SUMMARY
A common organizational feature of nervous systems is the existence of groups of neurons that share common traits but can be divided into individual subtypes based on anatomical or molecular features. We elucidate the mechanistic basis of neuronal diversification processes in the context of C. elegans ventral cord motor neurons which share common traits that are directly activated by the terminal selector UNC-3. Diversification of motor neurons into different classes, each characterized by unique patterns of effector gene expression, is controlled by distinct combinations of phylogenetically conserved, class-specific transcriptional repressors. These repressors are continuously required in postmitotic neurons to prevent UNC-3, which is active in all neuron classes, from activating class-specific effector genes in specific motor neuron subsets via discrete cis-regulatory elements. The strategy of antagonizing the activity of broadly acting terminal selectors of neuron identity in a subtype-specific fashion may constitute a general principle of neuron subtype diversification.
INTRODUCTION
The systematic classification of neurons into individual types and subtypes has been a central goal of neuroscience since the days of Ramon y Cajal. In vertebrate nervous systems, neuron type classification schemes are based on anatomical, electrophysiological and functional features and are currently accumulating more granularity with the advent of more refined molecular classification schemes (Macosko et al., 2015; Nelson et al., 2006; Tasic et al., 2016; Zeisel et al., 2015). One core organizational principle has already been evident for a long time: Neurons can be grouped together based on a number of shared molecular, functional and developmental traits, but can be subdivided into individual subtypes based on subtype-specific traits. For example, serotonergic neurons in the vertebrate central nervous system are a group of neurons in the Raphe nuclei of the brainstem defined by their usage of the same neurotransmitter and their specification by the transcription factor PET1, but they fall into specific subtypes based on anatomical, functional and molecular features (Gaspar and Lillesaar, 2012; Jensen et al., 2008; Okaty et al., 2015). Similarly, midbrain neurons that utilize dopamine as a neurotransmitter are a group of neurons that are specified by a common set of transcription factors, but can be also further subdivided into individual subtypes based on electrophysiological criteria and molecular features (Poulin et al., 2014; Roeper, 2013). These examples point to a bipartite scheme of neuron classification and subtype specification: A common factor specifies a group of neurons with shared features, but additional regulatory mechanisms must then operate to diversify neurons into individual subtypes. The mechanistic basis of such diversification processes is not well understood and, more generally, the existence and extent of diversification of neuronal subtypes remains ill-defined in complex vertebrate nervous systems.
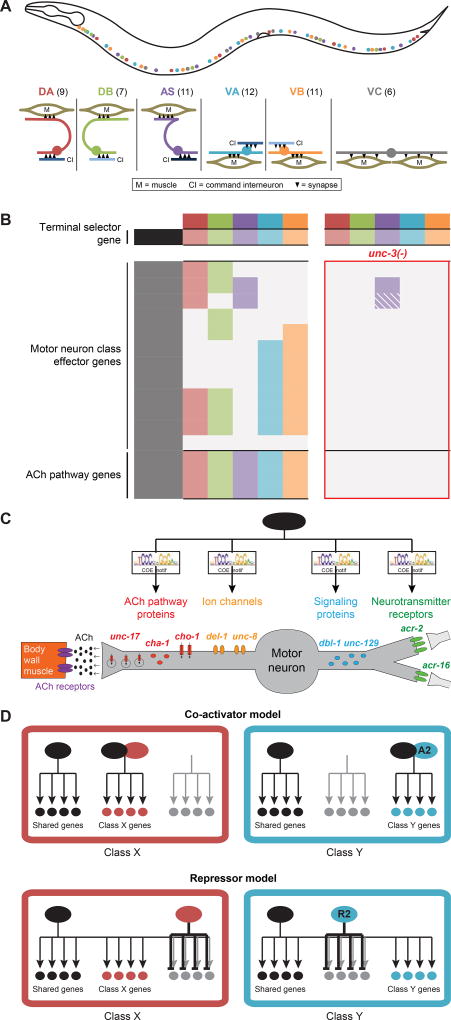
We study this problem in the nervous system of the nematode Caenorhabditis elegans. The 302 neurons of its nervous system are exceptionally well described on an anatomical level (White et al., 1986) and also on a molecular level (www.wormbase.org). Motor neurons (MNs) in the C. elegans ventral nerve cord (VNC) are a paradigm for the neuron subtype diversification problem described above. The VNC is populated by cholinergic and GABAergic MNs that innervate body wall muscles. As illustrated in Fig.1A, cholinergic MNs can be further divided into six classes (subtypes), the embryonically generated DA and DB and the post-embryonically generated AS, VA, VB and VC classes (Von Stetina et al., 2006; White et al., 1976). The DA, DB, VA, VB and AS (but not VC) classes not only share the common trait of being cholinergic (Duerr et al., 2008), but their identities are also specified by the same transcription factor, the COE/EBF-type transcription factor UNC-3 (Kratsios et al., 2011). DA, VA and AS can be grouped together based on additional shared anatomical and functional traits, as can DB and VB classes (White et al., 1976). For example, DA and VA are required for backward locomotion, while DB and VB classes control forward locomotion (Chalfie et al., 1985).
Figure 1. C. elegans ventral nerve cord motor neurons.
A: Cholinergic MN classes in C. elegans VNC and their relative positions and morphology. Numbers in brackets represent the number of individual MNs in each class.
B: Unique combinations of effector gene expression define each unc-3-expressing MN class (as represented by different colors). As with the shared genes of the ACh pathway, class-specific genes are unc-3-dependent since their expression is lost in unc-3 mutants (bounded by red margin). Color-filled rectangles represent expression in the corresponding class; grey rectangles represent absence of expression; superimposed diagonal stripes indicate dim expression.
C: UNC-3 is a terminal selector which coordinately regulates the expression of class-specific as well as shared effector genes in cholinergic MNs. This co-regulation is achieved via phylogenetically conserved binding sites, termed COE motifs, found within the cis-regulatory region of these genes.
D: The co-activator versus repressor models of regulation: hypothetical models of how UNC-3 selectively controls the expression of class-specific genes. Black and color-filled circles indicate gene is expressed; grey-filled circles indicate gene is not expressed. Class-specific co-activators (A1, A2) and repressors (R1, R2).
Each individual MN class can also be distinguished at the molecular level by their class-specific expression of a specific combination of effector genes that define the unique properties of each individual MN class. These effectors include neurotransmitter receptors, gap junction proteins, signaling molecules, and ion channels (Fig.1B,C). Importantly, the terminal selector gene unc-3 is known to be required not only for the expression of shared traits of these cholinergic MNs (e.g. of proteins that synthesize and package acetylcholine (ACh)), but also for the expression of the MN class-specific features (Fig.1C). Such necessity is not only revealed by the loss of expression of class-specific effector genes in unc-3 mutants, but is also reflected by the presence and requirement of UNC-3 binding sites in these genes (Kratsios et al., 2011). This poses an intriguing conundrum that is analogous to the situation found in many other neuron subtypes with shared traits: How can a transcription factor that activates shared traits in a group of neurons, also activate specific traits only in subsets of these neurons? For example, how does PET1 activate serotonin pathway genes in all vertebrate serotonergic neurons, but also activate the expression of a host of other genes only in a subtype-specific manner (Okaty et al., 2015; Wyler et al., 2016)? Understanding the mechanistic basis of this problem will provide a major step towards understanding how a brain can generate a diverse set of neuron subtypes.
Two different scenarios can, in principle, be envisioned for how such diversification processes work: The activator that controls shared traits in several neurons, like UNC-3 in C. elegans VNC MNs, requires class-specific cofactors to activate class-specific features (upper panels in Fig.1D). Alternatively, the shared regulatory factor is capable of activating all features, including class-specific features, but is prevented from doing so via class-specific repressor proteins (lower panel in Fig.1D). These two models make specific genetic predictions: A loss of direct co-activator function would lead to loss of expression of class-specific features, while a loss of direct repressor function would lead to ectopic expression of class-specific features. We tested these predictions by performing genetic screens for mutants in which the expression of class-specific features is altered. These screens, as well as the testing of candidate genes, reveal a remarkably homogenous picture for how cholinergic MN classes diversify: MN class-specific repressor proteins act in distinct classes to prevent UNC-3 from activating subsets of class-specific effector genes. These class-specific repressors include several previously known transcription factors and also a newly identified Zn finger transcription factor, BNC-1. All repressors are phylogenetically conserved. We propose that the mechanism that we define here may constitute a broadly applicable principle of neuron identity diversification across phylogeny.
RESULTS
Genetic screens for motor neuron class-specific regulators
To test the different models for MN identity specification (Fig.1D), we screened for mutants in which MN class-specific effector genes are either misexpressed (supporting the repressor model) or lost in specific MN classes (supporting the co-activator model). We initially chose two different reporter transgenes for our genetic screens: (a) a reporter for unc-129, a TGFβ-family member gene which is a direct target of UNC-3 (Kratsios et al., 2015) expressed in DA and DB classes of cholinergic VNC MNs (but not VA, VB and AS classes, where unc-3 actively promotes cholinergic identity yet is apparently not sufficient to induce unc-129 expression); (b) a reporter for unc-53, a novel unc-3-dependent gene encoding a cytoskeletal protein expressed in DA and AS, but not VA, DB or VB MN classes (Fig.4E,F). We mutagenized these reporter strains with ethyl methanesulfonate (EMS) and screened ~70,760 haploid genomes for expression defects of the unc-129 reporter and ~4500 haploid genomes for expression defects of the unc-53 reporter. In the ensuing section, we describe three distinct complementation groups identified from these screens.
