Abstract
Inhibiting the protein neddylation pathway using the NEDD8-activating enzyme inhibitor MLN4924 represents an attractive anticancer strategy having been demonstrated to exhibit promising anticancer efficacy and with tolerable levels of toxicity; however, the mechanism by which MLN4924 inhibits cell proliferation in human esophageal squamous cell carcinoma (ESCC) cells requires further investigation. The present study revealed that MLN4924 treatment led to G2 cell cycle arrest and enhanced the protein stability of Wee1-like protein kinase and cyclin dependent protein kinase inhibitor 1A and B and p27. Furthermore, MLN4924 induced DNA damage and sensitized esophageal cancer cells to cisplatin by enhancing apoptosis. These findings extend the understanding of the function and mechanism of MLN4924 in ESCC and provide further evidence for the future development of neddylation inhibitors in clinical trials of esophageal cancer therapy, either alone or in combination.
Keywords: esophageal squamous cell carcinoma, neddylation, MLN4924, Cullin-RING ligase
Introduction
Esophageal squamous cell carcinoma (ESCC) is a prominent subtype of esophageal cancer and has the fourth-highest incidence of cancer-associated mortality in China, 2015 (1). Besides surgery, the current treatment options for esophageal cancer include chemoradiotherapy and chemotherapy, which is often accompanied by high toxicity and acquired therapeutic resistance (2). It is therefore urgent to identify novel drugs and regimens for the treatment of esophageal cancer.
Protein neddylation is a novel regulatory post-translational modification pathway of, in which ubiquitin-like molecule neural precursor cell expressed, developmentally downregulated 8 (NEDD8) was added to substrate proteins via a sequential enzymatic process, which includes the NEDD8-activating enzyme E1 (NAE), NEDD8-conjugating enzyme E2 and substrate-specific NEDD8-E3 ligases (3–5). The best-characterized substrates of neddylation are cullin family proteins (6), which are important skeleton protein of Cullin-RING E3 ligases (CRLs) (6). Previous studies demonstrated that the hyperactivated neddylation pathway and abnormally activated CRL E3 ligases serve important functions in disease progression, including poorer patient overall survival rates (7–11). These results indicate that the neddylation pathway is a potential anticancer target.
MLN4924 has been shown to function as a potent and selective inhibitor of NEDD8-activating enzyme (NAE) (3); previous studies revealed that MLN4924 inhibits cancer cell growth in vitro and in vivo by inactivating CRLs and causing the accumulation of substrates (12–16). Previously, MLN4924 exhibited attractive pharmacodynamic effects in phase I studies in patients with advanced solid tumors, relapsed/refractory lymphoma and metastatic melanoma for its tolerable safety profile (17–19). MLN4924 may function as a novel chemosensitizer or radiosensitizer in multiple types of cancer (20–24). These results indicate that targeting the neddylation pathway may represent an attractive anticancer strategy, either alone or in combination with other therapies.
To further explore the effect and mechanism of MLN4924 on esophageal cancer cell growth alone or in combination with cisplatin (CDDP), a standard chemotherapy drug for patients with esophageal cancer, the cell cycle arrest induced by MLN4924 treatment and the stability of cell-cycle associated proteins, including cyclin-dependent kinase inhibitor 1 (CDKN1A; also known as p21), CDKN1B (also known as p27) and Wee1-like protein kinase (Wee1) were investigated. In addition, MLN4924 and CDDP have been demonstrated to induce DNA damage, therefore the combinational effect on DNA damage and apoptosis was also examined.
Materials and methods
Cell lines, culture and reagents
Human ESCC EC1 and Kyse450 cell lines were kindly gifted from Professor Zhao (Zhengzhou University, Zhengzhou, China) and cultured in Dulbecco's modified Eagle's medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Biochrom, Ltd., Cambridge, UK) at 37°C with 5% CO2. MLN4924 was synthesized and prepared as previously described (7,25).
Cell viability
Cell viability was detected using an ATPLite Luminescence Assay kit (PerkinElmer, Inc., Waltham, MA, USA) as previously described (7,25). Briefly, cells were seeded in 96-well plates (3×103 cells/well) and treated with dimethyl sulfoxide (DMSO; 0.1%) or MLN4924 (0.0, 0.1, 0.2, 0.3, 0.4, 0.6, 0.8, 1.0 µM). Cell proliferation was determined at 24, 48 and 72 h with indicated concentration (0.0, 0.1, 0.2, 0.3, 0.4, 0.6, 0.8, 1.0 µM) using the assay kit according to the manufacturer's instructions.
