Abstract
Cell adhesion molecule (CADM) genes encode immunoglobulin superfamily molecules, which are involved in cell-cell adhesion in a number of human epithelia. Through the maintenance of epithelia, CADM genes protect against malignant conversion and metastasis. Whilst numerous in vitro studies have investigated the molecular characteristics of CADM1 and CADM4 and in vivo studies have investigated CADM1 and CADM4 expression in a number of tumor types, the roles of CADM1 and CADM4 have yet to be elucidated. Therefore, in the present study, CADM1 and CADM4 expression levels were evaluated using immunohistochemistry staining in 208 patients with breast cancer and compared with clinicopathological factors. CADM1 and CADM4 expression levels were negative in 160 (76.9%) and 166 (79.8%) of the 208 cases, respectively. The lack of expression in these cases was associated with advanced tumor stage, suggesting that inactivation of CADM1 and CADM4 promotes breast cancer development. The prognostic role of CADM1 and CADM4 in breast cancer was also evaluated and the expression of CADM1 and CADM4 were not associated with cancer-specific survival or overall survival rate in the cohort of patients in the present study. Whilst these results suggested that CADM1 and CADM4 possess tumor suppressive roles, further functional experiments are required to address the important mechanisms involving CADM1 and CADM4.
Keywords: cell adhesion molecule 1, cell adhesion molecule 4, tumor suppressor gene, breast cancer
Introduction
Breast cancer remains the leading cause of cancer mortality in females in developing and developed regions (1,2). Recurrence and poor survival rates are still reported for certain patients diagnosed with stage I breast cancer (3–5). In addition to surgical resection, there are specific treatments administered to patients with breast cancer, including sentinel lymph node biopsy followed by surgery, radiation therapy, chemotherapy, hormone therapy and targeted therapy. Personalized therapies are administered depending on the type and stage of the cancer and the expression status of estrogen receptor (ER), progesterone receptor (PgR) and human epidermal growth factor receptor 2 (HER2). Although personalized therapy based on the intrinsic subtype of breast cancer has progressed, further understanding of the molecular biology of the genes involved in breast cancer development is required.
Cell adhesion molecule (CADM) genes include 4 subfamilies, which are CADM1, CADM2, CADM3 and CADM4. These genes encode an immunoglobulin superfamily molecule that is involved in the maintenance of cell-cell adhesion in a variety of human epithelial cells and therefore protects against malignant conversion (6,7). It has been established that CADM1 is located on chromosome 11q23.2 and functions as a tumor suppressor gene in non-small cell lung cancer (8). CADM1 expression is preferentially lost in invasive lung adenocarcinoma lesions compared with those that are non-invasive (9). Previous studies have identified that the expression of CADM1 is frequently inactivated in numerous types of cancer, including breast cancer (10–14). The loss of CADM1 and protein 4.1B expression leads to the development and progression of breast cancer, particularly in invasion and metastasis, using primary breast cancer specimens and breast cancer cell lines (15). Low or aberrant expression of CADM1 in tumors is significantly associated with lymphovascular invasion and advanced tumor stages, demonstrating the potential for CADM1 expression to be used as a prognostic biomarker that may aid effective personalized therapy.
CADM4 is located on chromosome 19q13.31 and investigation into its role in tumor development is ongoing. Previous studies have demonstrated that CADM4 may function as a tumor suppressor in renal clear cell carcinoma and that it is inactivated in breast and colon cancer (16,17). Although several previous studies have demonstrated the tumor suppressive roles of CADM1 and CADM4, further studies are required to evaluate the expression levels in clinical samples.
The present study reports the association of CADM1 and CADM4 expression levels with clinicopathological factors, including survival rate in patients with breast cancer, evaluated using immunohistochemistry (IHC) staining.
Materials and methods
Patients and tissue samples
Primary invasive breast cancer samples were obtained from patients that underwent surgical resection without systemic drug therapy prior to surgery at the Department of Organ Regulatory Surgery at Fukushima Medical University Hospital (Fukushima, Japan). This cohort consisted of 208 patients who were recruited at the time of surgery between January 2002 and December 2005.
