Abstract
Alternative splicing is a tightly regulated biological process by which the number of gene products for any given gene can be greatly expanded. Genomic variants in splicing regulatory sequences can disrupt splicing and cause disease. Recent developments in sequencing technologies and computational biology have allowed researchers to investigate alternative splicing at an unprecedented scale and resolution. Population-scale transcriptome studies have revealed many naturally occurring genetic variants that modulate alternative splicing and consequently influence phenotypic variability and disease susceptibility in human populations. Innovations in experimental and computational tools such as massively parallel reporter assays and deep learning have enabled the rapid screening of genomic variants for their causal impacts on splicing. In this review, we describe technological advances that have greatly increased the speed and scale at which discoveries are made about the genetic variation of alternative splicing. We summarize major findings from population transcriptomic studies of alternative splicing and discuss the implications of these findings for human genetics and medicine.
Main Text
Introduction
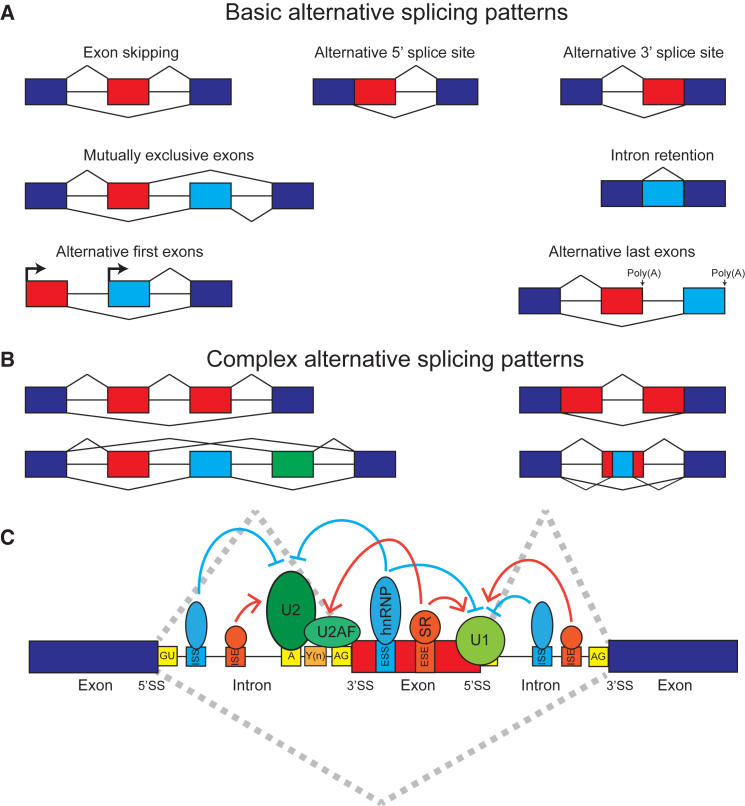
Pre-mRNA splicing is a conserved biological process in which introns within nascent RNA molecules are removed and exons are ligated to form mature mRNA products.1 Through alternative choices of exons and splice sites during splicing—a process known as alternative splicing—a single gene can produce multiple mRNA isoforms that dramatically diversify the transcriptome and the proteome.2 Although the human genome has only approximately 20,000 protein-coding genes,3 the unique mRNA isoforms generated from each gene can be more than ten times that number.4 Nearly all multi-exon human genes are alternatively spliced.5, 6 The basic patterns of alternative splicing include exon skipping, alternative 5′ and 3′ splice sites, mutually exclusive exons, intron retention, and alternative splicing coupled with alternative first or last exons (Figure 1A). Beyond these basic patterns involving binary choices of exons or splice sites during splicing, many complex alternative splicing patterns exist in the transcriptome7 (see Figure 1B for examples). In extreme cases, the combinatorial choices of multiple alternatively spliced regions can generate tens of thousands of mRNA isoforms from a single gene.8 The resulting mRNA isoforms can have distinct regulatory properties in the cell, such as localization, stability, and translational efficiency, and can be translated into stable protein isoforms with divergent structures and functions.9, 10 Therefore, alternative splicing provides a powerful mechanism for expanding the regulatory and functional repertoire of eukaryotic organisms.
Figure 1.
A Primer on Alternative Splicing
(A and B) Basic (A) and complex (B) patterns of alternative splicing. Dark-blue boxes represent constitutively spliced exons. Red, light-blue, and green boxes represent alternatively spliced exons.