Clonogenic assay
A total of 500 cells were seeded into 6-well plates in triplicate, treated with DMSO (0.1%), MLN4924 (0.05 µM), CDDP (MedChem Express Co., Ltd., Shanghai, China) (1 µg/ml) or MLN4924 (0.05 µM) + CDDP (1 µg/ml) and then incubated for 12 days. The colonies were fixed with 4% paraformaldehyde for 30 min at room temperature and stained with crystal violet for 30 min at room temperature. The colonies were counted under light field of inverted microscope at ×100 magnification, and captured using Gel Doc™ XR+ Gel Documentation System (Bio-Rad Laboratories, Inc., Shanghai, China). Colonies comprising 50 cells or more were counted.
Cell cycle analysis
Cells treated with MLN4924 at indicated concentrations were harvested, fixed in 70% ethanol at −20°C and then stained with 50 µg/ml propidium iodide (PI) containing 30 µg/ml RNase A (both from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C for 30 min and analyzed for cell-cycle profile by flow cytometry (Becton Dickinson FACScan; Becton-Dickinson, San Jose, CA, USA). Data were analyzed with ModFit LT 3.1 software (Verity Software House, Inc., Topsham, ME, USA).
Western blotting
EC1 and Kyse450 were treated with MLN4924 (0.1, 0.3, 0.6 µM) or DMSO (0.1%) for 72 h. Then the protein was extracted using radioimmunoprecipitation buffer (Beyotime Institute of Biotechnology, Jiangsu, China) and protein concentration was determined using BCA kit (Beyotime Institute of Biotechnology). When detected, 50 µg proteins were loaded per lane with 2% SDS. The proteins were transferred to polyvinylidene fluoride membrane and blocked with 5% skim milk for 2 h at room temperature. The protein was detected using antibodies against p21 (cat. no. 2947), p27 (cat. no. 3686), Wee1 (cat. no. 4936), cyclin B (cat. no. 12231), Phospho-Histone H3 (Ser10; cat. no. 3377), origin recognition complex (ORC1; cat. no. 4731), DNA replication factor Cdt1 (CDT1; cat. no. 8064), serine/threonine-protein kinase Chk1 (CHK1; cat. no. 2360), CHK2 (cat. no. 2662), p-histone H2AX at Ser139 (γH2AX; cat. no. 9718), cleaved caspase-3 (cat. no. 9664), cleaved poly(ADP-ribose) polymerase (PARP; cat. no. 5625), GAPDH (cat. no. 2118) (all from Cell Signaling Technology, Inc., Danvers, MA, USA) and p-CHK1 (cat. no. ab58567), p-CHK2 (cat. no. ab59408) (both from Abcam, Cambridge, UK). All the primary antibodies were diluted in 1:1,000 and incubated at 4°C overnight. Secondary antibodies peroxidase-conjugated goat anti-mouse IgG (cat. no. ZB-2305) and peroxidase-conjugated goat anti-rabbit IgG (cat. no. ZB-2301) were purchased from ZGSB-Bio, Inc., (Beijing, China). The membrane was incubated with according secondary antibodies (1:3,000) for 2 h at room temperature. Then the membrane was detected using an ECL Kit (Beyotime Institute of Biotechnology).
Apoptosis detection
Cells were treated with the indicated concentration of MLN4924 (0.3 µM), CDDP (1.6 µg/ml) and MLN4924 (0.3 µM) + CDDP (1.6 µg/ml) for 72 h. Apoptosis was determined with the Annexin V-Fluorescein Isothiocyanate/PI Apoptosis kit and CaspGLOW Fluorescein Active Caspase-3 Staining kit (both from BioVision, Inc., Milpitas, CA, USA), according to the manufacturer's instructions. Using Annexin V-Fluorescein Isothiocyanate/PI, apoptosis and the activity of caspase-3 were detected using a flow cytometer, and results were analyzed using Summit 6.1 software (BD Biosciences, Franklin Lakes, NJ, USA).