Detailed backgrounds for each tissue donor were collected, including age, sex, clinical stage and hormone status (Table I). Tumor histopathology was classified according to the Union for International Cancer Control (UICC) tumor-node-metastasis classification (the 7th classification) (18,19). Written informed consent was obtained from all participants. This study was approved by the Institutional Review Board of Fukushima Medical University (Fukushima, Japan).
Table I.
Clinicopathological characteristics of patients with primary breast cancer.
| Characteristics | n (%) |
|---|---|
| Age (years) | |
| Average (range) | 55.3 (31–88) |
| <50 | 82 (39) |
| ≥50 | 126 (61) |
| Sex | |
| Male | 0 (0) |
| Female | 208 (100) |
| Metastatic lymph nodes | |
| 0 | 128 (62) |
| 1–3 | 56 (27) |
| ≥4 | 24 (11) |
| Lymphovascular invasion | |
| − | 84 (40) |
| + | 124 (60) |
| Hormone status | |
| +/+ and +/− | 157 (75) |
| −/− | 51 (25) |
| Human epidermal growth factor receptor 2 | |
| + | 16 (8) |
| − | 192 (92) |
| Triple negative | 33 (16) |
| pT stage | |
| 1 | 116 (56) |
| 2 | 83 (40) |
| 3 | 3 (1) |
| 4 | 6 (3) |
| Pathological stage | |
| I | 82 (39) |
| II | 120 (58) |
| III | 6 (3) |
| Local recurrence | |
| − | 202 (97) |
| + | 6 (3) |
| Remote recurrence | |
| − | 192 (92) |
| + | 16 (8) |
Total number of patients, 208. Hormone status, estrogen and progesterone receptor status; TNM, tumor-node-metastasis; pT, pathological tumor stage.
IHC analysis
The aforementioned breast cancer tissue samples were fixed in 10% formalin, embedded in paraffin, cut into 4 µm sections and stained with hematoxylin and eosin (H&E) and other primary antibodies. Rabbit polyclonal antibodies against CADM1 (1:500, C-18, generated by the Division of Molecular Pathology, Institute of Medical Science, The University of Tokyo) and CADM4 (1:500, Bc-2, generated by the Division of Molecular Pathology, Institute of Medical Science, The University of Tokyo) were used as described previously (20). Antibodies used for IHC staining were as follows: Anti-ER (1:500, cat. no., MA5-13191; Dako; Agilent Technologies GmbH, Waldbronn, Germany), anti-PgR (1:500, cat. no., MA5-12581; Dako; Agilent Technologies GmbH). For HER2 status, the Histofine® Simple Stain HER2 mono assay kit was used (cat. no. 427041; Nichirei Biosciences, Inc., Tokyo, Japan). Analyses of ER, PgR and HER2 were performed by IHC staining according to the manufacturer's protocol. Sections were deparaffinized in xylene and hydrated using a graded series of ethanol at room temperature. Subsequently the sections were washed 3 times in PBS and endogenous peroxidase was blocked with 0.3% in methanol for 30 min at room temperature. Antigens were retrieved by autoclaving the sections on slides in 0.01 M pH 6.0 citrate buffer for 10 min at 121°C. Subsequent to washing in PBS, the sections were incubated in primary antibody overnight at 4°C. A further wash in PBS was followed by treatment with the secondary antibody [K1491, Dako EnVision kit/horseradish peroxidase (HRP)] for 30 min at room temperature and diaminobenzidine (K1491, Dako EnVision kit/HRP) was used for staining detection (both from Dako: Agilent Technologies GmbH). Finally, the sections were counterstained with hematoxylin. Expression of these proteins was evaluated using optical microscopy (BX43; Olympus Corporation, Tokyo, Japan) at ×400 magnification.