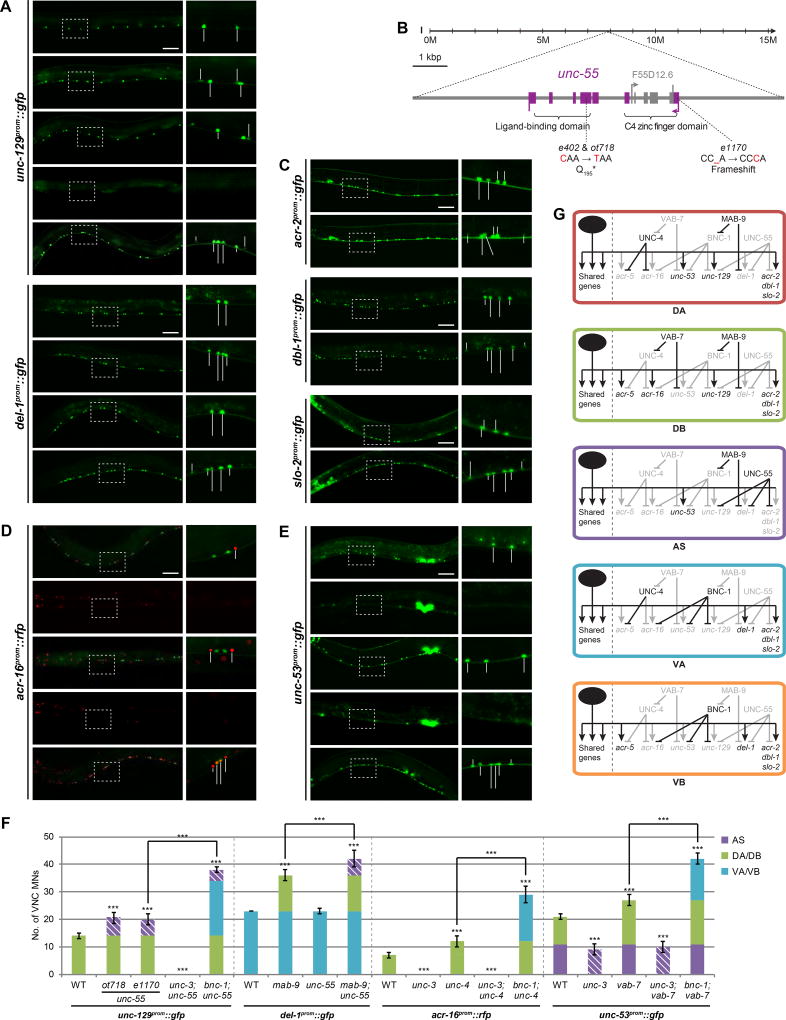
Figure 4. Additional class-specific repressors control motor neuron identity.
A: In ot718 animals, unc-129 is derepressed in AS MNs (phenocopied by the unc-55 mutant allele, e1170) and this phenotype is unc-3-dependent. ot718 fails to complement e1170 and is thus confirmed to be an unc-55 mutant allele. del-1 expression is unaffected in unc-55 mutants, but is derepressed in AS MNs in mab-9; unc-55 double mutants – indicating a redundant role of these two genes. The individual defects of bnc-1, mab-9, and unc-55 mutants are additive since the expression of unc-129 in bnc-1; unc-55 and del-1 in mab-9; unc-55 double mutants is no longer class-specific. WT and mab-9 mutant images repeated from Fig.2A and Fig.3F, respectively; all images show anterior half of worm; all scale bars = 50 µm.
B: unc-55 gene locus.
C: DA/DB/VA/VB-specific and unc-3-dependent effector genes acr-2, dbl-1, and slo-2 are derepressed in AS MNs in unc-55 mutants. See Fig.S3A for quantification.
D: In unc-4 mutants, acr-16 is derepressed in DA MNs in an unc-3-dependent manner. Additive effect of double mutant bnc-1; unc-4 causes acr-16 to no longer be as class-specific.
E: In vab-7 mutants, unc-53 is derepressed in DB MNs in an unc-3-dependent manner. Additive effect of double mutant bnc-1; vab-7 causes unc-53 to no longer be as class-specific.
F: Quantification for A, D, and E; error bars show SD. Unpaired t-tests were performed for all mutants compared to WT unless otherwise indicated; ***p < 0.001; n ≥ 13.
G: Genetic model depicting the interactions of repressors on class-specific effector genes in VNC MNs. In AS MNs, acr-16 and acr-5 are likely repressed by as yet unidentified AS-specific repressors, perhaps redundantly with UNC-55.
A phylogenetically conserved Zn finger transcription factor regulates motor neuron class-specific gene expression
From the screen for unc-129::gfp expression defects, we isolated a mutant allele, ot721, in which unc-129 is ectopically expressed in VNC MNs other than those of the DA and DB classes (Fig.2A,B). Another mutant allele, ot763, was retrieved in which unc-53::gfp is also ectopically expressed in VNC MNs (Fig.2G,H). ot721 and ot763 display similar phenotypes; in both mutant strains, ectopic expression of unc-129 is observed in VA and VB MNs, with similar penetrance and expressivity. ot721 and ot763 fail to complement each other, suggesting that both mutant alleles affect the same gene (Fig.2A,B).
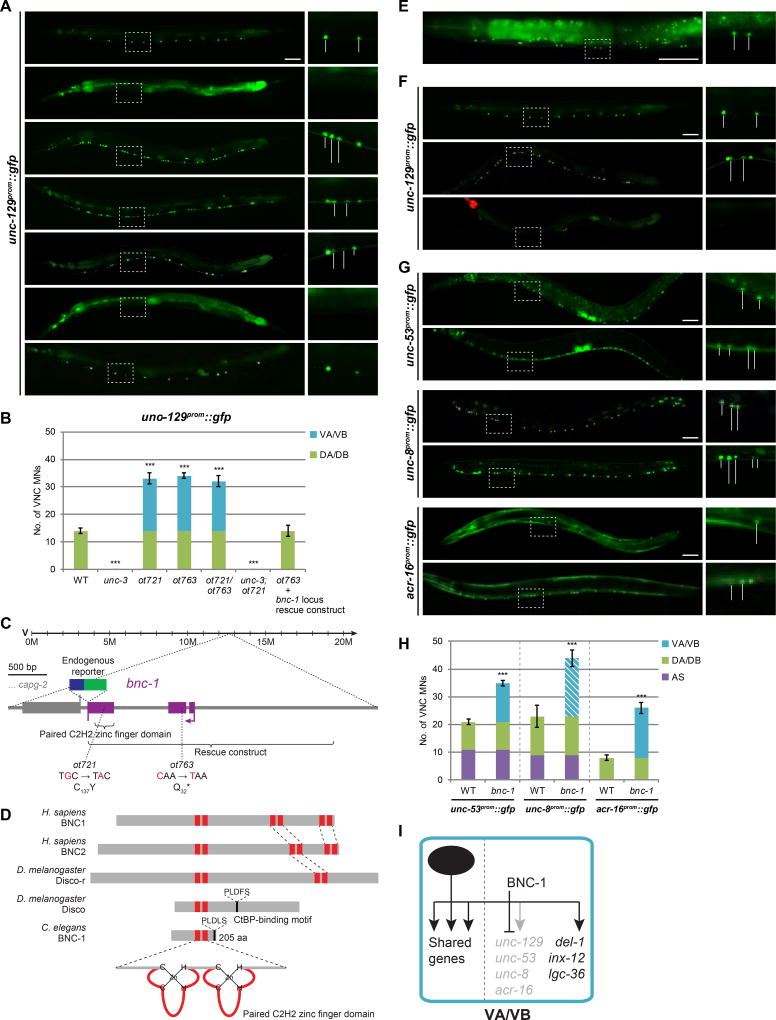
Figure 2. bnc-1 is a class-specific regulator of motor neuron identity.
A: unc-129 is expressed in DA/DB MNs (class member neurons within white bounding rectangle is magnified in inset on the right) and is regulated by unc-3. In ot721 and ot763 animals (which do not complement each other), unc-129 is derepressed in VA/VB MNs in an unc-3-dependent manner. This phenotype is rescued by the genomic locus of a novel C. elegans gene, bnc-1 (basonuclin 1). Wildtype (WT); all scale bars = 50 µm.
B. Quantification for A; error bars show standard deviation (SD). Unpaired t-tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 13.
C: bnc-1 gene locus.
D: BNC-1 protein and its orthologs.
E: Endogenous bnc-1 locus tagged with mNG shows remarkably specific expression in VA/VB MNs (except for VB1) in the VNC (see Fig.S4A for additional expression elsewhere). Gut auto-fluorescence (*); stitched together from two images of the same worm; showing anterior half of worm.
F: bnc-1 cDNA driven by an unc-3 promoter in DA/DB/VA/VB MNs in bnc-1 mutants rescues the unc-129 derepression phenotype in VA/VB, and is sufficient to ectopically repress unc-129 in DA/DB MNs. WT image repeated from A; red pharyngeal expression marks unc-3prom::bnc-1 transgene.
G: Expression of the unc-3-dependent class-specific effector genes unc-53 (DA/AS), unc-8 (DA/DB/AS) and acr-16 (DB) are derepressed in VA/VB MNs of bnc-1 mutants.
H: Quantification for G; error bars show SD. Superimposed diagonal stripes indicate dim expression. Unpaired t-tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 13.
I: Genetic model depicting BNC-1 repressing DA/DB-specific effector genes in VA/VB MNs.
Through our previously described mapping and whole-genome sequencing pipeline (see Supplemental Experimental Procedures), we localized the molecular lesions of ot721 and ot763 to a previously uncharacterized C2H2 Zn finger-encoding gene (Fig.S2A,B). This gene represents the sole ortholog of a Drosophila transcription factor called Disco and two paralogous human genes called Basonuclin 1 and 2 (BNC1 and BNC2; Fig.2C,D) (Vanhoutteghem et al., 2011) and we therefore named the C. elegans locus bnc-1. Disco and C. elegans BNC-1 are likely repressor proteins based on the presence of a binding site for the corepressor protein CtBP and the ability of the fly Disco protein to physically interact with CtBP (Patel, 2007).
The ot721 allele is defined by a mutation of a critical Zn coordinating cysteine in the second of the two C2H2 Zn fingers of BNC-1, whereas ot763 is defined by a premature stop codon before the Zn finger domains, thus constituting a likely null allele. The unc-129 expression defect of the bnc-1(ot763) mutant can be rescued by: (a) a genomic sequence that exclusively encompasses the bnc-1 locus (1.8 kb upstream of the start codon to the stop codon); (b) a construct in which the bnc-1 locus cDNA is specifically driven in cholinergic MNs under control of the unc-3 promoter (Fig.2A,B,F; Fig.S1A).
We analyzed the expression of several different class-specific MN genes in bnc-1 mutants and found a coherent theme: All MN class-specific genes that are normally not expressed in VA and VB become ectopically expressed in VA and VB MNs of bnc-1 mutants. Apart from the DA/DB-specific unc-129 gene, this ectopic VA and VB expression applies to the gene reporters of unc-53 (DA/AS-specific); the DEG/ENaC channel unc-8 (DA/DB/AS-specific); and the nicotinic ACh receptor (nAChR) acr-16 (DB-specific) (Fig.2G,H). VA/VB-specific (del-1 and inx-12) or DB/VB-specific (acr-5) genes are not affected in bnc-1 mutants, and neither is the overall morphology of VA and VB MNs. Moreover, expression of genes shared by all cholinergic MNs (e.g. ACh pathway genes) is unaffected in bnc-1 mutants (Fig.S1B–F). Taken together, bnc-1 appears to repress genes that are expressed in neither VA nor VB classes, and these MNs adopt a “mixed” identity in bnc-1 mutants due to derepression of these genes in VA and VB MNs (summarized in Fig.2I; Fig.8A).