Immunofluorescence staining
EC1 and Kyse450 cells were treated with MLN4924 using 0.0, 0.3, 0.6 or 1.0 µM and fixed with 4% paraformaldehyde for 30 min at room temperature, permeabilized using 0.2% Triton X-100 for 10 min at room temperature, and incubated with γH2AX primary antibody (cat. no. 9718; 1:200) at 4°C overnight and Alexa Fluor-488 goat anti-rabbit immunoglobulin G (H+L) secondary antibody (cat. no. A0423; 1:500) (Beyotime Institute of Biotechnology) for 2 h at room temperature. The nuclei were stained with DAPI (Beyotime Institute of Biotechnology). Images were captured using a fluorescence microscope (magnification, ×200; Olympus BX-51; Olympus Corp., Tokyo, Japan).
Cycloheximide (CHX) chase assay
Kyse450 and EC1 cells were pretreated with 0.6 µM MLN4924 for 18 h to increase the basal protein level of proteins. Cells were then washed with PBS to remove residual drug and divided into 2 groups, which were further treated with 50 µg/ml CHX (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) or CHX+MLN4924 (0.6 µM) for indicated times and then collected for western blot analysis, as aforementioned.
Statistical analysis
For Figs. 1A, 3B and 4A, the statistical significance of differences between groups was assessed using SPSS 17 software (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance followed by a least-significant difference post hoc test was used to compare between multiple groups. For Fig. 4B, the statistical significance of differences between CDDP and MLN+CDDP was assessed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA. Unpaired Student's t-test was used for the comparison of parameters between groups. P<0.05 was considered to indicate a statistically significant difference. Data is presented as the mean ± standard.
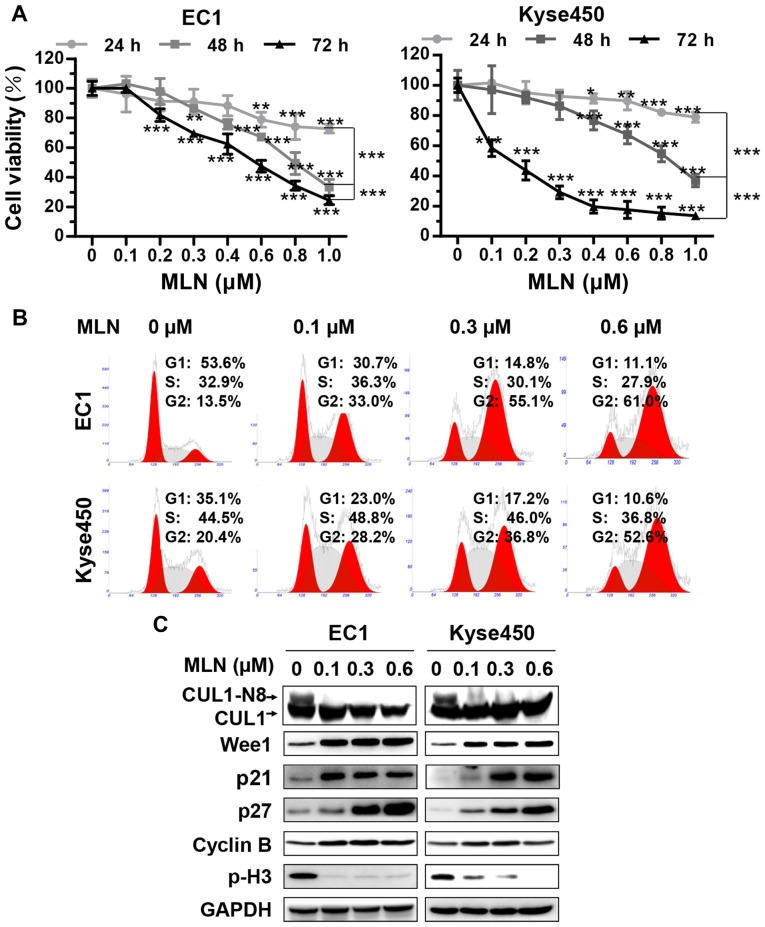
Figure 1.
MLN triggered G2 cell cycle arrest in esophageal cancer cells. (A) MLN suppressed the proliferation of esophageal cancer cells. Cells were seeded on a 96-well plate and treated with MLN at indicated concentrations. Cell viability was measured at the time-points indicated using the ATPLite assay. *P<0.05, **P<0.01, ***P<0.001 vs. 0 µM unless marked otherwise; n=3. Error bars, standard deviation. (B) MLN induced G2/M cell cycle arrest. EC1 and Kyse450 cells were treated with MLN at different concentration for 24 h, followed by propidium iodide staining and FACS analysis for cell cycle profile. (C) MLN induced accumulation of Wee1, p21, p27, cyclin B and decrease of p-H3. EC1 and Kyse450 cells were treated with MLN at indicated concentration for 72 h, and subjected to western blotting. GAPDH served as a loading control. p21, cyclin-dependent kinase inhibitor 1A; p27, cyclin-dependent kinase inhibitor 1B; Wee1, Wee1-like protein kinase; CHX, cyclohexamide; MLN, MLN4924.