Assessment of IHC stain
The stain signals of CADM1 and CADM4 protein levels were detected in the membranes in normal mammary epithelial cells. Cytoplasmic immunoreactivity without membrane staining was defined as aberrant expression. Membranous staining of CADM1 or CADM4 was evaluated by calculating the percentage of cancer cells with membrane expression in the entire area of invasive and non-invasive lesions. The tumors or lesions were then scored as previously described (15). Tumors were defined with scores of 1 (11–30% cells with membrane expression), 2 (31–60%) or 3 (61–100%) as positive staining for CADM1 or CADM4 expression and tumors with a score of 0 (0–10%) as negative staining. ER, PgR and HER2 expression levels were evaluated semi-quantitatively with scores representing the ratio of the number of positive staining cells compared with negative cells, as previously described (21). Assessment of the staining was performed blindly by two independent investigators, including an experienced pathologist (Dr Akiteru Goto from Akita University, who is the pathologist and Dr Motonobu Saito from Fukushima Medical University). Discordance was resolved by discussion.
Statistical analysis
Statistical analysis was performed using JMP v10 software (SAS Institute, Inc., Cary, NC, USA). The Fisher's exact test and χ2 test was used to examine the association between CADM1/4 expression levels and clinicopathological parameters. The Kaplan-Meier method using GraphPad Prism v6.0 (GraphPad Software, Inc., La Jolla, CA, USA) and log rank test were used to plot overall survival and cancer specific curves. P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics and CADM1 and CADM4 expression levels in breast cancer
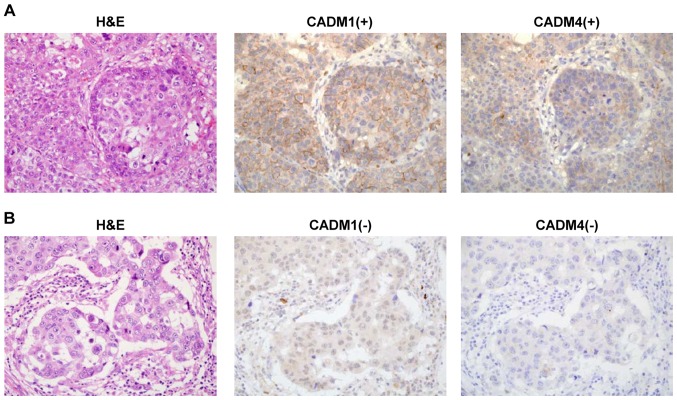
CADM1 and CADM4 expression levels were evaluated using IHC staining in 208 patients with breast cancer (Fig. 1A and B). The characteristics of these patients are presented in Table I. This cohort included 33 (16%) patients with triple negative breast cancer and primarily consisted of stage I and II disease. CADM1 expression was positive in 48 patients (23.1% with scores of 1, 2 or 3) and negative in 160 patients (76.9% with a score of 0). CADM4 expression was positive in 42 patients (20.2%, with scores of 1, 2 or 3) and negative in 166 patients (79.8% with a score of 0; Table II).
Figure 1.
Representative images of CADM1 and CADM4 immunohistochemistry staining with H&E staining in breast cancer tissue. (A) Tissue with positive CADM1 and CADM4 staining. (B) Tissue with negative CADM1 and CADM4 staining. Magnification, ×400. H&E, haematoxylin and eosin; CADM, cell adhesion molecule.
Table II.
CADM1 and CADM4 expression status in the current cohort.
| IHC results | CADM1 | CADM4 |
|---|---|---|
| Negative | ||
| 0 | 160 | 166 |
| Positive | ||
| 1 | 34 | 29 |
| 2 and 3 | 14 | 13 |
Total number of patients, 208. IHC staining scores of 1 (11–30% cells with membrane expression), 2 (31–60%) and 3 (61–100%) as positive staining (+) and tumors with score of 0 (0–10%) as negative staining (−) for CADM1 and CADM4 expression. IHC, immunohistochemistry; CADM, cell adhesion molecule.