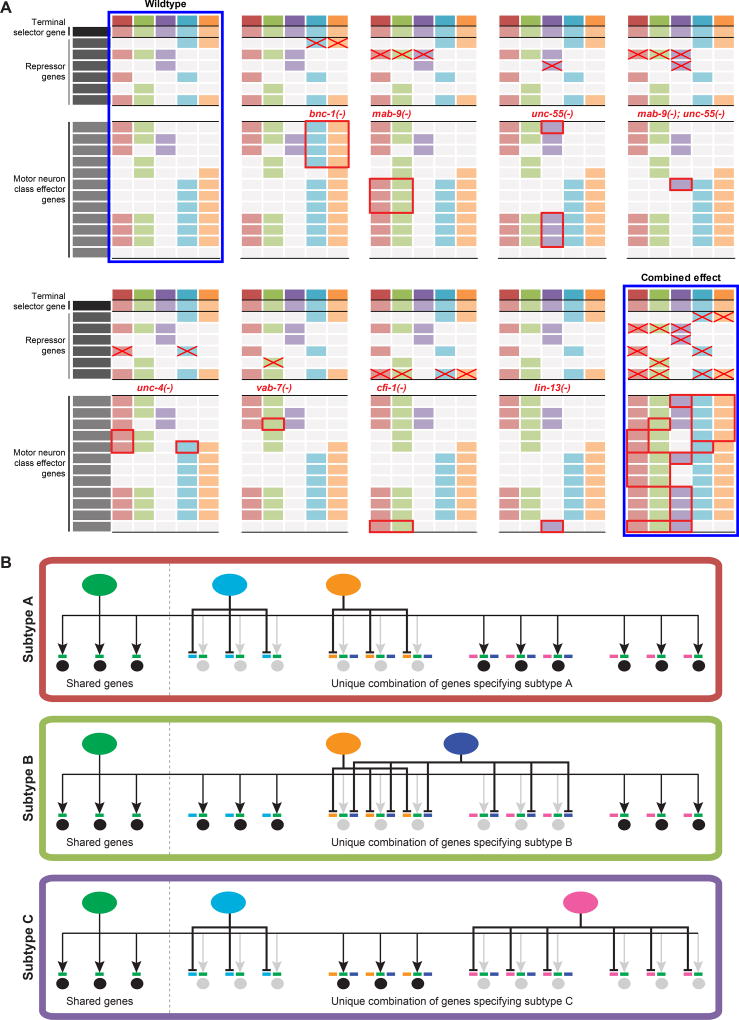
Figure 8. Proposed general principle of motor neuron diversification.
A: Repressor mutant effects on class-specific effector genes in VNC MNs. Only derepression effects are shown within red bounding rectangles. Red crosses indicate absence of a repressor in its mutant. The hypothetical combined effects of all repressor mutants predict the loss of the unique combinations of class-specific effector genes which define MN classes, leading to the loss of MN diversity (compare the first and last tables bounded by blue margins).
- subtype-specific repressors counteract in parallel the transcriptional activity of the broadly acting terminal selector
- each subtype possesses a unique combination of repressors to control the unique effector gene profile that defines the subtype identity
- repressors work via cognate binding motifs adjacent to the activator motif within the cis-regulatory region of the effector gene
- subtype-specific expression of an effector gene is determined by its combination of repressor binding motifs
- continuous presence of these trans-acting factors are required to maintain subtype identity throughout the lifetime of the neuron.
See Fig.S6A for a similar model detailed with the incorporation of results from this study.
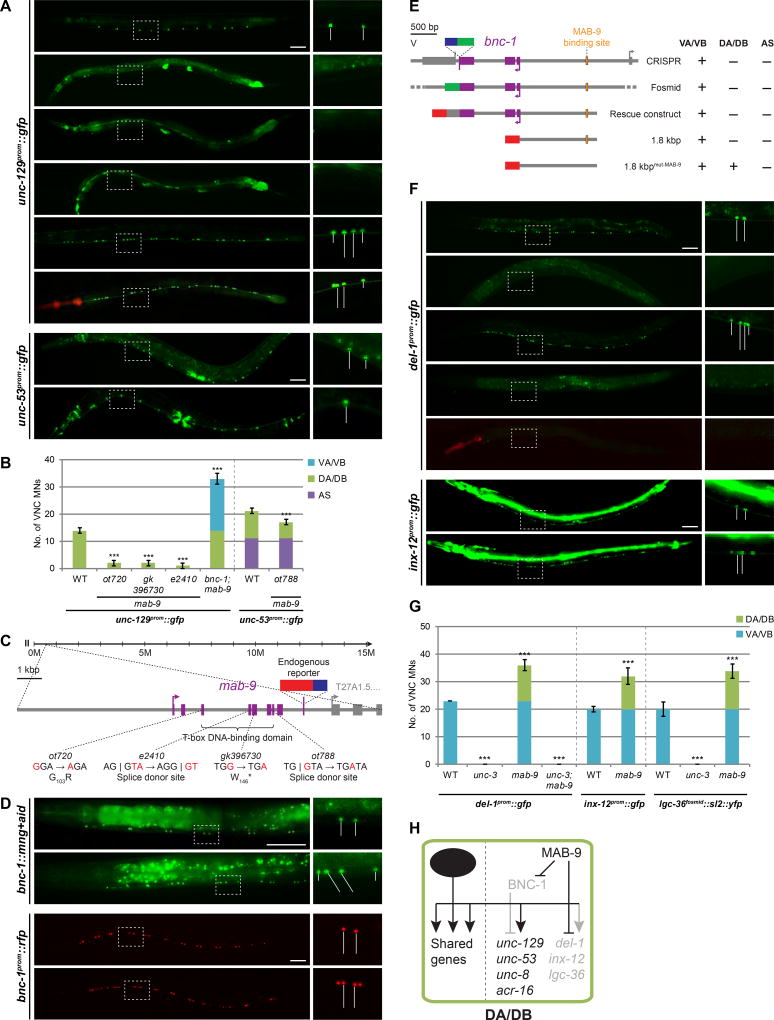
By tagging the bnc-1 locus with mNeonGreen (mNG) using CRISPR/Cas9-based genome engineering, we find that bnc-1 displays a remarkably specific expression pattern. In both males and hermaphrodites, bnc-1 is almost exclusively expressed in VA and VB MNs (Fig.2E), indicating that it functions cell-autonomously. Weaker expression is also observed in the SABV neuron pair (Fig.S4A). The protein localizes exclusively to nuclei. bnc-1 expression is initiated around the postembryonic birth of VA and VB MNs at the late first larval (L1) stage and is maintained throughout the life of the animals. A similar expression pattern is observed with successively smaller reporter gene constructs: (a) a fosmid reporter in which bnc-1 is tagged at the C-terminus; (b) the aforementioned rescue construct that contains sequences 1.8 kb upstream of the start codon all the way to the stop codon; (c) a promoter fusion construct that only contains the sequence 1.8 kb upstream of the start codon (Fig.3D,E; Fig.S1G,H).
Figure 3. mab-9 is a class-specific regulator of motor neuron identity.
A: Expression of unc-129 is lost in mab-9(ot720) animals (phenocopied by gk396730 and e2410). In the double mutant, the phenotype of bnc-1 is epistatic to that of mab-9, indicating that MAB-9 represses bnc-1. mab-9 cDNA driven by the unc-3 promoter in mab-9 mutants rescues the unc-129 expression loss phenotype in DA/DB, and is sufficient to ectopically derepress unc-129 in VA/VB MNs. Similarly, unc-53 expression in DA MNs is lost (albeit only in the anterior half of the VNC) in mab-9(ot788). WT images repeated from Fig.2A,G; red pharyngeal expression marks unc-3prom::mab-9 transgene; all scale bars = 50 µm.
B: Quantification for A; error bars show SD. Unpaired t-tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 13.
C: mab-9 gene locus. The endogenous reporter is not visible in the VNC.
D: bnc-1 is derepressed in DA/DB MNs in mab-9 mutants. WT image repeated from Fig.2E. A bnc-1 promoter 1.8 kb upstream of the start codon recapitulates the VA/VB-specific expression of endogenous bnc-1. Mutation (mut-) of MAB-9 binding sites causes bnc-1 to be derepressed in DA/DB MNs. All rfp represents mchopti+h2b. See Fig.S1H for quantification.
E: bnc-1 fluorescent reporters of the endogenous gene locus (CRISPR; see Fig.2E), a fosmid (see Fig.S1G), the rescue construct used in Fig.2A,B (see Fig.S1G), and the abovementioned 1.8 kb promoter (see Fig.3D) express specifically in VA/VB MNs. Deletion of MAB-9 binding sites (white-filled vertical rectangle) causes bnc-1 to be derepressed in DA/DB MNs. See Fig.S1H for quantification.
F: del-1 is expressed in VA/VB MNs and is regulated by unc-3. In mab-9 mutants, del-1 is derepressed in DA/DB MNs in an unc-3-dependent manner. The unc-3prom::mab-9 transgene rescues this phenotype, and is sufficient to ectopically repress del-1 in VA/VB MNs. Similarly, unc-3-dependent inx-12 which is VA/VB-specific is also derepressed in DA/DB MNs in mab-9 mutants.
G: Quantification for F and the derepression of unc-3-dependent, VA/VB-specific lgc-36 in DA/DB MNs in mab-9 mutants (see Fig.S1I for images); error bars show SD. Unpaired t-tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 13.
H: Genetic model depicting MAB-9 repressing BNC-1 as well as VA/VB-specific effector genes in DA/DB MNs.
The ectopic gene expression in VA and VB MNs of bnc-1 mutants depends on unc-3, since in bnc-1; unc-3 double mutants the ectopic as well as wildtype (DA and DB) expression of the unc-3 target unc-129 is lost (Fig.2A,B). We conclude that bnc-1 genetically acts to prevent unc-3 from activating target genes in VA and VB, and henceforth that bnc-1 loss results in VA and VB MNs acquiring a “mixed” identity where unc-3 can now activate target genes that are normally expressed in a more restricted, class-specific manner.