Figure 3.
Treatment with MLN induced DNA damage. γH2AX foci were determined by immunofluorescence. EC1 and Kyse450 cells were treated with MLN at indicated concentrations. γH2AX foci were determined by immunofluorescence. (A) Representative images; and (B) statistical results. Cells with more than 10 foci were defined as the positive. ***P<0.001 vs. 0 µM; n=3. Error bars, standard deviation. (C) The expression of γH2AX and DNA damage-associated proteins were determined by western blotting. EC1 and Kyse450 cells were treated with MLN at indicated concentrations for 72 h. Cell extracts were subjected to western blotting. GAPDH served as a loading control. γH2AX, phosphorylated histone H2AX; MLN, MLN4924; ORC1, origin recognition complex 1; CDT1, DNA replication factor Cdt1; tCHK1, total serine/threonine-protein kinase Chk1; pCHK1; phosphorylated CHK1.
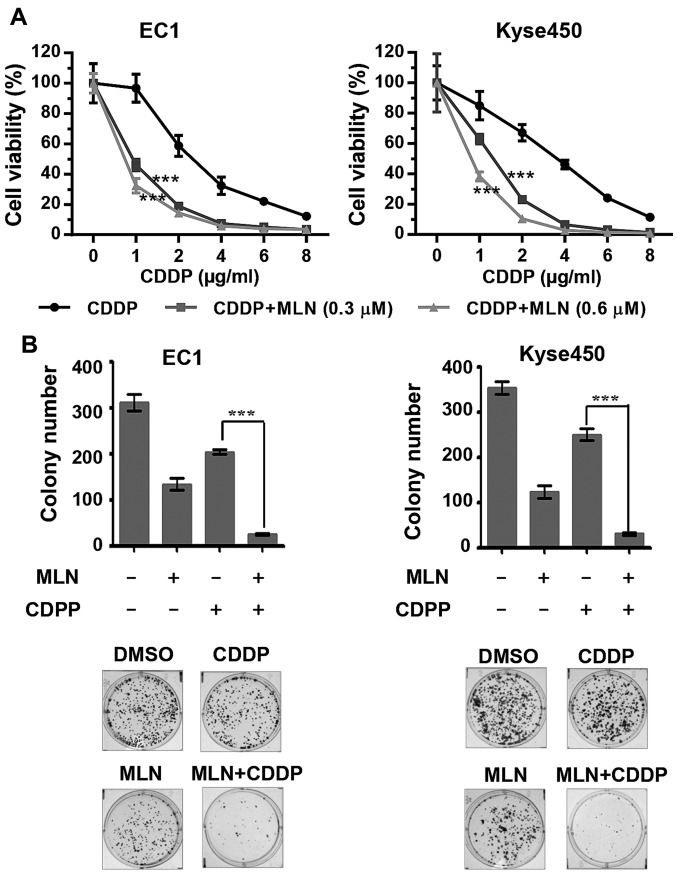
Figure 4.
MLN enhances cytotoxicity of CDDP. (A) EC1 and Kyse450 cells were treated with MLN and different concentrations of CDDP for 72 h. Cell viability was measured at the indicated time-points using the ATPLite assay kit. The data was normalized using the mean of DMSO in the CDDP group and MLN in the MLN+CDDP group. Analysis of variance and least-significant difference post hoc test were used for the comparison of parameters between CDDP and CDDP+MLN (0.3 µM) or CDDP+MLN (0.6 µM). (B) The combination of CDDP with MLN suppressed colony formation in esophageal cancer cells. The results of statistical analysis are shown in the upper panels and representative images are shown in the lower panels. ***P<0.001 CDDP vs. CDDP+MLN; n=3. Error bars, standard deviation. CDDP, cisplatin; DMSO, dimethyl sulfoxide; MLN, MLN4924.