Associations between CADM1 and CADM4 expression levels and clinicopathological factors in breast cancer
CADM1 and CADM4 expression levels were not associated with age, number of metastatic lymph nodes, lymphovascular invasion, local recurrence or remote recurrence (Table III). However, the lack of expression of CADM4 was significantly reduced in patients who were ER and PgR negative compared with those who were ER and/or PgR positive (hormone status; P=0.004). Notably, the lack of expression of CADM4 was significantly reduced in the triple negative breast cancer cases compared with other all cases (P=0.008). In addition, there was a no statistically significant association between CADM1 and CADM4 expression and depth of invasion (pT stage; P=0.041 and P=0.039, respectively) and pathological stage (P=0.038 and P=0.029, respectively).
Table III.
Clinicopathological characteristics of patients with primary breast cancer and expression of CADM1 and CADM4.
| CADM1, n (%) | CADM4, n (%) | |||||
|---|---|---|---|---|---|---|
| Characteristics | + (n=48) | − (n=160) | P-value | + (n=42) | − (n=166) | P-value |
| Age (years) | 0.738 | 0.857 | ||||
| <50 | 20 (42) | 62 (39) | 14 (33) | 60 (36) | ||
| ≥50 | 28 (58) | 98 (61) | 28 (67) | 106 (64) | ||
| Metastatic lymph nodes | 0.137 | 0.083 | ||||
| 0 | 34 (71) | 94 (59) | 30 (71) | 99 (60) | ||
| 1–3 | 12 (25) | 44 (28) | 11 (26) | 44 (27) | ||
| ≥4 | 2 (4) | 22 (13) | 1 (3) | 23 (13) | ||
| Lymphovascular invasion | 0.617 | 0.863 | ||||
| − | 21 (44) | 63 (39) | 17 (40) | 70 (42) | ||
| + | 27 (56) | 97 (61) | 25 (60) | 96 (58) | ||
| Hormone status | 0.570 | 0.004 | ||||
| +/+ or +/− | 38 (79) | 119 (74) | 24 (57) | 133 (80) | ||
| −/− | 10 (21) | 41 (26) | 18 (43) | 33 (20) | ||
| Her2 | 0.126 | 1 | ||||
| Positive | 1 (2) | 15 (9) | 3 (7) | 13 (8) | ||
| Negative | 47 (98) | 145 (91) | 39 (93) | 153 (92) | ||
| Triple negative | 0.367 | 0.008 | ||||
| Yes | 10 (21) | 23 (14) | 13 (31) | 20 (12) | ||
| No | 38 (79) | 137 (86) | 29 (69) | 146 (88) | ||
| pT stage | 0.041 | 0.039 | ||||
| 1 | 35 (73) | 81 (50) | 31 (74) | 83 (50) | ||
| 2 | 13 (27) | 70 (44) | 10 (24) | 75 (45) | ||
| 3 | 0 | 3 (2) | 0 | 3 (2) | ||
| 4 | 0 | 6 (4) | 1 (2) | 5 (3) | ||
| Pathological stage | 0.038 | 0.029 | ||||
| I | 26 (54) | 56 (35) | 24 (57) | 58 (35) | ||
| II | 22 (46) | 98 (61) | 17 (41) | 103 (62) | ||
| III | 0 | 6 (4) | 1 (2) | 5 (3) | ||
| Local recurrence | 1 | 1 | ||||
| − | 47 (98) | 155 (97) | 41 (98) | 161 (97) | ||
| + | 1 (2) | 5 (3) | 1 (2) | 5 (3) | ||
| Remote recurrence | 0.372 | 0.314 | ||||
| − | 46 (96) | 146 (91) | 37 (88) | 156 (94) | ||
| + | 2 (4) | 14 (9) | 5 (12) | 10 (6) | ||
P-values were calculated using Fisher's exact test for age, lymphovascular invasion, hormone status, her2, triple negative, local recurrence and remote recurrence or a χ2 test for metastatic lymph node, pT stage and pathological stage. HER2, Human epidermal growth factor receptor 2; CADM, cell adhesion molecule; hormone status, estrogen and progesterone receptor status; pT, pathological tumor stage.