The T-box transcription factor mab-9 represses VA and VB identity features in DA and DB motor neurons
Another allele retrieved from our screens, ot720, displayed a fundamentally distinct phenotype from bnc-1 mutants: unc-129 expression in DA and DB MNs is lost in these animals (Fig.3A,B). By whole-genome sequencing, we identified the molecular lesion of ot720 as a missense mutation in the previously identified T-box gene mab-9, the C. elegans ortholog of vertebrate TBX20 (Fig.S2C) (Woollard and Hodgkin, 2000). Animals carrying a previously described splice site mutation, e2410, and a premature stop codon identified in the Million Mutation Project (Thompson et al., 2013), gk396730, in the mab-9 locus show the same loss of unc-129 expression as ot720 animals. From our screen for defects in unc-53 expression, we identified yet another mab-9 allele, ot788, in which animals lose unc-53 expression in DA MNs (Fig.3A,B; Fig.S2D). mab-9 was previously described to be expressed in DA and DB MNs (Fox et al., 2005).
Loss of expression of class-specific effector genes is consistent with the UNC-3 co-activator model (Fig.1D). However, based on: (a) the repressor function of mab-9 orthologs in other species (Formaz-Preston et al., 2012; Kaltenbrun et al., 2013); (b) the presence of an Engrailed homology motif in MAB-9 (Copley, 2005) (c) the above-described repressive effect of bnc-1 on unc-129 and unc-53 expression, we considered the alternative possibility that mab-9 acts as a repressor of bnc-1 expression in DA and DB MNs. As a consequence, derepression of bnc-1 in mab-9 mutants may then result in loss of unc-129 and unc-53 expression. In support of this notion, genetically, we indeed find that in mab-9; bnc-1 double mutants, the loss of unc-129 expression in mab-9 mutants depends on bnc-1 (Fig.3A,B). Moreover, expression of bnc-1 is derepressed in DA and DB MNs in mab-9 mutants. This repressive effect of mab-9 appears direct since deletion of predicted MAB-9 binding sites upstream of the bnc-1 locus results in ectopic expression of bnc-1 in DA and DB MNs (Fig. 3D,E; Fig.S1H).
However, mab-9 not only represses the regulatory factor bnc-1, it also represses three unc-3-dependent VA/VB-specific effector genes in DA and DB MNs, namely the DEG/ENaC channel del-1, the innexin inx-12, and the GABA receptor lgc-36 (Fig.3F,G; Fig.S1I; summarized in Fig.3H; Fig.8A). Hence, in analogy to our observations in bnc-1 mutants, we considered the possibility that the ectopic expression of VA and VB genes in DA and DB MNs of mab-9 mutants is a reflection of these genes now being available for activation by unc-3. We indeed find that ectopic del-1 expression is abolished in mab-9; unc-3 double mutants (Fig.3F,G). Since expression of DB/VB-specific acr-5 is unaffected in mab-9 mutants (Fig.S1J), we conclude that mab-9 targets only genes that are expressed in neither DA nor DB MNs.
Lastly, we asked whether mab-9 is not only required, but also sufficient to block VA and VB gene expression. To this end, we ectopically expressed mab-9 cDNA in VA and VB MNs using the unc-3 promoter and find that VA/VB-specific del-1 is indeed turned off in these transgenic animals. Moreover, DA/DB-specific unc-129 becomes ectopically expressed in VA and VB MNs, apparently because ectopic mab-9 expression represses bnc-1, the VA/VB-specific repressor of unc-129 (Fig.3A,F; Fig.S1K).
The COUP-TF orphan nuclear receptor unc-55 represses DA/DB- and VA/VB-specific identity features in the AS motor neuron class
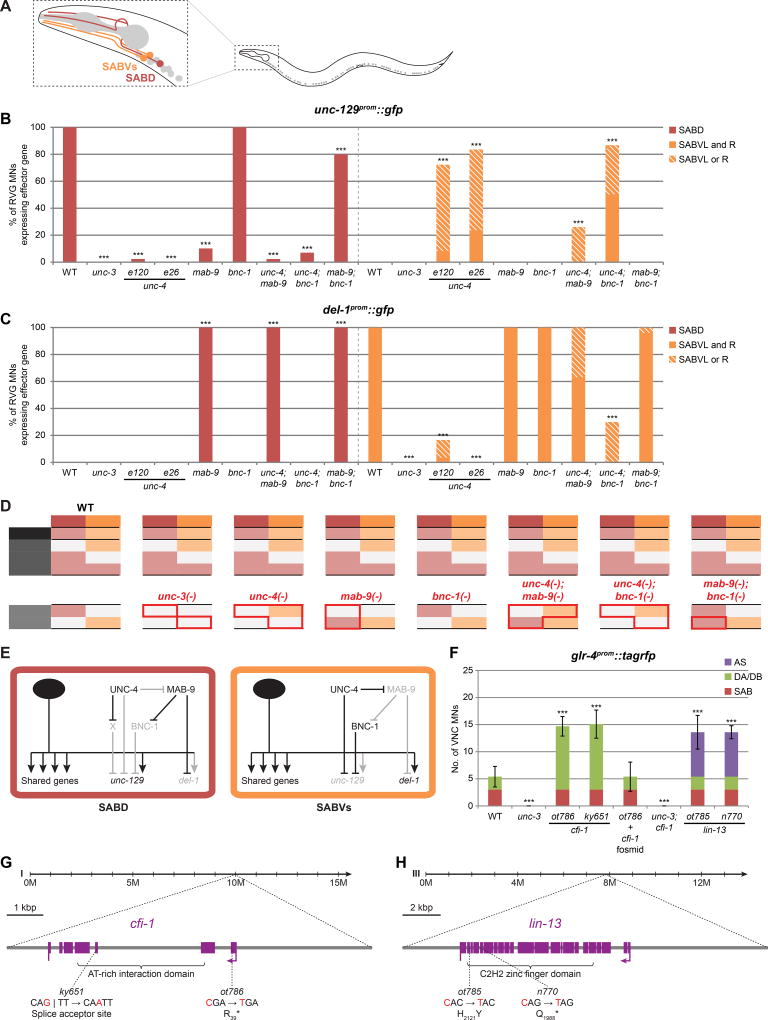
The third complementation group isolated from our screens is defined by a single allele, ot718 (Fig.4A,F). ot718 animals display an uncoordinated locomotory “coiler” phenotype that resembles a number of different unc mutants, including unc-55 mutants (Shan et al., 2005; Walthall and Plunkett, 1995; Zhou and Walthall, 1998). ot718 indeed fails to complement the unc-55 allele e1170 and harbors a premature stop codon in the fourth exon of the unc-55 locus (as does the previously non-sequenced e402 allele) (Fig.4B). unc-55 encodes an orphan nuclear receptor of the COUP-TF type and is expressed in AS MNs (and GABAergic VD) where its function was not previously examined (Shan et al., 2005; Walthall and Plunkett, 1995).
We find that the expression of four unc-3-dependent effector genes, all expressed in DA and DB but not AS MNs (TGFβ family members unc-129 and dbl-1, nAChR acr-2, and potassium channel slo-2), becomes derepressed in AS MNs of unc-55 mutants (Fig.4A,C,F; Fig.S3A; summarized in Fig.4G; Fig.8A). This ectopic expression depends on unc-3 as unc-55; unc-3 double mutants display no derepression (Fig.4A,F). VA/VB-specific del-1 is not derepressed in AS MNs of unc-55 mutants. Since mab-9 which represses del-1 in DA and DB is also expressed in AS MNs (Fox et al., 2005), we tested whether unc-55 and mab-9 may work redundantly to repress del-1 in AS MNs. Indeed, del-1 is derepressed in AS MNs in unc-55; mab-9 double mutants (Fig.4A,F). We note that the expression of both DB-specific acr-16 and DB/VB-specific acr-5 is unaffected in unc-55 mutants (Fig.S3B). Altogether, it appears that genes that are derepressed in AS in unc-55 mutants normally need to be expressed in the DA class (which is morphologically most similar to AS (White et al., 1976)).
unc-4 and vab-7 are additional repressors that control motor neuron class identity
bnc-1, mab-9 and unc-55 represent repressors that counteract unc-3 in a subset of MN classes. We set out to test whether other previously described repressors may operate in a similar manner. Specifically, the Prd-type homeobox gene unc-4 (UNCX in vertebrates) has previously been shown to repress DB/VB features in DA/VA MNs; these features include synaptic connectivity pattern (White et al., 1992), DB/VB-specific expression of nAChR acr-5 (derepressed in DA/VA MNs of unc-4 mutants; Fig.S3C,D) (Kratsios et al., 2011; Winnier et al., 1999), and DB-specific expression of nAChR acr-16 (derepressed in DA MNs of unc-4 mutants; Fig.4D,F) (summarized in Fig.4G; Fig.8A). Both acr-5 and acr-16 are unc-3-dependent genes, and unc-4 also appears to counteract the activity of unc-3 in DA and VA MNs (Fig.4D,F). We note that the expression of DA/DB-specific unc-129 is unaffected in unc-4 mutants (Fig.S3D) and conclude that unc-4 targets only genes that are normally expressed in neither DA nor VA classes.
The homeobox gene vab-7, the ortholog of Drosophila Eve and vertebrate EVX, was previously shown to be expressed in DB MNs and to repress unc-4 expression in this class (Esmaeili et al., 2002). However, effector genes under the control of vab-7 had not been described. In analogy to mab-9 which represses both a regulatory factor and effector genes, we find that vab-7 also counteracts unc-3 activity to repress DA/AS-specific unc-53 in DB MNs (Fig.4E,F) (summarized in Fig.4G; Fig.8A). Taken together, unc-4 and vab-7 operate along similar lines as bnc-1, mab-9 and unc-55, albeit with different spectra of MN class specificity. Lastly, we tested whether the mutant defects of these repressors which act in distinct classes are additive in nature. To this end, we built bnc-1; unc-55, mab-9; unc-55, bnc-1; unc-4, and bnc-1; vab-7 double mutants, and find that their defects are indeed additive (Fig.4A,D–F).