(C) Alternative splicing is regulated by an extensive protein-RNA interaction network involving cis elements within the pre-mRNA and trans-acting factors that bind to these cis elements. The most essential splicing signals within the pre-mRNA are the 5′ splice site (5′SS), 3′ splice site (3′SS), branch site (A), and polypyrimidine tract (Y(n)). The 5′ and 3′ splice sites have highly conserved GU and AG dinucleotides as the first and last two nucleotides of the intron, respectively. The U1 snRNP complex recognizes the 5′ splice site, and the U2 snRNP complex recognizes the branch site. The U2AF proteins recognize the 3′ splice site and polypyrimidine tract. Exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs) are pre-mRNA cis regulatory motifs that recruit various RNA-binding proteins (e.g., SR and hnRNP proteins) to regulate alternative splicing.
Alternative splicing is regulated in a cell-type- and developmental-stage-specific manner.11 This regulation is orchestrated through an extensive protein-RNA interaction network involving cis elements within the pre-mRNA and trans-acting factors that bind to these cis elements12 (Figure 1C). The most conserved cis splicing elements include the 5′ and 3′ splice sites that define the boundary of an intron with its upstream and downstream exon, respectively, as well as the branch site and polypyrimidine tract upstream of the 3′ splice site. These elements are recognized by the core splicing machinery (the spliceosome) and play an essential role in defining exon and intron identity.12 In addition to these core elements, auxiliary cis elements in exons or flanking introns can act as splicing enhancer or silencer elements to promote or repress exon splicing via their interactions with trans-acting splicing regulators, in particular RNA-binding proteins (RBPs).13 For example, cell-type-specific splicing regulators, such as ESRP, CELF, MBNL, RBFOX, and PTB family members, control the alternative splicing profiles and cell identities of epithelial, muscle, and neuronal cells by interacting with their cognate cis elements within the pre-mRNA to produce cell-type-specific isoforms.11
Alternative splicing is frequently affected by human genetic variants and disease mutations. A large fraction of human disease mutations disrupt splice site signals or splicing enhancer or silencer elements within the pre-mRNA, leading to the production of aberrant mRNA and protein products.14 It has been estimated that such cis splicing mutations constitute 15%–60% of human disease mutations.15 Additionally, mutations disrupting trans-acting splicing regulators cause a wide spectrum of diseases by globally compromising the splicing of many downstream target genes.16 Through decades of genetic and medical research, the role of aberrant splicing as a primary cause of Mendelian diseases has been firmly established and extensively reviewed.15, 17 However, until recently, much less was known and appreciated about the extent of naturally occurring alternative splicing variation among human individuals and how alternative splicing affects phenotypic variability and disease susceptibility in human populations.
Recent developments in genomic technologies and computational tools have enabled transcriptome-wide studies of alternative splicing at an unprecedented scale and resolution.5, 6 New data depict an expanding landscape of alternative splicing variation across human tissues and populations. Here, we describe technological advances that have markedly increased the speed and scale at which discoveries are made about the genetic variation of alternative splicing. We review population-scale transcriptome studies that have revealed alternative splicing to be a primary causal mechanism underlying genome-wide association study (GWAS) signals of complex traits and diseases. We highlight innovative experimental and computational approaches that enable the rapid discovery and characterization of genomic variants that alter splicing. Finally, we discuss the clinical applications of these findings as well as their implications for future genetic and medical research.
Technologies for High-Throughput Analysis of Alternative Splicing
The conventional molecular biology approach to the quantification of alternative splicing is reverse transcription polymerase chain reaction (RT-PCR).18 In the late 1990s, sequencing of expressed sequence tags (ESTs), which are fragments of full-length mRNAs, revealed widespread alternative splicing in eukaryotic organisms.19 The development of splicing-sensitive microarrays in the mid-2000s allowed researchers to examine global splicing regulatory programs across tissues, cellular states, and species.20 Notably, all three types of technologies have been used to discover the association between genotypes and alternative splicing patterns in human populations.21 However, these technologies have low throughput (RT-PCR and ESTs), have high noise (ESTs and splicing microarray), or are limited to known splicing events (RT-PCR and splicing microarray).19, 20
Powered by high-throughput second-generation DNA sequencers, the advent of RNA sequencing (RNA-seq) in the late 2000s transformed many aspects of biomedical research, including studies of transcriptome complexity and alternative splicing.22 Because of their massively parallel nature, state-of-the-art high-throughput sequencers are now able to generate billions of short sequence reads in a single run.23 Sequencing mRNAs with these sequencers allows the discovery of novel genes and mRNA isoforms, the estimation of gene expression levels, and the quantitation of alternative splicing events.22 Three landmark papers in 2008 demonstrated the use of RNA-seq for characterizing alternative splicing in mammalian tissues.5, 6, 24 Since then, RNA-seq has rapidly eclipsed microarray as the standard approach for transcriptome profiling. Currently, RNA-seq data for over 70,000 human samples have been deposited into public repositories,25 and the number continues to rise at a rapid pace.