Results
Targeting neddylation by MLN4924 inhibited the proliferation of ESCC cells
To assess the anticancer function of MLN4924 on esophageal cancer cells, the present study assessed the effect of MLN4924 on the proliferation of the ESCC EC1 and Kyse450 cell lines by ATPLite cell viability assay. MLN4924 inhibited the cell proliferation of the 2 cell lines in a time- and dose-dependent manner (Fig. 1A), which indicated that MLN4924 could serve as an attractive anticancer drug for patients with ESCC.
Neddylation inhibition with MLN4924 triggered G2 cell cycle arrest of ESCC cells
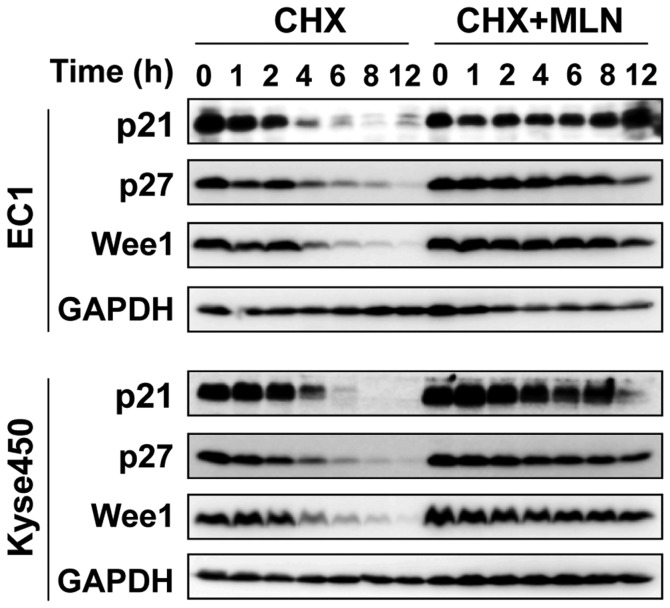
To investigate the mechanism of cell growth inhibition, the effect of MLN4924 on cell cycle progression was evaluated using PI staining. MLN4924 induced a marked G2-M cell cycle arrest in the 2 treated cell lines in a dose-dependent manner (Fig. 1B). MLN4924 also induced accumulation of p21, p27, inhibitor of G2-M phase transition Wee1 (26) and an increase in cyclin B, accompanied by a decrease in the level of phosphohistone H3 (p-H3), a hallmark of M-phase cells (27), indicating that MLN4924-treated cells were arrested at the G2 phase (Fig. 1C). Furthermore, following the blocking of protein synthesis using CHX, the protein stability of p21, p27 and Wee1 were markedly increased by MLN4924 treatment (Fig. 2), indicating that G2-phase cell cycle arrest may be attributable to the accumulation of cell cycle-associated proteins.
Figure 2.
MLN enhanced the protein stability of p21, p27 and Wee1. EC1 and Kyse450 cells were pretreated with MLN and then divided into CHX or CHX+MLN groups. Cell proteins were collected at indicated time and analyzed by western blotting. p21, cyclin-dependent kinase inhibitor 1A; p27, cyclin-dependent kinase inhibitor 1B; Wee1, Wee1-like protein kinase; CHX, cyclohexamide; MLN, MLN4924.
MLN4924 induces DNA damage response
Activation of the DNA damage response is an important property of cytotoxic drugs; the expression of γH2AX was therefore determined to detect DNA double-strand breaks by immunofluorescence and western blotting. MLN4924 induced sustained γH2AX foci in the EC1 and Kyse450 cells in a dose-dependent manner (Fig. 3A and B), which was further confirmed by western blotting (Fig. 3C). MLN4924 treatment increased the level of CDT1 and ORC1 proteins in a dose-dependent manner (Fig. 3C). These results implied that the accumulation of DNA-replication licensing proteins CDT1 and ORC1, known CRL substrates (28), partially contributed to the induction of DNA damage by MLN4924 treatment.
MLN4924 enhances the cytotoxicity of CDDP to ESCC cells
To assess the function of MLN4924 as a novel chemosensitizer to increase the anti-ESCC activity of CDDP, ESCC EC1 and Kyse450 cell lines were treated with MLN4924, CDDP or the 2 together. Results demonstrated that MLN4924 significantly enhanced the cytotoxicity of CDDP and inhibited cell viability (Fig. 4A; P<0.001) and clonogenic cell survival (Fig. 4B). The combination of MLN4924 with CDDP markedly enhanced apoptosis compared with either single agent, which is evident by the increased percentage of Annexin V-positive (Fig. 5A) or caspase-3 active cells (Fig. 5B) and evaluated expression of cleaved-PARP (Fig. 5C).