A similar repressor logic operates in SAB motor neurons in the head
Moving outside the VNC, we asked whether the MN diversification process of a different anatomical region of the C. elegans nervous system operates by a similar principle. Specifically, we considered the SAB MNs of the head, whose cell bodies are located in the retrovesicular ganglion (RVG), from which they send processes to innervate head muscle. Based on anatomical and molecular features, SAB MNs can be divided into two distinct subclasses: ventral SABVs (consisting of SABVL and R) and a single dorsal SABD (Fig.5A). Like in VNC MNs, unc-3 is required for the expression of genes that are: (a) shared by all three SAB MNs; (b) exclusively in SABVs (del-1) or SABD (unc-129) (Kratsios et al., 2015; White et al., 1986). We find that a subset of repressors that counteract unc-3 in VNC MNs also do so in SAB MNs. Specifically, in mab-9 mutants, SABV-specific del-1 is derepressed in SABD, while SABD-specific unc-129 expression is lost. The latter effect, as in VNC MNs, is due to ectopic expression of bnc-1 in mab-9 mutants because: (a) in mab-9; bnc-1 double mutants, unc-129 expression in SABD is restored (Fig.5B–D); (b) bnc-1 is derepressed in SABD of mab-9 mutants (Fig.S4A) (summarized in Fig.5E).
Figure 5. A repressor regulatory logic also operates in head SAB motor neurons.
A: SAB MNs in the RVG and their relative positions and morphology.
B: unc-129 expression is specific to SABD and is unc-3-dependent. This expression is lost in mab-9 mutants in a bnc-1-dependent manner, indicating that MAB-9 represses bnc-1 which represses unc-129. Similarly, unc-129 expression in SABD is lost in unc-4 mutants. This effect is likely also indirect with UNC-4 possibly repressing an unidentified repressor of unc-129 (note that e26 disrupts UNC-4 interaction with the Groucho-like corepressor UNC-37). This repressor is not bnc-1 as the bnc-1 mutant phenotype is not epistatic to that of unc-4. On the other hand, unc-129 is derepressed in SABVs (albeit incompletely) in unc-4 mutants in a mab-9-dependent manner, indicating that UNC-4 represses mab-9. As in SABD, mab-9 is likely repressing bnc-1 which represses unc-129. This predicts that bnc-1 mutants would phenocopy those of unc-4 – which is not the case. UNC-4 might independently to, and redundantly with bnc-1 be repressing unc-129 in SABVs. Fischer’s exact tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 25.
C: del-1 expression is specific to SABVs and is unc-3-dependent. In mab-9 mutants, del-1 is derepressed in SABD. In unc-4 mutants, del-1 expression is lost in SABVs in a mab-9-dependent manner, indicating that UNC-4 represses mab-9 which represses del-1. Fischer’s exact tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 20.
D: Effect of repressor mutants on class-specific effector genes in SAB MNs. Note expression pattern of repressor genes (see Fig.S4A). Red bounding lines highlight changes in effector gene expression in repressor mutants compared to WT.
E: Genetic model depicting the interactions of repressors on class-specific effector genes in SAB MNs.
F: glr-4 is expressed in SAB MNs in an unc-3-dependent manner, but expression is largely absent in VNC MNs. In cfi-1(ot786), glr-4 is derepressed in DA/DB MNs (phenocopied by ky651) in an unc-3-dependent manner. This phenotype is rescued by a fosmid containing the cfi-1 locus. Additionally, in lin-13(ot785), glr-4 is derepressed in AS MNs (phenocopied by n770). Error bars show SD; unpaired t-tests were performed for all mutants compared to WT; ***p < 0.001; n ≥ 13. See Fig.S4C,D for images.
G: cfi-1 gene locus.
H: lin-13 gene locus. Not shown are four regulatory mutations in ot785.
unc-4 also affects SAB subclass specification in a repressive manner that is conceptually similar to its function in VNC MNs. Specifically, in unc-4 mutants, we observe a derepression of unc-129 in SABVs, suggesting that unc-4 counteracts unc-3 similarly in SABVs as it does in VNC MNs. In addition, del-1 expression is lost in SABVs; this appears to be a consequence of mab-9 derepression in SABVs of unc-4 mutants, since del-1 expression is restored in unc-4; mab-9 double mutants (Fig.5B–D; see also Fig.S4A) (summarized in Fig.5E). Based on the molecular criteria tested here, we conclude that in the absence of unc-4 and mab-9, respectively, SABVs and SABD adopt a “mixed” identity. Altogether, we find that the repressor logic we identified by probing MN class diversification in C. elegans VNC also operates in another anatomical region of the nervous system to generate MN diversity.
Two additional repressors in VNC MNs repress the SAB-specific gene glr-4
It is notable that unc-3 directly drives the expression of glr-4, a glutamate receptor (GluR) gene, in all three SAB but not in VNC MNs (Kratsios et al., 2015). Despite all precedents detailed thus far, we did not observe consistent derepression of glr-4 in VNC MNs of any of the repressor mutants described above (Fig.S4B). We therefore conducted another genetic screen to identify possible repressors of glr-4 expression. From a screen of ~1500 haploid genomes for glr-4::tagrfp derepression in VNC MNs, we identified two alleles, ot785 and ot786. Specifically, ot785 animals show glr-4 derepression in AS MNs, while in ot786 animals, glr-4 becomes derepressed in DA and DB MNs (Fig.5F; Fig.S4C,D). Through whole-genome sequencing, we found that ot786 animals harbor an early stop codon in the first exon of the ARID-type transcription factor cfi-1. An independent cfi-1 allele, ky651 (Shaham and Bargmann, 2002), phenocopies the ot786 mutant phenotype which can be rescued with fosmid DNA that contains the cfi-1 locus (Fig.5F,G; Fig.S4C). The second allele, ot785 defines a distinct locus; it is a missense mutation in the lin-13 gene, a Zn finger transcription factor (Melendez and Greenwald, 2000). An independent lin-13 allele, n770, displays the same glr-4 derepression phenotype (Fig.5F,H; Fig.S4D). We did not further pursue the characterization of lin-13 henceforth (see Fig.S4D for reason).
cfi-1 was previously shown to be expressed in a neuron class-specific manner and involved in specific neuronal fate decisions (Shaham and Bargmann, 2002; Zhang et al., 2014). In VNC MNs, we observed cfi-1 expression in cholinergic DA, DB, VA and VB (and also GABAergic DD and VD) but not AS or VC, nor in SAB or DA9 MNs where glr-4 is normally expressed (Fig.S2E,F). Genetic double mutant analysis shows that the derepression of glr-4 expression in VNC MNs of cfi-1 mutants requires unc-3 (Fig.5F; Fig.S4C). We note that genes expressed either in subsets of cfi-1-expressing neurons (i.e., acr-5 in DB, VB; del-1 in VA, VB; inx-12 in VA,VB MNs) or more broadly (i.e., unc-17 in all cholinergic VNC MNs) are not affected in cfi-1 mutants (Fig.S4G). We thus conclude that cfi-1 targets only genes whose expression is specific to SAB MNs of the RVG and not VNC MNs.
UNC-3 activity is counteracted by repressor proteins through discrete repressive cis-regulatory elements
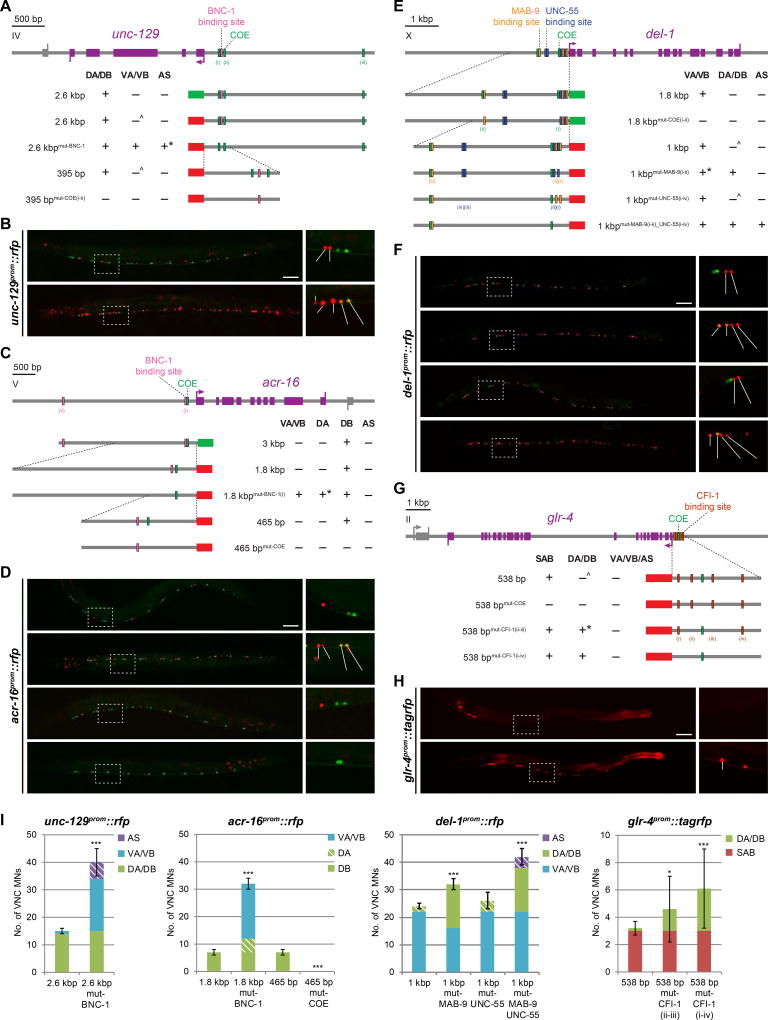
To explore the mechanistic basis of the antagonistic effects of the repressors described above, we dissected the cis-regulatory control region of the following unc-3-dependent, MN class-specific, terminal effector genes: TGFβ family member unc-129, DEG/EnaC channel del-1, nAChR acr-16, and GluR glr-4. Sequences 5’ to the start codon of these genes all contain UNC-3 binding sites (COE motifs). Previous work has already demonstrated that the COE motifs in del-1, unc-129, and glr-4 are required for their expression (shown again in Fig.6) (Kratsios et al., 2015; Kratsios et al., 2011), and mutation of the COE motif in acr-16 demonstrates the same requirement for this gene (Fig.6C,D,I).
Figure 6. Repressor binding sites adjacent to COE motif mediate class specificity of motor neuron effector gene expression.
White-filled vertical rectangles represent mutated sites. Expression observed in corresponding MN class (+); absence of expression (−); although largely absent, weak expression in a small number of MNs occasionally observed (^); unexpected result to be addressed (*). All scale bars = 50 µm.