Although typical RNA-seq experiments analyze polyadenylated (polyA+) mRNAs from whole cells or bulk tissue, the RNA-seq workflow is versatile enough to allow diverse types of applications that can obtain transcriptome information at a more fine-grained level.26 For example, RNA-seq analysis of non-polyadenylated (polyA−) RNAs enables the discovery and quantitation of polyA− non-coding RNAs, including circular RNAs created by back-splicing events.27, 28 Isolation and sequencing of RNAs from distinct subcellular fractions have been used for characterizing the subcellular localization of mRNA isoforms as well as co-transcriptional splicing of nascent RNAs on chromatin.29, 30, 31 Single-cell RNA-seq has become an increasingly popular approach to studying the transcriptome, including alternative splicing, at the individual-cell level.32, 33 Finally, although Illumina sequencers generate only short sequence reads, specialized protocols for library preparation can be used for inferring full-length mRNA isoforms with the use of Illumina RNA-seq data. Tilgner et al. developed a “synthetic long read” RNA-seq approach for use with Illumina sequencers.34 The principle behind this method is to generate RNA-seq libraries from a given sample separated into many small pools. Each pool contains a small number of RNA molecules (approximately 1,000 or fewer), and the assumption is that for most genes, no more than one molecule per gene is present in each pool. Then, short reads from each pool can be assembled into full-length transcripts by de novo sequence assembly algorithms. Using this approach, the authors identified novel mRNA isoforms and determined that certain distant alternatively spliced exons tend to co-occur in full-length mRNA molecules, whereas others tend to be spliced in a mutually exclusive manner. A caveat to this approach is that it is limited by the same issues of de novo assembly with short reads, primarily mis-assemblies and repetitive sequences.35 Moreover, the assumption of one RNA molecule per gene in each pool might not hold true for highly expressed genes.
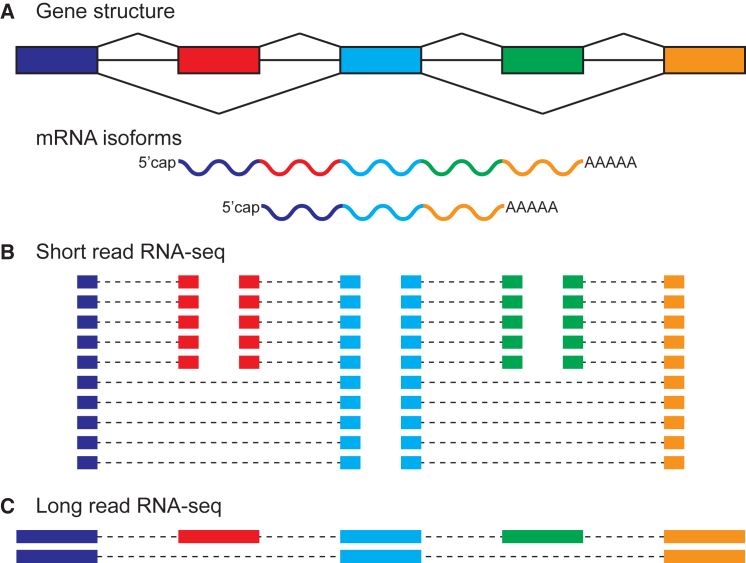
Ultimately, the interest in sequencing full-length mRNA transcripts has led to a renaissance of long-read mRNA sequencing, now using third-generation DNA sequencers most notably from Pacific Biosciences (PacBio)36 and Oxford Nanopore Technologies.37 For example, PacBio isoform sequencing (Iso-Seq) has successfully identified many novel transcripts and alternative splicing events in tissues and cell types with well-characterized transcriptomes,38, 39 whereas Nanopore RNA-seq has been used for determining exon connectivity and full-length mRNAs in complex alternatively spliced genes with thousands of distinct isoform products.40 The strengths of third-generation long-read RNA-seq are in their long read lengths, which allow the direct resolution of isoform structure and the interrogation of repetitive RNA sequences, whereas their main weaknesses are their higher error rates and lower throughput (Figure 2). For the purpose of analyzing alternative splicing, the higher error rates are tolerable because aligners can leverage the long read lengths to align reads to exons and splice junctions. However, the smaller read number due to the lower throughput is a major bottleneck for the accurate quantitation of isoform abundance. A hybrid approach of combining long, error-prone reads from third-generation sequencers with short, accurate reads from second-generation sequencers has been developed for correcting sequencing errors and obtaining isoform quantitation from long reads.38 From a historical perspective, the data of third-generation long-read RNA-seq resemble those of EST sequencing, and computational methods developed for EST data have proven useful for PacBio and Nanopore RNA-seq data.41
Figure 2.