Figure 5.
MLN increases CDDP-induced apoptosis of esophageal cancer cells. EC1 and Kyse450 cells were treated with 1.6 µg/ml CDDP alone or in combination with 0.3 µM MLN for 72 h. (A) Apoptosis was determined by the Annexin V-FITC/PI double-staining analysis. (B) Caspase 3 activity was analyzed by flow cytometry and (C) cleaved PARP were detected by western blotting. GAPDH served as a loading control. CDDP, cisplatin; FITC, fluorescein isothiocyanate; PI, propidium iodide; PARP, poly (ADP-ribose) polymerase; MLN, MLN4924.
Discussion
Previous studies demonstrated that MLN4924 inhibited the growth of a broad panel of cancer cells in vitro and in vivo (3,8,15,29–31). On the basis of its promising anticancer efficacy and tolerated toxicity, MLN4924 has undergone preclinical or phase I clinical trials for multiple human malignancies (17–19). Mechanistically, MLN4924 blocks cullin neddylation, inactivates CRL, induces the accumulation of tumor-suppressive CRL substrates and causes DNA damage, cell cycle arrest, cellular senescence and apoptosis in a tumor cell-specific manner (3,7,15,29,31). To date, the detailed mechanism of MLN4924 on cell proliferation inhibition in human ESCC cells requires further investigation. The present study demonstrated that MLN4924 could induce G2 cell cycle arrest in ESCC cells. At the same time, MLN4924 significantly enhanced the protein stability of p21, p27 and Wee1, which is in accordance with previous studies (9,15), where they indicate that all of the 3 proteins serve an important function in MLN4924-induced G2 cell cycle arrest (23).
G2/M-phase arrest is a crucial response to DNA damage in the majority of cancer cells (32). The present study found that MLN4924 treatment triggered DNA damage response in ESCC cells, as demonstrated by the appearance of γH2AX. Further analysis revealed that 2 well-known CRL substrates, CDT1 and ORC1, were accumulated in MLN4924-treated ESCC cells; their overexpression was known to trigger DNA damage response (3,11,29).
CDDP is a first-line chemotherapeutic agent for the treatment of ESCC (2). However, the toxicity and the development of resistance often limit its efficacy (33). Previously, a series of studies revealed that MLN4924 could be used as a sensitizer to chemotherapy: MLN4924 has been demonstrated to overcome resistance to a range of chemotherapeutics, including CDDP (20,21), carboplatin (21,34), bleomycin (35), doxorubicin (35) and etoposide (35) resistance in ovarian tumor cell lines in vitro and in vivo. MLN4924 significantly augmented the cytotoxicity of CDDP against CDDP-resistant cells in cervical carcinoma xenografts (22). The results of the present study revealed that the addition of MLN4924 significantly improved the efficacy of CDDP by enhancing the incidence of apoptosis in ESCC cells. These results indicate that MLN4924 could be used alone or in combination for the treatment of human ESCC in future studies.
The present study demonstrated that inactivating the neddylation pathway by MLN4924 suppressed the growth of esophageal cancer cells by triggering G2 cell cycle arrest and inducing DNA damage. Furthermore, the combination of MLN4924 and CDDP more effectively inhibited cell growth compared with treatment with either single drug by enhancing apoptosis in esophageal cancer cells. These results extend the understanding of the function of MLN4924 and propose its use in the treatment of esophageal cancer in single treatment or in combination.
Acknowledgements
The authors thank Professor Guoqiang Zhao from Zhengzhou University (Zhengzhou, China) for kindly providing the EC1 and Kyse450 cell lines. The present study was supported by the National Natural Science Foundation Grant of China (grant nos. 81001102, 81101894 and 81672421), the Outstanding Young Talent Research Fund of Zhengzhou University (grant no. 51999223), the Research Foundation of Education Bureau of Henan, China (grant no. 15A310024) and the Student's Platform for Innovation and Entrepreneurship Training Program of Zhengzhou University (grant no. 2016xjxm341).
