theory. The JNKs are members of the MAPK family and are activated by MAP2 kinases MKK-4 and MKK-7. This subsequently translocates to the nucleus where it can regulate the activity of multiple transcription factors. At the post-translational level, JNK enhances its transcriptional activity by double-phosphorylating Ser-63 and Ser-73 sites in the c-jun activation region, and eventually affects cell cycle, apoptosis, cell stress, and other physiological and pathological processes (44). According to the KEGG pathways and co-expression patterns identified in the present study, the putative CRLS1 JNK/c-Jun MAPK signaling pathway was demonstrated (Fig. 4A). CRLS1 prevents the phosphorylation of MKK-4 and MKK-7 by dephosphorylation and reducing its transcription by c-jun by suppressing the activation of JNK. The p38 signaling pathway (45) is activated through phosphorylation by the upstream MKK-3, which then translocates into the nucleus to interact with downstream protein kinases, chromosome remodeling and transcription factors, including mitogen-and stress-activated protein kinase 1/2 (MSK1/2), MYC associated factor X (MAX) and ETS transcription factor (ELK-1). Similarly, the putative CRLS1 p38 MAPK signaling pathway indicates the role of CRLS1 in tumors (Fig. 4B). CRLS1 inhibits the phosphorylation of MKK-3 and p38 by dephosphorylation, and reduces the transcription of MSK1/2, MAX/GADD153 and ELK-1 by suppressing the activation of p38. ERKs were the first subclass to be investigated and are also the most important members of the MAPK family (46). The ERK signaling pathway is activated by growth factors and mitotic stimuli, which are closely associated with cell proliferation, differentiation and transcription. ERK is activated continuously, and ultimately promotes cell proliferation and malignant transformation. The ERK cascade contains a typical three-level MAPK sequence activation process. Raf belongs to the MAPKKK family, which phosphorylates and activates MEK1/2 (MAPKK), and subsequently activates ERK1/2 by double phosphorylation. The same reaction model can also be applied to the CRLS1 ERK MAPK signaling pathway (Fig. 4C). CRLS1 prevents the phosphorylation of ERK by dephosphorylation, and reduces the transcription of MAP kinase-interacting kinase 1/2 and Elk-1 by suppressing the activation of ERK.
Figure 4.
MAPK signal pathway involved in CRLS1 co-expression genes by KEGG pathway analysis. (A) JNK/c-JUN MAPK signaling pathway. (B) p38 MAPK signaling pathway. (C) ERK MAPK signaling pathway. Arrows indicate promotion. T-shaped arrows represent an inhibitory effect. Genes in brackets represent CRLS1 co-expression genes. +p, phosphorylation; -p, dephosphorylation; CRLS1, cardiolipin synthase 1; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; JNK, c-Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; TNF, tumor necrosis factor; IL1, interleukin 1; TNFR, TNF receptor; CASP, caspase; TRAF2, TNFR-associated factor 2; MEKK1, MAPK kinase kinase kinase 1; HPK1, hematopoietic progenitor kinase 1; MLK3, mixed lineage kinase 3; MKK, MPAK kinase.
In terms of seleno-amino acid metabolism, KEGG pathways analysis via DAVID revealed that SEPHS1/2 is a CRLS1-co-expression gene involved in tumorigenesis. In the 1970s and 1980s, clinical data was collected examining the correlation between selenomethionine (Se-Met) and cancer incidence in various tumor types, including lung, skin and colon cancer (47). Several studies have identified selenium and Se-Met as potential chemopreventive agents against prostate cancer (48,49). SEPHS1/2 belongs to the selenophosphate synthetase 1 family, class II subfamily, which predominantly synthesizes selenophosphate from selenide and ATP (catalytic activity: ATP + selenide + H2O=AMP + selenophosphate+phosphate). Studies have also shown that SEPHS affects cell viability upon ionization radiation through the modulation of p53, which further suggests that SEPHS and its reaction product selenophosphate may be involved in inhibiting cancer progression in a p53-dependent manner and offers potential as a novel anticancer measure (50,51). However, the antitumor mechanisms of selenophosphate and SEPHS described above have not been validated in the field of lung cancer. By contrast, the co-expression analysis theory indicates a similar role for CRLS1, which may be confirmed by further investigations of NSCLC-related seleno-amino acid metabolism and the p53 pathway.