Strengths and Weaknesses of Short-Read and Long-Read RNA-Seq
(A) Schematic diagram of an alternatively spliced gene that generates two distinct mRNA isoforms. The first, middle, and last exons are constitutive exons. The second and fourth exons are alternative exons. The two alternative exons are co-spliced such that the long isoform contains all five exons and the short isoform contains only the first, middle, and last exons.
(B) Short-read RNA-seq generates many reads, enabling the accurate quantitation of individual alternative exons, but the long-range coupling between the two alternative exons is lost.
(C) Long-read RNA-seq captures the long-range coupling between alternative exons and identifies the correct full-length mRNA isoforms, but the limited number of reads reduces the precision of isoform quantitation.
Beyond sequencing, imaging is emerging as a powerful technology for transcriptome analysis with spatiotemporal resolution. Sequential fluorescence in situ hybridization (seqFISH)42 and multiplexed error-robust fluorescence in situ hybridization (MERFISH)43 are imaging-based methods for single-cell transcriptomics and can quantify hundreds of target transcripts at the single-molecule level with spatial resolution. These methods integrate single-molecule fluorescence in situ hybridization with a barcoding scheme to distinguish hundreds of transcripts simultaneously. Each target transcript has a predefined sequential fluorescent barcode, which is used for identifying the transcript via cycles of hybridization with different fluorescent probes. Currently, seqFISH and MERFISH have primarily been applied to gene-level quantification, but with customizable probes, these approaches are in principle applicable to isoform analysis.
Quantifying Alternative Splicing by Using RNA-Seq Data
Because of the popularity of Illumina RNA-seq, many computational tools have been developed for estimating mRNA isoform expression and quantifying alternative splicing variation with the use of short-read RNA-seq data.44, 45 These tools fall into two broad categories according to their strategies for data analysis.
The first category represents transcript-based tools that seek to estimate the abundances and relative proportions of full-length mRNA isoforms by using short-read RNA-seq data. This approach typically involves aligning short reads to a reference genome or transcriptome and then estimating the abundances of mRNA isoforms by using an expectation-maximization algorithm.46, 47 Recent innovations in pseudo-alignment algorithms have led to alignment-free RNA-seq transcript quantitation with significantly improved speed and computational efficiency.48, 49 Isoform proportions can then be inferred from the estimated abundances of all mRNA isoforms of a given gene. A drawback of the transcript-based approach is that inferring the abundance of full-length mRNA isoforms from short reads is non-trivial, and the results are sensitive to the choice of transcript annotations.45 Moreover, for genes with multiple alternatively spliced regions, it is not straightforward to attribute change in the abundance of mRNA isoforms to differential splicing regulation at specific exons or splice sites.