Associations between CADM1 and CADM4 expression levels and prognosis in breast cancer
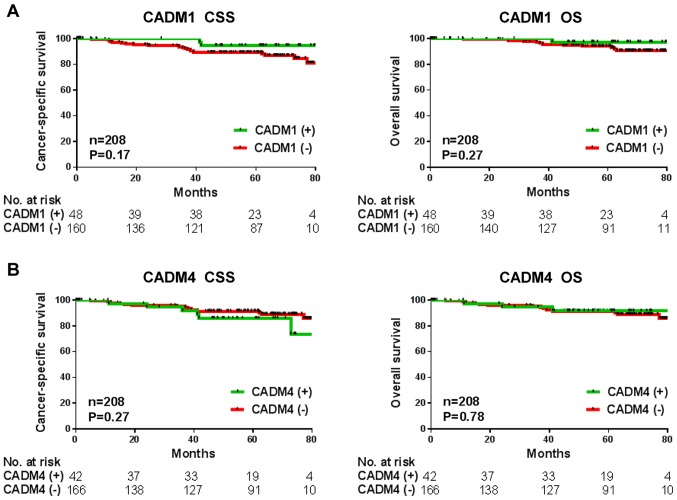
To investigate whether CADM1 and CADM4 may be a potential prognostic biomarker in the cohort in the present study, Kaplan-Meier analysis was performed. Date of tumor recurrence or mortality were considered the end points for cancer-specific survival and overall survival rates. The patients with decreased CADM1 expression levels had poorer cancer-specific and overall survival rates (P=0.17 and P=0.27, respectively), but this was not significant in the current cohort (Fig. 2A). There was also no association observed between decreased CADM4 levels and cancer-specific or overall survival rates (P=0.27 and P=0.78, respectively; Fig. 2B).
Figure 2.
Kaplan-Meier survival analysis of CADM1 and CADM4 tumor expression levels in the cohort of patients with breast cancer. (A) Kaplan-Meier estimate of CSS and OS according to CADM1 expression. (B) Kaplan-Meier estimate of CSS and OS according to CADM4 expression. CCS, cancer-specific survival; OS, overall survival; CADM, cell adhesion molecule.
Discussion
In the present study, CADM1 and CADM4 expression levels were evaluated in patients with breast cancer and compared with clinicopathological factors. The decreased expression levels of CADM1 and CADM4 were significantly associated with advanced tumor progression (pT stage and pathological stage) in this breast cancer cohort (Table III). It has been reported that decreased expression of CADM1 correlates with local invasion, lymph node metastasis, lymphovascular invasion (15) and poor prognosis in breast cancer (22). In addition, lower expression of CADM4 also correlates with poor prognosis (17). These previous studies suggest that CADM1 and CADM4 possess important tumor suppressive roles in breast cancer and support the results of the current study.
Although inactivation of CADM1 via promoter methylation has been observed in breast cancer (15), further investigation into the molecular aspects is required. CADM1 has an important role in tumor development through suppressing epithelial-mesenchymal transition and oncogenic signaling (23). In addition to the current study which demonstrates the loss of CADM1 and CADM4 are associated with breast cancer progression, a recent study revealed that loss of CADM1 expression was detected in patients with metastasis compared with those without, suggesting that CADM1 may serve an important role in preventing the progression of metastasis (22). As CADM4 is involved in cell-to-cell adhesion, loss of CADM4 may also be associated with metastasis through this function. However, the role of CADM4 in cancer progression remains to be fully elucidated. Whilst the association between hormone status and CADM1 and CADM4 expression levels remains unknown, the results of the present study identify that the loss of expression of CADM1 and CADM4 are less frequent in ER and PgR negative cases, particularly as the loss of CADM4 expression was not significantly associated with triple negative breast cancer cases in the present study. Therefore, further studies are required to investigate these associations.