In conclusion, using in silico approaches, the differential expression of CRLS1 in NSCLC and normal tissue was assessed in the present study, which revealed that the expression level of CRLS1 in lung cancer was lower, compared with that in normal lung tissues. In addition, a high expression of CRLS1 was found to be associated with improved OS in all patients with NSCLC and lung Ade. The meta-analysis for CRLS1-related co-expression analysis revealed that this molecule may be important for tumor-associated lipid metabolic processes, and its anti-oncogene position was also assessed, involving the MAPK signaling pathway. The above conclusions were obtained using various tools, software and large-scale databases, notably in silico rather than wet laboratory data. The results of the in silico experiments generally require confirmation in laboratory experiments. However, this remains an effective experimental approach for novel developments in understanding carcinogenesis and makes it convenient for further wet laboratory investigations.
References
- 1.van der Knaap JA, Verrijzer CP. Undercover: Gene control by metabolites and metabolic enzymes. Genes Dev. 2016;30:2345–2369. doi: 10.1101/gad.289140.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saadi H, Seillier M, Carrier A. The stress protein TP53INP1 plays a tumor suppressive role by regulating metabolic homeostasis. Biochimie. 2015;118:44–50. doi: 10.1016/j.biochi.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Hostetler K. Chapter 6 polyglycerophospholipids: Phosphatidylglycerol, diphosphatidylglycerol and bis, (monoacylglycero) phosphate. New Compr Biochem. 1982;4:215–261. doi: 10.1016/S0167-7306(08)60010-1. [DOI] [Google Scholar]
- 4.Hatch GM. Cell biology of cardiac mitochondrial phospholipids. Biochem Cell Biol. 2004;82:99–112. doi: 10.1139/o03-074. [DOI] [PubMed] [Google Scholar]
- 5.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 6.Garrido C, Galluzzi L, Brunet M, Puig PE, Didelot C, Kroemer G. Mechanisms of cytochrome C release from mitochondria. Cell Death Differ. 2006;13:1423–1433. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- 7.Ott M, Zhivotovsky B, Orrenius S. Role of cardiolipin in cytochrome c release from mitochondria. Cell Death Differ. 2007;14:1243–1247. doi: 10.1038/sj.cdd.4402135. [DOI] [PubMed] [Google Scholar]
- 8.Hostetler KY, Van den Bosch H, Van Deenen LL. Biosynthesis of cardiolipin in liver mitochondria. Biochim Biophys Acta. 1971;239:113–119. doi: 10.1016/0005-2760(71)90201-3. [DOI] [PubMed] [Google Scholar]
- 9.Lu B, Xu FY, Jiang YJ, Choy PC, Hatch GM, Grunfeld C, Feingold KR. Cloning and characterization of a cDNA encoding human cardiolipin synthase (hCLS1) J Lipid Res. 2006;47:1140–1145. doi: 10.1194/jlr.C600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Houtkooper RH, Akbari H, van Lenthe H, Kulik W, Wanders RJ, Frentzen M, Vaz FM. Identification and characterization of human cardiolipin synthase. FEBS Lett. 2006;580:3059–3064. doi: 10.1016/j.febslet.2006.04.054. [DOI] [PubMed] [Google Scholar]
- 11.Abdollahi A, Omranipour R. Is increase of homocysteine, anti-cardiolipin, anti-phospholipid antibodies associated with breast tumors? Acta Med Iran. 2015;53:681–685. [PubMed] [Google Scholar]
- 12.Schvartsman G, Ferrarotto R, Massarelli E. Checkpoint inhibitors in lung cancer: Latest developments and clinical potential. Ther Adv Med Oncol. 2016;8:460–473. doi: 10.1177/1758834016661164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santarpia M, Giovannetti E, Rolfo C, Karachaliou N, González-Cao M, Altavilla G, Rosell R. Recent developments in the use of immunotherapy in non-small cell lung cancer. Expert Rev Respir Med. 2016;10:781–798. doi: 10.1080/17476348.2016.1182866. [DOI] [PubMed] [Google Scholar]