The second category represents event-based tools that seek to directly quantify individual alternative splicing events by using RNA-seq data. In this approach, alternative splicing events are discovered from RNA-seq data, reads aligned to specific exons or splice junctions are counted, and appropriate statistical methods are used for quantifying alternative splicing and detecting differential splicing between distinct biological conditions. A widely used metric in event-based analyses is percent spliced in (PSI or ψ), which represents the percentage of a gene’s mRNA transcripts that include a specific exon or splice site.50 For a given alternative splicing event, the PSI value can be calculated from the counts of RNA-seq reads supporting specific exons or splice junctions.50, 51 Many popular computational tools for RNA-seq analysis of alternative splicing are event based (MISO,50 SpliceTrap,52 rMATS,51 and MAJIQ,7 to name a few). These tools differ in their definitions of alternative splicing events (basic versus complex), read-counting procedures, and statistical methods for quantifying and determining differential alternative splicing. Nonetheless, for the same set of alternative splicing events, these tools tend to produce highly concordant PSI estimates.53 Given that the PSI value represents a proportion estimated from read counts, the confidence interval of the PSI estimate is dependent on the overall RNA-seq read coverage for an event of interest, such that a higher coverage leads to a more reliable PSI estimate. This is a critical issue in RNA-seq analysis of alternative splicing, and studies have shown that modeling the confidence interval of PSI values on the basis of RNA-seq read counts improves downstream statistical inference.50, 51, 54 Interestingly, a hybrid approach leveraging full-length transcript quantitation for event-based analysis has been employed in a tool called SUPPA.53 This tool first runs alignment-free transcript quantitation software to estimate the abundance of mRNA isoforms and then converts these estimates to alternative splicing quantitation at the event level. With the use of pseudo-alignment algorithms,48, 49 this approach is fast and scalable to large datasets. However, it is restricted to pre-existing transcript annotations and cannot discover or quantify novel alternative splicing events. This issue is a limitation for analyzing genetic variation of alternative splicing, given that genomic variants can generate novel alternative splicing events in individual transcriptomes.55, 56
Computational Approaches for Discovering Genetic Associations of Alternative Splicing
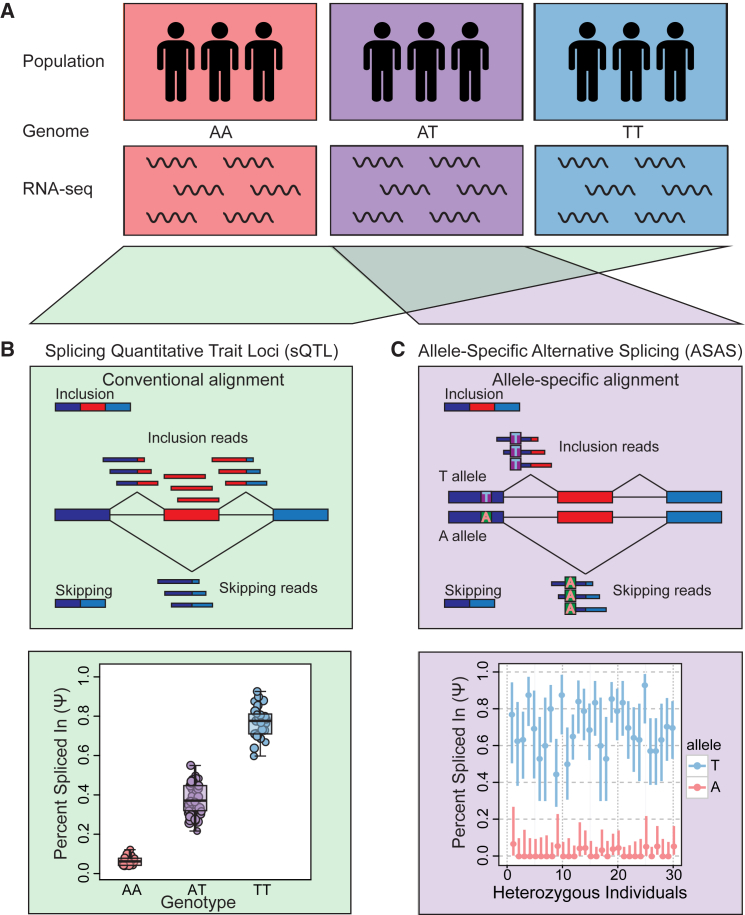
With the continued increase in capacity and reduction in cost of high-throughput sequencers, generating RNA-seq datasets across many individuals in a population has become feasible (Figure 3A). Such population-scale RNA-seq datasets enable transcriptome-wide studies to associate genotypes with alternative splicing variation. Splicing quantitative trait locus (sQTL) analysis is a commonly used approach for discovering genetic variants associated with alternative splicing (Figure 3B).57, 58, 59 QTL analyses involve correlating genotypes with quantifiable phenotypes (traits). In sQTL analysis, the quantitative profiles of alternative splicing (e.g., PSI values) are treated as traits and tested for association with genotypes. Several computational methods have been developed for identifying sQTLs from population-scale genotype and RNA-seq data.57, 58, 59, 60, 61 Zhao et al. developed GLiMMPS, a computational method that identifies sQTLs at the event level by associating the PSI values of individual alternative splicing events with genotypes across the population. An important feature of GLiMMPS is that it uses a generalized linear mixed model to model the confidence interval of the PSI value in each individual as a function of RNA-seq coverage, which leads to improved accuracy over competing statistical models that treat the PSI value as a point estimate.57 Monlong et al. developed sQTLseekeR, a computational method that identifies sQTLs at the transcript level.59 sQTLseekeR treats the relative abundances of all alternatively spliced isoforms of a gene as a vector and uses a distance-based approach to test for association with genotypes. Because this method is applicable to any number of isoforms, it can detect sQTLs arising from both simple and complex alternative splicing events. Notably, the sQTL approach can be used to test for the association between any alternative splicing event and any SNP in cis or trans.62 cis-sQTL analyses could pinpoint genetic variants affecting cis splicing regulatory elements. On the other hand, trans-sQTL analyses can potentially identify hotspots where a SNP at a single genomic locus affects the alternative splicing of numerous genes across the genome. Such trans-sQTL hotspots have the potential to reveal known or novel regulators of alternative splicing.