A,B: Mutation of BNC-1 binding site results in unc-129 derepression in VA/VB and AS MNs. *The BNC-1 site harbors two UNC-55 site consensus motifs with one mismatch in each. These two repressors may be sharing the same site in this case. del-1prom::gfp marks VA/VB MNs.
C,D: Mutating the COE motif causes acr-16 expression to be lost. When the BNC-1 binding site is mutated, acr-16 becomes derepressed in VA/VB and DA MNs. *The BNC-1 site does not resemble that of UNC-4 nor is there an UNC-4 site near that of BNC-1. This result nonetheless supports the notion that effector genes are repressed at the cis-regulatory level to achieve class specificity. 1.8 kb reporter image repeated from Fig.4D; del-1prom::gfp marks VA/VB MNs.
E,F: Mutation of two MAB-9 binding sites adjacent to the COE motif proximal to the start codon results in del-1 derepression in DA/DB MNs. *Expression in anterior VA/VBs (from VA2/VB3 and upward) is at times repressed – hinting at finer subclass regulation. No derepression is observed when all four UNC-55 sites are mutated, but deletion of both validated MAB-9 and all four UNC-55 sites results in del-1 derepression in DA/DB and AS MNs. unc-129prom::gfp marks DA/DB MNs.
G,H: Mutating the two CFI-1 binding sites flanking the COE motif causes glr-4 to be derepressed in DA/DB MNs, albeit in a variable manner. This effect is potentiated when all four sites are mutated.
I: Quantification for B, D, F, and H; error bars show SD. Superimposed diagonal stripes indicate dim expression. At least three independent transgenic lines were assessed although only the most representative line is shown. Unpaired t-tests were performed comparing each line with its corresponding non-mutated control; ***p < 0.001; *p < 0.05; n ≥ 10.
Each of these four effector genes also harbors, in relatively close proximity to the COE motif (15 to 60 bp), predicted binding sites for any of the four presumptive repressor proteins: BNC-1, MAB-9, UNC-55, and CFI-1 (Fig.6). These binding site predictions are based on in vitro-determined binding site motifs using C. elegans proteins (BNC-1 and CFI-1) or on motifs determined for orthologs in other species (MAB-9 and UNC-55) (Table S1, related to Fig.6). We find that in every case tested, mutation of these binding sites results in the predicted derepression of the respective reporter genes. Specifically, mutation of the predicted BNC-1 binding site in the unc-129 5’ upstream region results in derepression of unc-129 reporter in VA and VB MNs (Fig.6A,B,I), phenocopying the effect of bnc-1 loss on wildtype unc-129 reporter (Fig.2A,B). Similarly, mutation of the predicted BNC-1 binding site in the acr-16 locus also results in ectopic VA and VB MN expression of the reporter (Fig.6C,D,I). Mutation of the predicted MAB-9 binding site in the del-1 promoter results in derepression in DA and DB MNs (Fig.6E,F, I), phenocopying the genetic removal of mab-9 (Fig.3F,G). Again, mirroring the genetic analysis that indicated a redundant function for mab-9 and unc-55 in repressing del-1 in AS (Fig.4A,F), only the deletion of both MAB-9 and UNC-55 binding sites results in del-1 reporter derepression in AS MNs (Fig.6E,F, I). Lastly, mutating predicted CFI-1 binding sites in the glr-4 cis-regulatory region results in ectopic DA and DB MN expression (Fig.6G,H,I), as in cfi-1 mutants (Fig.5F; Fig.S4C).
We conclude that the overall logic in the function of the repressors lies in these proteins binding to sites that are in proximity to UNC-3 binding sites, thereby blocking the activating effects of UNC-3 on the expression of these effector genes. Removal of either the repressor binding site or the repressor itself allows UNC-3 to activate its target.
Repressor proteins are continuously required to counteract UNC-3 activity
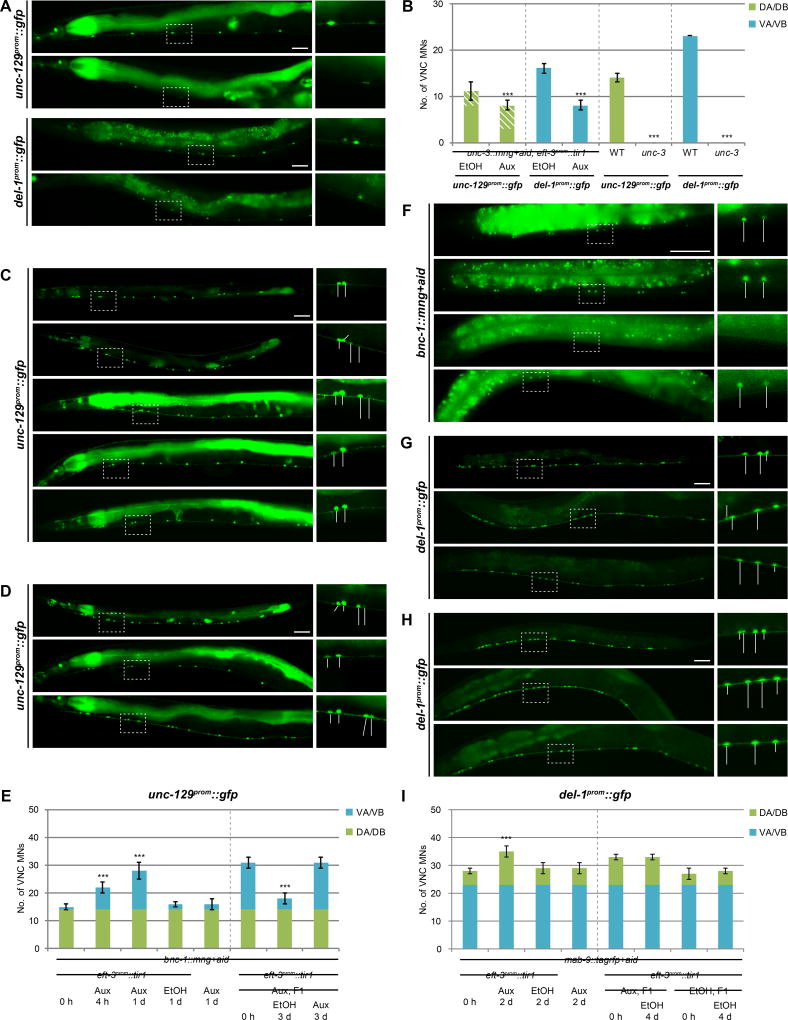
Are MN class-specific repressors only transiently required to, for example, establish a repressive chromatin environment or are they continuously required to prevent activation of genes by UNC-3? We generated conditional alleles of the repressors, based on an auxin-inducible degron (AID) system that can remove tagged proteins in a temporally controlled fashion (Zhang et al., 2015). We first used this system by tagging the unc-3 locus using CRISPR/Cas9-mediated genome engineering. AID-tagging resulted in a slight reduction of UNC-3 activity, even without the addition of auxin; however, postdevelopmental addition of auxin significantly augmented these mild defects, demonstrating that UNC-3 is continuously required to maintain effector gene expression in VNC MNs (Fig.7A,B).
Figure 7. BNC-1 and MAB-9 are continuously required to postdevelopmentally maintain motor neuron class identity.
A: Postdevelopmental UNC-3 degradation by treating young adult, conditional unc-3 worms with auxin for 3 days, results in reduction of unc-129 and del-1 expression. All scale bars = 50 µm.
B: Quantification for A; error bars show SD. UNC-3 function is slightly disrupted when fused to AID. Superimposed diagonal stripes indicate dim expression. Unpaired t-tests were performed comparing each auxin-treated condition with its corresponding EtOH control; ***p < 0.001; n ≥ 13. Data of unc-129 and del-1 expression in WT and unc-3 mutants repeated from Fig.2B and Fig.3G, respectively.
C: Postdevelopmental BNC-1 degradation by treating conditional bnc-1 worms at the L4 stage (just before entry into adulthood) with auxin for 1 day results in unc-129 derepression in VA/VB MNs.
D: First-generation progenies (F1) of conditional bnc-1 worms constitutively grown on auxin phenocopy bnc-1 mutants in terms of unc-129 derepression. When auxin is removed at the L4 stage, repression of unc-129 in VA/VB MNs is restored after 3 days.
E: Quantification for C and D; error bars show SD. BNC-1 function is slightly disrupted when fused to AID. Unpaired t-tests were performed comparing each experimental condition with its corresponding untreated control; ***p < 0.001; n ≥ 13.
F: AID-tagged BNC-1 is degraded within 1 h upon auxin treatment. Upon auxin removal, BNC-1 expression is restored within 6 hrs.
G: Postdevelopmental MAB-9 degradation by treating young adult conditional mab-9 worms with auxin for 2 days results in del-1 derepression in DA MNs. MAB-9 function in DB MNs is disrupted non-conditionally when fused to AID.
H: F1 conditional mab-9 worms constitutively grown on auxin phenocopy mab-9 mutants in terms of del-1 derepression. When auxin is removed at the L4 stage, repression of del-1 in DB MNs is not restored even after 4 days, likely due to the hypomorphic nature of this mab-9 allele.
I: Quantification for G and H; error bars show SD. Unpaired t-tests were performed comparing each experimental condition with its corresponding untreated control; ***p < 0.001; n ≥ 13.
We then proceeded to tag the endogenous bnc-1 and mab-9 loci with AID. In both cases, we find that continuous treatment with auxin results in the expected loss of function phenotypes for the respective loci. Removal of either repressor protein at postdevelopmental stages alone also resulted in derepression of effector genes (unc-129 in the case of bnc-1::mng+aid and del-1 in the case of mab-9::tagrfp+aid) (Fig.7C–I). We conclude that BNC-1 and MAB-9 are indeed continuously required to prevent UNC-3 from ectopically activating MN class-specific genes.
Conversely, we asked whether genes that are ectopically activated by UNC-3 in the absence of BNC-1 or MAB-9, can be shut off following late supply of BNC-1 or MAB-9. In other words, do repressors need to be present during the initial activation phase or can they exert their effect even after activation has occurred? Using the AID-tagged bnc-1 allele, we find that ectopic unc-129 expression due to BNC-1 removal via constitutive supply of auxin can be diminished after auxin removal, which restores BNC-1 and thereby allows it to counteract UNC-3-mediated gene activation (Fig.7D,E). In conclusion, we have shown that the continuous requirement of repressor proteins in postmitotic neurons is required to maintain neuronal identity.