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Yam PC, Tong D, Law S. Comparisons of sixth and seventh edition of the American Joint Cancer Committee staging systems for esophageal cancer. Ann Surg Oncol. 2014;21:583–588. doi: 10.1245/s10434-013-3335-5. [DOI] [PubMed] [Google Scholar]
- 3.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 4.Enchev RI, Schulman BA, Peter M. Protein neddylation: Beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abidi N, Xirodimas DP. Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic. Endocr Relat Cancer. 2015;22:T55–T70. doi: 10.1530/ERC-14-0315. [DOI] [PubMed] [Google Scholar]
- 6.Siergiejuk E, Scott DC, Schulman BA, Hofmann K, Kurz T, Peter M. Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. EMBO J. 2009;28:3845–3856. doi: 10.1038/emboj.2009.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P, Hu T, Liang Y, Li P, Chen X, Zhang J, Ma Y, Hao Q, Wang J, Zhang P, et al. Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5 axis in human esophageal cancer cells. Clin Cancer Res. 2016;22:4145–4157. doi: 10.1158/1078-0432.CCR-15-2254. [DOI] [PubMed] [Google Scholar]
- 8.Hua W, Li C, Yang Z, Li L, Jiang Y, Yu G, Zhu W, Liu Z, Duan S, Chu Y, et al. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro Oncol. 2015;17:1333–1343. doi: 10.1093/neuonc/nov066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Wang M, Yu G, Chen P, Li H, Wei D, Zhu J, Xie L, Jia H, Shi J, et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer Inst. 2014;106:dju083. doi: 10.1093/jnci/dju083. [DOI] [PubMed] [Google Scholar]
- 10.Xie P, Zhang M, He S, Lu K, Chen Y, Xing G, Lu Y, Liu P, Li Y, Wang S, et al. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat Commun. 2014;5:3733. doi: 10.1038/ncomms4733. [DOI] [PubMed] [Google Scholar]
- 11.Gao Q, Yu GY, Shi JY, Li LH, Zhang WJ, Wang ZC, Yang LX, Duan M, Zhao H, Wang XY, et al. Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget. 2014;5:7820–7832. doi: 10.18632/oncotarget.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo KL, Ho IL, Shi CS, Wu JT, Lin WC, Tsai YC, Chang HC, Chou CT, Hsu CH, Hsieh JT, et al. MLN4924, a novel protein neddylation inhibitor, suppresses proliferation and migration of human urothelial carcinoma: In vitro and in vivo studies. Cancer Lett. 2015;363:127–136. doi: 10.1016/j.canlet.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Wan J, Zhu J, Li G, Zhang Z. Radiosensitization of human colorectal cancer cells by MLN4924: An inhibitor of NEDD8-activating enzyme. Technol Cancer Res Treat. 2016;15:527–534. doi: 10.1177/1533034615588197. [DOI] [PubMed] [Google Scholar]
- 14.Lan H, Tang Z, Jin H, Sun Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci Rep. 2016;6:24218. doi: 10.1038/srep24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Luo Z, Pan Y, Wang W, Zhou X, Jeong LS, Chu Y, Liu J, Jia L. Targeting protein neddylation with an NEDD8-activating enzyme inhibitor MLN4924 induced apoptosis or senescence in human lymphoma cells. Cancer Biol Ther. 2015;16:420–429. doi: 10.1080/15384047.2014.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han K, Wang Q, Cao H, Qiu G, Cao J, Li X, Wang J, Shen B, Zhang J. The NEDD8-activating enzyme inhibitor MLN4924 induces G2 arrest and apoptosis in T-cell acute lymphoblastic leukemia. Oncotarget. 2016;7:23812–23824. doi: 10.18632/oncotarget.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatia S, Pavlick AC, Boasberg P, Thompson JA, Mulligan G, Pickard MD, Faessel H, Dezube BJ, Hamid O. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Invest New Drugs. 2016;34:439–449. doi: 10.1007/s10637-016-0348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah JJ, Jakubowiak AJ, O'Connor OA, Orlowski RZ, Harvey RD, Smith MR, Lebovic D, Diefenbach C, Kelly K, Hua Z, et al. Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin Cancer Res. 2016;22:34–43. doi: 10.1158/1078-0432.CCR-15-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarantopoulos J, Shapiro GI, Cohen RB, Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD, Faessel HM, et al. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin Cancer Res. 2016;22:847–857. doi: 10.1158/1078-0432.CCR-15-1338. [DOI] [PubMed] [Google Scholar]