Figure 3.
Strategies for Discovering Genetic Associations of Alternative Splicing
(A) A population of individuals is genotyped, and their transcriptomes are subject to RNA-seq.
(B) Splicing quantitative trait locus (sQTL) analysis. For a given exon, the splicing level (PSI value) is measured for each individual on the basis of RNA-seq reads aligned to distinct mRNA isoforms. The PSI values are treated as quantitative traits and tested for association with genotypes across all individuals for the identification of significant sQTLs.
(C) Allele-specific alternative splicing (ASAS) analysis. Splicing levels (PSI values) are measured in an allele-specific manner for individuals who are heterozygous for a given SNP. For each individual, a PSI measurement can be obtained for each allele on the basis of allele-specific reads aligned to distinct mRNA isoforms. Reproducible allelic differences in PSI values across multiple heterozygous individuals provide evidence for significant ASAS events.
Allele-specific alternative splicing (ASAS) analysis is a complementary approach to sQTL analysis for discovering genetic variants associated with alternative splicing (Figure 3C). ASAS analysis aims to identify differential alternative splicing between mRNA transcripts expressed from two haplotypes of an individual. This approach involves using heterozygous SNPs present in mRNAs to assign RNA-seq reads to two alleles and then testing for differential splicing between the two alleles. Such an allele-specific strategy has been applied to different types of alternative RNA processing mechanisms, including alternative splicing.63, 64, 65 Compared with the sQTL approach, the ASAS approach is unique in that the two alleles are exposed to an identical cellular environment; thus, their splicing differences in the individual can be attributed to cis genetic effects. However, for the ASAS approach to work, a heterozygous SNP must be expressed outside of the alternatively spliced region to enable allele-specific read assignment while being sufficiently close to the alternative splicing event to be detected on the same RNA-seq read with this event. As a result of this limitation, certain events might not be accessible with the ASAS approach using short-read RNA-seq data; however, recent work has explored the use of long-read RNA-seq for identifying ASAS events.66 In an interesting extension of the conventional ASAS approach applied to RNA-seq data of polyA+ mRNAs, Hsiao et al. integrated ASAS analysis with polyA+ and polyA− RNA-seq data for distinct subcellular compartments (cytosolic and nuclear).67 By examining the allelic ratio of RNA-seq reads from mature cytosolic polyA+ mRNAs or from nuclear polyA− RNAs representing spliced-out products, the authors were able to identify both exonic and intronic variants affecting alternative splicing.