UNC-3 is required for maintenance of bnc-1 and cfi-1 expression
How is the continuous expression and hence continuous activity of the repressors controlled? As some transcription factors can both activate and repress, we wondered if the repressors positively autoregulate and hence maintain their own expression. Using bnc-1 as a potential paradigm, we find that VA/VB-specific expression of the bnc-1 5’ regulatory sequence is unaffected in bnc-1 mutants (Fig.S5A). Since effector genes require unc-3 for their continuous expression, we then asked whether unc-3 may also be required for continuous expression of the repressor genes. Again using bnc-1 as an example, we find that VA/VB-specific bnc-1 expression is initiated in unc-3 mutants at the late L1 stage, shortly after the birth of VA and VB MNs, but is diminished by the fourth larval (L4) stage (Fig.S5B,D). Similarly for cfi-1, in unc-3 mutants, the cfi-1 fosmid reporter is expressed normally in DA and DB MNs at L1 but loses its expression at L4 stage in cholinergic VNC MNs (Fig.S5C,D). These observations indicate that expression of class-specific repressors is initiated independently of unc-3, but then requires unc-3 to be maintained, allowing repressor proteins to in turn counteract the activity of the terminal selector` to specify MN class identity.
DISCUSSION
We have described here a coherent theme for cholinergic MN class diversification in the VNC of the nematode C. elegans. Analyzing five distinct cholinergic MN classes, we described the function of seven distinct repressor proteins that counteract the activity of a broadly acting, master-regulatory transcription factor, UNC-3 to sculpt class-specific gene expression programs (schematized in Fig.S6A). Hence, unique, class-specific combinations of these repressors endow individual MN classes with specific molecular features that define their specific properties. At least some and perhaps all of the repressors function by directly repressing effector genes that are normally expressed in other MN classes. The specificity of expression of an effector gene thus depends on the unique combination of repressor binding sites located in proximity to the effector gene locus. Therefore, the MN classes of animals lacking gene activity for any of the seven repressors described display a “mixed” molecular identity, which is perhaps reflective of a more ancient MN “ground state”.
All transcriptional repressors described here sculpt the expression of effector genes whose protein products are continuously operating in a mature neuron (e.g. ion channels or neurotransmitter receptors). On the other hand, the impact of these repressor proteins on features that are programmed earlier during the differentiation of MNs, namely their axonal trajectories and synaptic specificity, are more varied. UNC-4 ensures the specificity of synaptic innervation of the VA MNs (White et al., 1992; Winnier et al., 1999) while VAB-7 is crucial for correctly instructing the direction of DB axonal projection (Esmaeili et al., 2002). Our own analysis, however, seems to indicate less of a role for BNC-1 and MAB-9 in dictating such features (Fig.S1C–F). Axonal projection and synaptogenesis are presumably controlled by earlier acting factors. How these as yet unidentified factors intersect with UNC-3 function, if they intersect at all, is presently not clear. In Drosophila, transcription factors have been identified to specifically control morphological, but not other molecular identity features of specific MN classes (Enriquez et al., 2015). These so-called “morphology transcription factors (mTFs)” may have counterparts in C. elegans as well.
A number of repressor proteins have been previously shown to be involved in neuronal patterning in vertebrate and invertebrate nervous systems (e.g. (Muhr et al., 2001; Pflugrad et al., 1997; Vallstedt et al., 2001; William et al., 2003) but their mechanism of action has been mostly left unclear, particularly in regard to their target effector genes, their interaction with gene activators (whose activity they must antagonize), and their timing of action. For instance, in the vertebrate spinal cord, HOX proteins of different paralog groups acting as repressors to mutually cross-repress have been shown to be important for specifying the various MN pool as well as columnar identities. Downstream of this developmental program though, only a handful of intermediate transcription factor targets have been characterized, while little is known about the terminal effector gene targets of HOX proteins in MNs (Dasen and Jessell, 2009; Jung et al., 2010). As another example, in invertebrates, UNC-4 acts as a repressor in the VA class of VNC MNs to prevent the adoption of VB-specific features (Pflugrad et al., 1997; Winnier et al., 1999). However, whether such repressor logic represented a common theme operating throughout all C. elegans MN classes was uncertain, and on a mechanistic level it was unclear how transcriptional repressors interact with activators to shape MN class-specific gene expression. Through our analysis of the cis-regulatory control regions of a number of effector genes, we reveal that class-specific genes integrate both positive and negative regulatory inputs. This stands in striking contrast to the mode of regulation of shared effector genes (e.g. ACh pathway genes) expressed in all MN classes, which require only positive regulatory input via the terminal selector UNC-3 and perhaps other, yet to be identified positive co-regulators (Fig.8B).
One important aspect of our findings is that repressor proteins that restrict gene expression in a neuron subtype-specific manner are continuously required to maintain the repressed state through the life of the respective neuron subtypes. It has only been appreciated during the past few years that transcriptional activators acting as terminal selectors to induce a specific differentiated state are also continuously required to maintain patterns of gene activation and hence neuronal identity (Deneris and Hobert, 2014). We corroborate this notion here for the terminal selector UNC-3 and then go on to show that at least some, and based on their continuous expression likely all repressor proteins that counteract UNC-3 are also continuously required to maintain such function. As such, these repressor proteins can themselves be thought of as terminal selectors in determining and maintaining MN class identity. Other scenarios would have been possible too; for example, repressor proteins could establish a chromatin state that prevents UNC-3 from activating its target genes and their activity could then become superfluous after the initiation event. However, as our conditional repressor alleles show, this does not appear to be the case. How the transcriptional repressors described here counteract UNC-3 activity is not clear. UNC-4 is known to act via the corepressor UNC-37, which recruits histone deacetylase complexes (Winnier et al., 1999), and based on the presence of an UNC-37 protein-binding site as well as functions of its orthologs in other species, MAB-9 is likely to act in the same manner. BNC-1 also contains a binding site for a corepressor, CtBP, that is thought to recruit histone deacetylases (Subramanian and Chinnadurai, 2003), but the molecular mode of UNC-55, VAB-7 and CFI-1 function is less clear.
We have shown here that the logic of selective repression of terminal selector-mediated gene activation also applies to the molecular diversification of MN classes that reside outside the VNC, namely the SAB MNs in the RVG. Cholinergic MN classes located in yet different neuronal ganglia in the C. elegans nervous system and controlled by terminal selector genes other than unc-3 face a similar diversification problem. For example, the homeobox gene unc-42 controls the identity of the six RMD MNs (Pereira et al., 2015) and the POU homeobox gene unc-86 controls the identity of the four URA MNs (Zhang et al., 2014). Like the SAB, the RMD and URA MNs can be subdivided into dorsal and ventral classes based on molecular and anatomical criteria. Perhaps in these cases, the terminal selector-type activity of UNC-42 and UNC-86 is also counteracted in a MN class-specific manner by yet to be identified transcriptional repressors. The repressor logic also applies to non-cholinergic MNs. In GABAergic VD class, it has been reported that UNC-55 counteracts the ability of the DD and VD MN terminal selector UNC-30 from inducing DD class genes in VD class MNs (Shan et al., 2005).
One unresolved question is how the MN class-specific expression of the regulatory factors described here is established. We postulate that the intersectional expression of transiently acting factors and their combined effect set up a transcriptional program which eventually turns on the terminal selector unc-3 in distinct MN classes. Similarly, the class-specific expression of the repressor genes is likely also brought about by transiently acting, lineage-specific cues. The discovery of these upstream factors and their regulatory logic and mechanism will further enhance our understanding of how distinct neuronal identities are specified.
Whether the repressor logic that involves selective repression of terminal selector targets, as defined here, may also apply to vertebrate neuron subtype diversification remains to be demonstrated. In progenitor fate specification, it has indeed been previously suggested that progenitor diversity is generated by the intersection of broad activating input (mediated by Sox) with progenitor-specific repressor proteins (Nishi et al., 2015). There are reasons to believe that diversification of postmitotic vertebrate neuron subtypes may follow a similar logic. Midbrain dopaminergic neurons are specified by the terminal selector NURR1, but its effect on select subtype-specific target genes appears to be antagonized by the homeobox gene OTX2 (Di Salvio et al., 2010). Similarly, in the glutamatergic photoreceptor system, NR2E3 antagonizes the activity of the photoreceptor terminal selector CRX in rods to repress the expression of cone-specific genes (Peng et al., 2005). However, in both cases, it is not clear whether repression of effector genes is direct or if continuous repressor activity is required. We hope that our findings will provide a conceptual framework upon which future studies on neuron diversification in other systems can be built.
EXPERIMENTAL PROCEDURES
C. elegans strains
For a complete list of C. elegans mutant and transgenic strains used in this study and descriptions of how they were generated, see Table S2 and Supplemental Experimental Procedures.
Bioinformatic analysis
The binding sites of UNC-3, BNC-1, MAB-9, UNC-55, and CFI-1 were predicted using FIMO (Find Individual Motif Occurrences)(Grant et al., 2011), which is one of the motif-based sequence analysis tools of the MEME (Multiple Expectation maximization for Motif Elicitation) bioinformatics suite (http://meme-suite.org/). See Table S1 for the sources of position weight matrices (PWM) of binding site consensus motifs used in this study. Many of these PWMs are catalogued in the CIS-BP (Catalog of Inferred Sequence Binding Preferences) database (http://cisbp.ccbr.utoronto.ca/)(Weirauch et al., 2014). For each analysis, the p-value threshold was set at < 0.001 and gradually decreased to increase the search stringency until the fewest number (but not zero) of predicted binding sites was obtained. Sequence conservation amongst Caenorhabditis species was assessed using the C. elegans BLAT (Basic local alignment search tool-Like Alignment Tool) search tool of the UCSC (University of California, Santa Cruz) Genome Browser bioinformatics suite (http://genome.ucsc.edu/).
Temporally controlled protein degradation
Conditional protein depletion using the auxin-inducible degradation system in C. elegans was first reported by Zhang et al., (2015). AID-tagged proteins are conditionally degraded when exposed to auxin in the presence of TIR1. To generate the experimental strains, conditional alleles of unc-3(ot837[unc-3::mng+aid]), bnc-1(ot845[bnc-1::mng+aid]), and mab-9(ot863[mab-9::TagRFP+aid]) were independently crossed with ieSi57[eft-3prom::tir1] which expresses TIR1 ubiquitously. The natural auxin, indole-3-acetic acid (IAA), was dissolved in ethanol (EtOH) to prepare 400 mM stock solutions which were stored at 4°C for up to one month. NGM (Nematode Growth Medium) agar plates with fully grown OP50 bacterial lawn were coated with the auxin stock solution to a final concentration of 4 mM and allowed to dry overnight at room temperature. To induce protein degradation, worms of the experimental strains were transferred onto the auxin-coated plates and kept at 20°C. To reverse the process by removing auxin or as controls, worms were transferred onto EtOH-coated plates instead. Auxin solutions, auxin-coated plates, and experimental plates were shielded from light.