- 20.Nawrocki ST, Kelly KR, Smith PG, Espitia CM, Possemato A, Beausoleil SA, Milhollen M, Blakemore S, Thomas M, Berger A, Carew JS. Disrupting protein NEDDylation with MLN4924 is a novel strategy to target cisplatin resistance in ovarian cancer. Clin Cancer Res. 2013;19:3577–3590. doi: 10.1158/1078-0432.CCR-12-3212. [DOI] [PubMed] [Google Scholar]
- 21.Jazaeri AA, Shibata E, Park J, Bryant JL, Conaway MR, Modesitt SC, Smith PG, Milhollen MA, Berger AJ, Dutta A. Overcoming platinum resistance in preclinical models of ovarian cancer using the neddylation inhibitor MLN4924. Mol Cancer Ther. 2013;12:1958–1967. doi: 10.1158/1535-7163.MCT-12-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin WC, Kuo KL, Shi CS, Wu JT, Hsieh JT, Chang HC, Liao SM, Chou CT, Chiang CK, Chiu WS, et al. MLN4924, a novel NEDD8-activating enzyme inhibitor, exhibits antitumor activity and enhances cisplatin-induced cytotoxicity in human cervical carcinoma: In vitro and in vivo study. Am J Cancer Res. 2015;5:3350–3362. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Zhang W, Yan Z, Liang Y, Li L, Yu X, Feng Y, Fu S, Zhang Y, Zhao H, et al. Radiosensitization by the investigational NEDD8-activating enzyme inhibitor MLN4924 (pevonedistat) in hormone-resistant prostate cancer cells. Oncotarget. 2016;7:38380–38391. doi: 10.18632/oncotarget.9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho IL, Kuo KL, Liu SH, Chang HC, Hsieh JT, Wu JT, Chiang CK, Lin WC, Tsai YC, Chou CT, et al. MLN4924 synergistically enhances cisplatin-induced cytotoxicity via JNK and Bcl-xL pathways in human urothelial carcinoma. Sci Rep. 2015;5:16948. doi: 10.1038/srep16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P, Hu T, Liang Y, Li P, Chen X, Zhang J, Ma Y, Hao Q, Wang J, Zhang P, et al. Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5 axis in human esophageal cancer cells. Clin Cancer Res. 2016;22:4145–4157. doi: 10.1158/1078-0432.CCR-15-2254. [DOI] [PubMed] [Google Scholar]
- 26.Sarcar B, Kahali S, Prabhu AH, Shumway SD, Xu Y, Demuth T, Chinnaiyan P. Targeting radiation-induced G(2) checkpoint activation with the Wee-1 inhibitor MK-1775 in glioblastoma cell lines. Mol Cancer Ther. 2011;10:2405–2414. doi: 10.1158/1535-7163.MCT-11-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 28.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oladghaffari M, Islamian JP, Baradaran B, Monfared AS. MLN4924 therapy as a novel approach in cancer treatment modalities. J Chemother. 2016;28:74–82. doi: 10.1179/1973947815Y.0000000066. [DOI] [PubMed] [Google Scholar]
- 30.Paiva C, Godbersen JC, Berger A, Brown JR, Danilov AV. Targeting neddylation induces DNA damage and checkpoint activation and sensitizes chronic lymphocytic leukemia B cells to alkylating agents. Cell Death Dis. 2015;6:e1807. doi: 10.1038/cddis.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Morgan MA, Sun Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid Redox Signal. 2014;21:2383–2400. doi: 10.1089/ars.2013.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark GR, Taylor WR. Analyzing the G2/M checkpoint. Methods Mol Biol. 2004;280:51–82. doi: 10.1385/1-59259-788-2:051. [DOI] [PubMed] [Google Scholar]
- 33.Florea AM, Büsselberg D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancers. 2011;3:1351–1371. doi: 10.3390/cancers3011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Sousa GF, Mde A Lima, Custodio DF, Freitas VM, Monteiro G. Chemogenomic study of carboplatin in saccharomyces cerevisiae: Inhibition of the NEDDylation process overcomes cellular resistance mediated by HuR and cullin proteins. PLoS One. 2015;10:e0145377. doi: 10.1371/journal.pone.0145377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan WW, Zhou JJ, Yu C, Xu Y, Guo LJ, Zhang HY, Zhou D, Song FZ, Fan HY. Ubiquitin E3 ligase CRL4(CDT2/DCAF2) as a potential chemotherapeutic target for ovarian surface epithelial cancer. J Biol Chem. 2013;288:29680–29691. doi: 10.1074/jbc.M113.495069. [DOI] [PMC free article] [PubMed] [Google Scholar]