Widespread Variation and Phenotypic Association of Alternative Splicing in Human Populations
In the last few years, population-scale RNA-seq datasets have been generated for diverse tissues and cell types (Table 1). Many of the initial RNA-seq studies were performed with lymphoblastoid cell lines (LCLs).64, 68, 69, 71, 76 LCLs are individual-specific immortalized cell lines created through the infection of human B cells with Epstein-Barr virus.77 These cell lines have been extensively characterized by large-scale genotyping efforts, such as the HapMap and 1000 Genomes projects.78, 79 Therefore, they provide readily available materials for studying the association between genetic variants and gene regulation, including alternative splicing. In two pioneering studies, Pickrell et al. and Montgomery et al. performed RNA-seq of LCLs from African and European populations.68, 69 In addition to identifying QTLs affecting overall gene expression levels (expression QTLs or eQTLs), both studies discovered over 100 sQTLs. The largest LCL RNA-seq dataset was generated by the Geuvadis (Genetic European Variation in Health and Disease) Consortium, which performed RNA-seq on 462 LCL samples from five populations from the 1000 Genomes Project.64 A major limitation of LCLs, however, is that they represent a single, relatively homogeneous cell type, whereas transcriptome regulation is strongly tissue and cell-type specific. More recently, population-scale RNA-seq studies have been applied to different tissues.62, 70, 72, 73, 74, 75 The most comprehensive effort to date is the GTEx (Genotype-Tissue Expression) Consortium,80, 81 which has released raw RNA-seq data along with whole-genome genotype data for over 10,000 tissue samples from 53 tissue sites (GTEx release V7), and this dataset continues to expand. Furthermore, induced pluripotent stem cells (iPSCs) are being explored for RNA-seq-based QTL studies as an alternative to LCLs and tissues.82 Not only would human iPSCs be able to replace LCLs as a source of individual-specific, continuously expandable biological materials, but these cells can also be differentiated in vitro into many mature cell types, thus circumventing the bottleneck of availability and access in tissue-based RNA-seq studies.
Table 1.
Population-Scale RNA-Seq Studies of Alternative Splicing Variation in Human Transcriptomes
| Study | Tissue or Cell Type | Sample Size | Summary |
|---|---|---|---|
| Montgomery et al.68 | LCLs | 60 | one of the first two population-scale transcriptome genetics studies to use RNA-seq; identified 110 sQTL events in a European population at a 0.01 permutation threshold |
| Pickrell et al.69 | LCLs | 69 | one of the first two population-scale transcriptome genetics studies to use RNA-seq; identified 187 genes with significant sQTLs in an African population at a 10% FDR, and many of these altered splicing by affecting cis splicing regulatory elements |
| Lappalainen et al.64 | LCLs | 462 | the largest population-scale RNA-seq dataset on LCLs; was generated by the Geuvadis project and included data on four European populations and one African population; identified 639 genes with trQTLs, where the genotype is significantly associated with the ratio of individual transcript level to total gene expression; found that genetic variation of gene expression levels and transcript isoform structure is equally common but largely controlled by independent causal variants |
| Battle et al.62 | whole blood | 922 | whole blood from the Depression Genes and Networks cohort; identified 1,370 genes with significant sQTLs at a 5% FDR; a total of 159 sQTLs were in high LD with trait- and disease-associated GWAS SNPs; the large sample size also allowed the identification of candidate trans-sQTLs |
| Fadista et al.70 | pancreatic islets | 89 | identified 371 sQTLs, including sQTLs in known T2D-associated loci or in genes associated with beta cell function and glucose metabolism |
| Li et al.71 | LCLs | 17 | RNA-seq study of a 17-individual, three-generation family; allowed the discovery of sQTLs controlled by rare variants; identified 261 sQTLs at a 50% FDR; found that sQTLs with large effects in the family were enriched with rare variants |
| GTEx Consortium72 | 43 tissues | 1,641 | data from the pilot phase of the GTEx project: 1,641 samples from 43 tissues across 175 individuals; identified an average of ∼1,900 and ∼250 sQTL genes per tissue with Altrans58 and sQTLseekeR,59 respectively; most sQTL genes were not eQTL genes; significant sQTLs tended to be shared among tissues, whereas tissue-specific sQTLs represented only 7%–21% of sQTLs, depending on the tissue type |
| Chen et al.73 | monocytes, neutrophils, and T cells | 197 | CD14+ monocytes, CD16+ neutrophils, and naive CD4+ T cells from up to 197 individuals; quantified splicing by using both PSI event-based measurements and relative abundances of transcript isoforms; identified over 2,000 genes with sQTLs at a 5% FDR in each of the three cell types |
| Pala et al.74 | leukocytes | 624 | included a total of 624 individuals from Sardinia; first sQTL study to integrate whole-genome and RNA-seq data of multiple families to discover common and rare variants affecting splicing; identified 6,768 sQTLs |
| Takata et al.75 | brain (prefrontal cortex) | 206 | identified 1,595 sQTLs in 1,341 unique genes; significant sQTLs were enriched with disease-associated GWAS loci, particularly loci associated with schizophrenia |
The following abbreviations are used: FDR, false discovery rate; T2D, type 2 diabetes; and trQTL, transcript ratio QTL.