- 14.Cufer T, Ovcaricek T, O'Brien ME. Systemic therapy of advanced non-small cell lung cancer: Major-developments of the last 5-years. Eur J Cancer. 2013;49:1216–1225. doi: 10.1016/j.ejca.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated Data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyorffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirlaw JT. The metabolism of tumors. Br Med J. 1931;1:74. doi: 10.1136/bmj.1.3653.74-a. [DOI] [Google Scholar]
- 21.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 22.Veech RL. The therapeutic implications of ketone bodies: The effects of ketone bodies in pathological conditions: Ketosis, ketogenic diet, redox states, insulin resistance and mitochondrial metabolism. Prostaglandins Leukot Essent Fatty Acids. 2004;70:309–319. doi: 10.1016/j.plefa.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Seyfried TN, Mukherjee P. Targeting energy metabolism in brain cancer: Review and hypothesis. Nutr Metab. 2005;2:30. doi: 10.1186/1743-7075-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 25.Galarraga J, Loreck DJ, Graham JF, DeLaPaz RL, Smith BH, Hallgren D, Cummins CJ. Glucose metabolism in human gliomas: Correspondence of in situ and in vitro metabolic rates and altered energy metabolism. Metab Brain Dis. 1986;1:279–291. doi: 10.1007/BF00999357. [DOI] [PubMed] [Google Scholar]
- 26.Baggetto LG, Clottes E, Vial C. Low mitochondrial proton leak due to high membrane cholesterol content and cytosolic creatine kinase as two features of the deviant bioenergetics of Ehrlich and AS30-D tumor cells. Cancer Res. 1992;52:4935–4941. [PubMed] [Google Scholar]
- 27.Bergelson LD, Dyatlovitskaya EV, Sorokina IB, Gorkova NP. Phospholipid compositon of mitochondria and microsomes from regenerating rat liver and hepatomas of different growth rate. Biochim Biophys Acta. 1974;360:361–365. doi: 10.1016/0005-2760(74)90067-8. [DOI] [PubMed] [Google Scholar]
- 28.Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 29.Hoch FL. Cardiolipins and biomembrane function. Biochim Biophys Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 30.Kiebish MA, Han X, Cheng H, Chuang JH, Seyfried TN. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: Lipidomic evidence supporting the Warburg theory of cancer. J Lipid Res. 2008;49:2545–2556. doi: 10.1194/jlr.M800319-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy S, El-Assaad W, Przybytkowski E, Joly E, Prentki M, Langelier Y. Saturated fatty acid-induced apoptosis in MDA-MB-231 breast cancer cells. A role for cardiolipin. J Biol Chem. 2003;278:31861–31870. doi: 10.1074/jbc.M300190200. [DOI] [PubMed] [Google Scholar]
- 32.Mcmillin JB, Dowhan W. Cardiolipin and apoptosis. Biochim Biophys Acta. 2002;1585:97–107. doi: 10.1016/S1388-1981(02)00329-3. [DOI] [PubMed] [Google Scholar]
- 33.Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: An update. Arch Toxicol. 2015;89:867–882. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
- 34.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Zhou Z, Lin H, Lv X, Fu J, Lin P, Zhu C, Wang H. Protein phosphatase 1A (PPM1A) is involved in human cytotrophoblast cell invasion and migration. Histochem Cell Biol. 2009;132:169–179. doi: 10.1007/s00418-009-0601-5. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Gong Z, Pan C, Xie DD, Tang JY, Cui M, Xu YF, Yao W, Pang Q, Xu ZG, et al. Metal-dependent protein phosphatase 1A functions as an extracellular signal-regulated kinase phosphatase. FEBS J. 2013;280:2700–2711. doi: 10.1111/febs.12275. [DOI] [PubMed] [Google Scholar]