Biomarkers that are capable of identifying patients at high risk of relapse following surgical resection may provide physicians with tools to aid in the diagnosis of cancer recurrence and administer optimal therapeutic strategies. Previous studies have demonstrated that CADM1 and CADM4 may be an effective prognostic biomarker in numerous types of malignant tumor, including breast cancer (17,22). Loss of CADM1 was significantly associated with poorer disease free survival and overall survival rates in patients with breast cancer as well as in a limited number of ER and PgR-positive patients (22). Furthermore, the loss of CADM4 expression was associated with poorer disease-free survival in patients with stage I or II breast cancer.
In conclusion, the present study aimed to validate the use of CADM1 and CADM4 as prognostic biomarkers, but neither was validated using the current cohort (Fig. 2). Future studies are required to further inform on this important field of research. The present study has a number of limitations. Firstly, the patients primarily had stage I or II breast cancer and this was a small sample size in this cohort and therefore are not sufficient to enable an effective analysis of patient prognosis. Secondly, the evaluation of CADM1 and CADM4 protein expression levels used IHC staining only. As CADM1 expression was regulated by promoter methylation or gene aberrations, further investigation of these alterations are required.
References
- 1.Katanoda K, Hori M, Matsuda T, Shibata A, Nishino Y, Hattori M, Soda M, Ioka A, Sobue T, Nishimoto H. An updated report on the trends in cancer incidence and mortality in Japan, 1958-2013. Jpn J Clin Oncol. 2015;45:390–401. doi: 10.1093/jjco/hyv002. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Chia SK, Speers CH, Bryce CJ, Hayes MM, Olivotto IA. Ten-year outcomes in a population-based cohort of node-negative, lymphatic, and vascular invasion-negative early breast cancers without adjuvant systemic therapies. J Clin Oncol. 2004;22:1630–1637. doi: 10.1200/JCO.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 4.Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. Oncologist. 2004;9:606–616. doi: 10.1634/theoncologist.9-6-606. [DOI] [PubMed] [Google Scholar]
- 5.Hanrahan EO, Valero V, Gonzalez-Angulo AM, Hortobagyi GN. Prognosis and management of patients with node-negative invasive breast carcinoma that is 1 cm or smaller in size (stage 1; T1a, bN0M0): A review of the literature. J Clin Oncol. 2006;24:2113–2122. doi: 10.1200/JCO.2005.02.8035. [DOI] [PubMed] [Google Scholar]
- 6.Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 2005;96:543–552. doi: 10.1111/j.1349-7006.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami Y. Functional cloning of a tumor suppressor gene, TSLC1, in human non-small cell lung cancer. Oncogene. 2002;21:6936–6948. doi: 10.1038/sj.onc.1205825. [DOI] [PubMed] [Google Scholar]
- 8.Kuramochi M, Fukuhara H, Nobukuni T, Kanbe T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura T, et al. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 9.Goto A, Niki T, Chi-Pin L, Matsubara D, Murakami Y, Funata N, Fukayama M. Loss of TSLC1 expression in lung adenocarcinoma: Relationships with histological subtypes, sex and prognostic significance. Cancer Sci. 2005;96:480–486. doi: 10.1111/j.1349-7006.2005.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui AB, Lo KW, Kwong J, Lam EC, Chan SY, Chow LS, Chan AS, Teo PM, Huang DP. Epigenetic inactivation of TSLC1 gene in nasopharyngeal carcinoma. Mol Carcinog. 2003;38:170–178. doi: 10.1002/mc.10156. [DOI] [PubMed] [Google Scholar]