Cell identification
The class(es) of fluorescently labeled MNs were identified based on combinations of the following factors: (i) co-localization with or exclusion from another reporter transgene of known class-specific expression, (ii) stereotypic positional pattern, either along the VNC/within the RVG, or relative to other MN classes, (iii) axonal morphology, (iv) developmental stage of generation, (v) number, and (vi) size.
Statistical analysis
For graphs of quantification data shown in all figures, values for those with error bars are expressed as mean ± SD and statistical analyses were performed using the unpaired t-test (two-tailed). For graphs without error bars, values are expressed as percentage (%) and statistical analyses were performed using Fisher’s exact test (two-tailed). Calculations were performed using the GraphPad QuickCalcs online software (http://www.graphpad.com/quickcalcs/). Differences with p < 0.001 were considered significant.
Supplementary Material
Acknowledgments
We thank Chi Chen for the generation of transgenic strains, the CGC for providing strains, the Million Mutation Project for providing the mab-9(gk396730) allele, the Transgeneome Project for providing the bnc-1 fosmid reporter, the Dernburg, Goldstein and Woollard labs for sharing reagents, Kelsey Roberts and Deandrea Ellis for help with the genetic screens, Tulsi Patel for providing the AID-tagged unc-3 allele, Lori Glenwinkel for providing bioinformatics expertise and Hynek Wichterle, Richard Mann and members of the Hobert lab for comments on this manuscript. This work was funded by the Howard Hughes Medical Institute, a K99 fellowship (K99NS084988) and a NINDS grant (R00NS084988) to P.K. and an NIH postdoctoral training grant to M.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHORS CONTRIBUTIONS
S.Y.K., P.K., and O.H. conceived the project. S.Y.K., P.K., M.H., and R.M. performed the experiments. S.Y.K., P.K., and M.H. designed the experiments and analyzed the data. S.Y.K and O.H. prepared the manuscript, with P.K. providing substantial input. O.H. supervised the project.
Supplemental Information includes six figures, two tables, and Supplemental Experimental Procedures.
References
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley RR. The EH1 motif in metazoan transcription factors. BMC Genomics. 2005;6:169. doi: 10.1186/1471-2164-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- Deneris ES, Hobert O. Maintenance of postmitotic neuronal cell identity. Nat Neurosci. 2014;17:899–907. doi: 10.1038/nn.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Acampora D, Prosperi R, Omodei D, Prakash N, Wurst W, Simeone A. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat Neurosci. 2010;13:1481–1488. doi: 10.1038/nn.2661. [DOI] [PubMed] [Google Scholar]
- Duerr JS, Han HP, Fields SD, Rand JB. Identification of major classes of cholinergic neurons in the nematode Caenorhabditis elegans. The Journal of comparative neurology. 2008;506:398–408. doi: 10.1002/cne.21551. [DOI] [PubMed] [Google Scholar]
- Enriquez J, Venkatasubramanian L, Baek M, Peterson M, Aghayeva U, Mann RS. Specification of individual adult motor neuron morphologies by combinatorial transcription factor codes. Neuron. 2015;86:955–970. doi: 10.1016/j.neuron.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili B, Ross JM, Neades C, Miller DM, 3rd, Ahringer J. The C. elegans even-skipped homologue, vab-7, specifies DB motoneurone identity and axon trajectory. Development. 2002;129:853–862. doi: 10.1242/dev.129.4.853. [DOI] [PubMed] [Google Scholar]
- Formaz-Preston A, Ryu JR, Svendsen PC, Brook WJ. The Tbx20 homolog Midline represses wingless in conjunction with Groucho during the maintenance of segment polarity. Dev Biol. 2012;369:319–329. doi: 10.1016/j.ydbio.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Fox RM, Von Stetina SE, Barlow SJ, Shaffer C, Olszewski KL, Moore JH, Dupuy D, Vidal M, Miller DM., 3rd A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics. 2005;6:42. doi: 10.1186/1471-2164-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P, Lillesaar C. Probing the diversity of serotonin neurons. Philos Trans R Soc Lond B Biol Sci. 2012;367:2382–2394. doi: 10.1098/rstb.2011.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P, Farago AF, Awatramani RB, Scott MM, Deneris ES, Dymecki SM. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lacombe J, Mazzoni EO, Liem KF, Jr, Grinstein J, Mahony S, Mukhopadhyay D, Gifford DK, Young RA, Anderson KV, et al. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron. 2010;67:781–796. doi: 10.1016/j.neuron.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbrun E, Greco TM, Slagle CE, Kennedy LM, Li T, Cristea IM, Conlon FL. A Gro/TLE-NuRD corepressor complex facilitates Tbx20-dependent transcriptional repression. J Proteome Res. 2013;12:5395–5409. doi: 10.1021/pr400818c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P, Pinan-Lucarre B, Kerk SY, Weinreb A, Bessereau JL, Hobert O. Transcriptional coordination of synaptogenesis and neurotransmitter signaling. Curr Biol. 2015;25:1282–1295. doi: 10.1016/j.cub.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci. 2011;15:205–214. doi: 10.1038/nn.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Greenwald I. Caenorhabditis elegans lin-13, a member of the LIN-35 Rb class of genes involved in vulval development, encodes a protein with zinc fingers and an LXCXE motif. Genetics. 2000;155:1127–1137. doi: 10.1093/genetics/155.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Hempel C, Sugino K. Probing the transcriptome of neuronal cell types. Curr Opin Neurobiol. 2006;16:571–576. doi: 10.1016/j.conb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Nishi Y, Zhang X, Jeong J, Peterson KA, Vedenko A, Bulyk ML, Hide WA, McMahon AP. A direct fate exclusion mechanism by Sonic hedgehog-regulated transcriptional repressors. Development. 2015;142:3286–3293. doi: 10.1242/dev.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Freret ME, Rood BD, Brust RD, Hennessy ML, deBairos D, Kim JC, Cook MN, Dymecki SM. Multi-Scale Molecular Deconstruction of the Serotonin Neuron System. Neuron. 2015;88:774–791. doi: 10.1016/j.neuron.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MK. Disconnected, a C2H2 Zinc Finger Protein has a role in Appendage Formation and Gene Regulation in Drosophila. Raleigh, North Carolina: North Carolina State University; 2007. [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, White JG, LeBoeuf B, Garcia LR, Alon U, et al. A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife. 2015;4 doi: 10.7554/eLife.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrad A, Meir JY, Barnes TM, Miller DM., 3rd The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development. 1997;124:1699–1709. doi: 10.1242/dev.124.9.1699. [DOI] [PubMed] [Google Scholar]
- Poulin JF, Zou J, Drouin-Ouellet J, Kim KY, Cicchetti F, Awatramani RB. Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep. 2014;9:930–943. doi: 10.1016/j.celrep.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeper J. Dissecting the diversity of midbrain dopamine neurons. Trends in neurosciences. 2013;36:336–342. doi: 10.1016/j.tins.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Shaham S, Bargmann CI. Control of neuronal subtype identity by the C. elegans ARID protein CFI-1. Genes Dev. 2002;16:972–983. doi: 10.1101/gad.976002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan G, Kim K, Li C, Walthall WW. Convergent genetic programs regulate similarities and differences between related motor neuron classes in Caenorhabditis elegans. Dev Biol. 2005;280:494–503. doi: 10.1016/j.ydbio.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Subramanian T, Chinnadurai G. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 2003;540:255–258. doi: 10.1016/s0014-5793(03)00275-8. [DOI] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell TM, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31:743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- Vanhoutteghem A, Bouche C, Maciejewski-Duval A, Herve F, Djian P. Basonuclins and disco: Orthologous zinc finger proteins essential for development in vertebrates and arthropods. Biochimie. 2011;93:127–133. doi: 10.1016/j.biochi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Von Stetina SE, Treinin M, Miller DM. The Motor Circuit. Int Rev Neurobiol. 2006;69:125–167. doi: 10.1016/S0074-7742(05)69005-8. [DOI] [PubMed] [Google Scholar]
- Walthall WW, Plunkett JA. Genetic transformation of the synaptic pattern of a motoneuron class in Caenorhabditis elegans. J Neurosci. 1995;15:1035–1043. doi: 10.1523/JNEUROSCI.15-02-01035.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN. Mutations in the Caenorhabditis elegans unc-4 gene alter the synaptic input to ventral cord motor neurons. Nature. 1992;355:838–841. doi: 10.1038/355838a0. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London B Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- William CM, Tanabe Y, Jessell TM. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130:1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
- Winnier AR, Meir JY, Ross JM, Tavernarakis N, Driscoll M, Ishihara T, Katsura I, Miller DM., 3rd UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes Dev. 1999;13:2774–2786. doi: 10.1101/gad.13.21.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard A, Hodgkin J. The caenorhabditis elegans fate-determining gene mab-9 encodes a T-box protein required to pattern the posterior hindgut. Genes Dev. 2000;14:596–603. [PMC free article] [PubMed] [Google Scholar]
- Wyler SC, Spencer WC, Green NH, Rood BD, Crawford L, Craige C, Gresch P, McMahon DG, Beck SG, Deneris E. Pet-1 Switches Transcriptional Targets Postnatally to Regulate Maturation of Serotonin Neuron Excitability. J Neurosci. 2016;36:1758–1774. doi: 10.1523/JNEUROSCI.3798-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhang F, Bhattacharya A, Nelson JC, Abe N, Gordon P, Lloret-Fernandez C, Maicas M, Flames N, Mann RS, Colon-Ramos DA, et al. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development. 2014;141:422–435. doi: 10.1242/dev.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015;142:4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HM, Walthall WW. UNC-55, an Orphan Nuclear Hormone Receptor, Orchestrates Synaptic Specificity among Two Classes of Motor Neurons in Caenorhabditis elegans. J Neurosci. 1998;18:10438–10444. doi: 10.1523/JNEUROSCI.18-24-10438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.