- 37.Su PH, Lin YW, Huang RL, Liao YP, Lee HY, Wang HC, Chao TK, Chen CK, Chan MW, Chu TY, et al. Epigenetic silencing of PTPRR activates MAPK signaling, promotes metastasis and serves as a biomarker of invasive cervical cancer. Oncogene. 2013;32:15–26. doi: 10.1038/onc.2012.29. [DOI] [PubMed] [Google Scholar]
- 38.Blanco-Aparicio C, Torres J, Pulido R. A novel regulatory mechanism of MAP kinases activation and nuclear translocation mediated by PKA and the PTP-SL tyrosine phosphatase. J Cell Biol. 1999;147:1129–1136. doi: 10.1083/jcb.147.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- 40.Bermudez O, Pagès G, Gimond C. The dual-specificity MAP kinase phosphatases: Critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299:C189–C202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 41.Caunt CJ, Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs): Shaping the outcome of MAP kinase signalling. FEBS J. 2013;280:489–504. doi: 10.1111/j.1742-4658.2012.08716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ham JE, Oh EK, Kim DH, Choi SH. Differential expression profiles and roles of inducible DUSPs and ERK1/2-specific constitutive DUSP6 and DUSP7 in microglia. Biochem Biophys Res Commun. 2015;467:254–260. doi: 10.1016/j.bbrc.2015.09.180. [DOI] [PubMed] [Google Scholar]
- 43.Huo Y, Rangarajan P, Ling EA, Dheen ST. Dexamethasone inhibits the Nox-dependent ROS production via suppression of MKP-1-dependent MAPK pathways in activated microglia. BMC Neurosci. 2011;12:49. doi: 10.1186/1471-2202-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeke A, Misheva M, Reményi A, Bogoyevitch MA. JNK signaling: Regulation and functions based on complex protein-protein partnerships. Microbiol Mol Biol Rev. 2016;80:793–835. doi: 10.1128/MMBR.00043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segalés J, Perdiguero E, Muñoz-Cánoves P. Regulation of muscle stem cell functions: A focus on the p38 MAPK signaling pathway. Front Cell Dev Biol. 2016;4:91. doi: 10.3389/fcell.2016.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 47.Schrauzer GN, White DA, Schnieder CJ. Cancer mortality correlation studies-III: Statistical associations with dietary selenium intakes. Bioinorg Chem. 1997;7:23–31. doi: 10.1016/S0006-3061(00)80126-X. [DOI] [PubMed] [Google Scholar]
- 48.Willis MS, Wians FH. The role of nutrition in preventing prostate cancer: A review of the proposed mechanism of action of various dietary substances. Clin Chim Acta. 2003;330:57–83. doi: 10.1016/S0009-8981(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 49.Nyman DW, Stratton M Suzanne, Kopplin MJ, Dalkin BL, Nagle RB, Jay Gandolfi A. Selenium and selenomethionine levels in prostate cancer patients. Cancer Detect Prev. 2004;28:8–16. doi: 10.1016/j.cdp.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Morey M, Corominas M, Serras F. DIAP1 suppresses ROS-induced apoptosis caused by impairment of the selD/sps1 homolog in Drosophila. J Cell Sci. 2003;116:4597–4604. doi: 10.1242/jcs.00783. [DOI] [PubMed] [Google Scholar]
- 51.Chung HJ, Yoon SI, Shin SH, Koh YA, Lee SJ, Lee YS, Bae S. p53-Mediated enhancement of radiosensitivity by selenophosphate synthetase 1 overexpression. J Cell Physiol. 2006;209:131–141. doi: 10.1002/jcp.20714. [DOI] [PubMed] [Google Scholar]