- 11.Honda T, Tamura G, Waki T, Jin Z, Sato K, Motoyama T, Kawata S, Kimura W, Nishizuka S, Murakami Y. Hypermethylation of the TSLC1 gene promoter in primary gastric cancers and gastric cancer cell lines. Jpn J Cancer Res. 2002;93:857–860. doi: 10.1111/j.1349-7006.2002.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen M, Fukushima N, Rosty C, Walter K, Altink R, Heek TV, Hruban R, Offerhaus JG, Goggins M. Aberrant methylation of the 5′ CpG island of TSLC1 is common in pancreatic ductal adenocarcinoma and is first manifest in high-grade PanlNs. Cancer Biol Ther. 2002;1:293–296. doi: 10.4161/cbt.84. [DOI] [PubMed] [Google Scholar]
- 13.Fukuhara H, Kuramochi M, Fukami T, Kasahara K, Furuhata M, Nobukuni T, Maruyama T, Isogai K, Sekiya T, Shuin T, et al. Promoter methylation of TSLC1 and tumor suppression by its gene product in human prostate cancer. Jpn J Cancer Res. 2002;93:605–609. doi: 10.1111/j.1349-7006.2002.tb01297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steenbergen RD, Kramer D, Braakhuis BJ, Stern PL, Verheijen RH, Meijer CJ, Snijders PJ. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y, Iwai M, Kawai T, Arakawa A, Ito T, Sakurai-Yageta M, Ito A, Goto A, Saito M, Kasumi F, Murakami Y. Aberrant expression of tumor suppressors CADM1 and 4.1B in invasive lesions of primary breast cancer. Breast Cancer. 2012;19:242–252. doi: 10.1007/s12282-011-0272-7. [DOI] [PubMed] [Google Scholar]
- 16.Jang SM, Han H, Jun YJ, Jang SH, Min KW, Sim J, Ahn HI, Lee KH, Jang KS, Paik SS. Clinicopathological significance of CADM4 expression, and its correlation with expression of E-cadherin and Ki-67 in colorectal adenocarcinomas. J Clin Pathol. 2012;65:902–906. doi: 10.1136/jclinpath-2012-200730. [DOI] [PubMed] [Google Scholar]
- 17.Jang SM, Sim J, Han H, Ahn HI, Kim H, Yi K, Jun YJ, Rehman A, Chung MS, Jang K, Paik SS. Clinicopathological significance of CADM4 expression in invasive ductal carcinoma of the breast. J Clin Pathol. 2013;66:681–686. doi: 10.1136/jclinpath-2012-201405. [DOI] [PubMed] [Google Scholar]
- 18.Sobin LH, Compton CC. TNM seventh edition: What's new, what's changed: communication from the international union against cancer and the American joint committee on cancer. Cancer. 2010;116:5336–5339. doi: 10.1002/cncr.25537. [DOI] [PubMed] [Google Scholar]
- 19.Sobin LH, Gospodarowicz MK, Wittekind Ch. International Union Against Cancer (UICC) TNM Classification of Malignant Tumors. 7th. Wiley-Blackwell; Oxford, UK: 2009. [Google Scholar]
- 20.Nagata M, Sakurai-Yageta M, Yamada D, Goto A, Ito A, Fukuhara H, Kume H, Morikawa T, Fukayama M, Homma Y, Murakami Y. Aberrations of a cell adhesion molecule CADM4 in renal clear cell carcinoma. Int J Cancer. 2012;130:1329–1337. doi: 10.1002/ijc.26160. [DOI] [PubMed] [Google Scholar]
- 21.Saito M, Matsuzaki M, Sakuma T, Katagata N, Watanabe F, Yamaguchi Y, Schetter AJ, Takenoshita S, Nomizu T. Clinicopathological study of non-palpable familial breast cancer detected by screening mammography and diagnosed as DCIS. Breast Cancer. 2014;21:140–145. doi: 10.1007/s12282-012-0389-3. [DOI] [PubMed] [Google Scholar]
- 22.Wikman H, Westphal L, Schmid F, Pollari S, Kropidlowski J, Sielaff-Frimpong B, Glatzel M, Matschke J, Westphal M, Iljin K, et al. Loss of CADM1 expression is associated with poor prognosis and brain metastasis in breast cancer patients. Oncotarget. 2014;5:3076–3087. doi: 10.18632/oncotarget.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda M, Kikuchi S, Maruyama T, Sakurai-Yageta M, Williams YN, Ghosh HP, Murakami Y. Tumor suppressor in lung cancer (TSLC)1 suppresses epithelial cell scattering and tubulogenesis. J Biol Chem. 2005;280:42164–42171. doi: 10.1074/jbc.M507136200. [DOI] [PubMed] [Google Scholar]