Using these large-scale datasets, researchers have begun to define the landscape, genetic architecture, and phenotypic association of alternative splicing variation in human populations (Table 1). Despite the differences in tissue and cell type, sample size, and sequencing depth, as well as the computational methods used for discovering sQTLs, several consensuses have emerged. These studies demonstrate that inheritable genetic variation of alternative splicing is widespread across diverse human tissues and cell types. Although sQTL SNPs tend to be enriched at the essential 5′ and 3′ splice sites,57, 69, 72 many sQTLs can be attributed to SNPs located outside of the splice site regions. These SNPs can modify splicing enhancer or silencer elements as well as known RBP binding sites in exonic or intronic regions.83 The approach of coupling sQTL results to GWAS signals has identified a large number of sQTLs in high linkage disequilibrium (LD) with previously identified GWAS SNPs (Table 1), suggesting that SNPs affecting alternative splicing could be the causal variants underlying a substantial fraction of GWAS signals for complex traits and diseases. For example, an RNA-seq study of 206 human brain (prefrontal cortex) tissues reported significant enrichment of sQTLs among GWAS disease loci, particularly for GWAS SNPs associated with schizophrenia.75 Similarly, an RNA-seq study of 89 pancreatic islets identified sQTLs in known type-2-diabetes-associated loci.70 One key question is whether sQTLs identified in these studies are the primary contributors to GWAS-associated traits and diseases or merely reflect the secondary effects of SNPs that affect phenotypes via other layers of gene regulation. To address this question, an elegant study by Li et al. integrated multiple datasets to analyze eight types of regulatory QTLs in a cohort of LCLs from an African population.84 The authors found that most sQTLs are independent of eQTLs, and sQTLs appear to have a comparable or even greater magnitude of effects on GWAS traits than eQTLs. These data suggest that splicing is a primary link between genetic variation and complex diseases, consistent with the prevalence of aberrant splicing as a primary cause of Mendelian diseases.15, 17
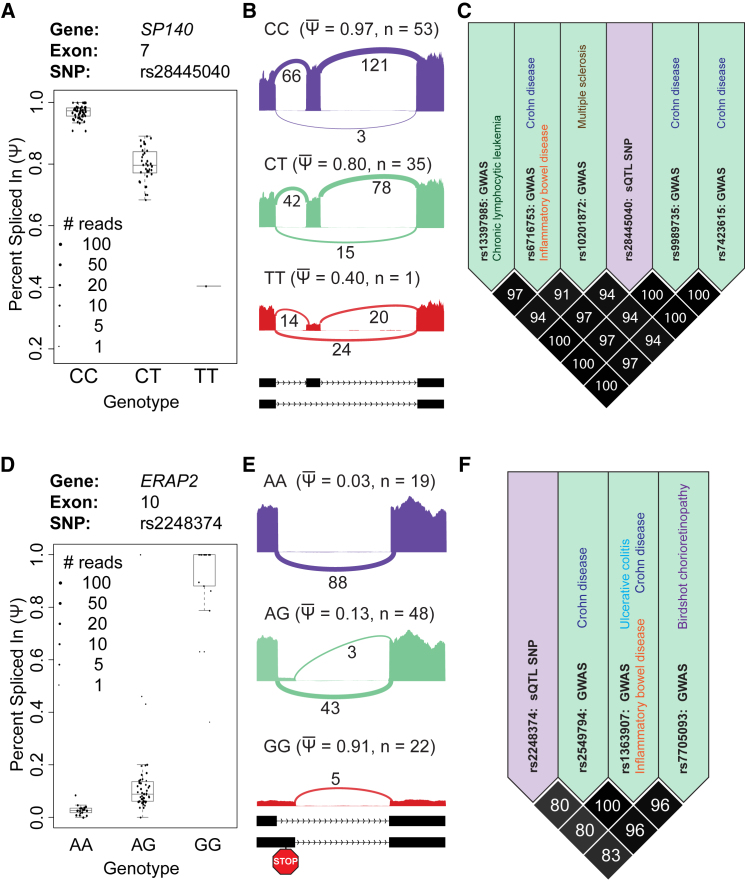
Two examples of sQTLs that correlate with GWAS signals are highlighted here. SP140 is a tissue-restricted gene with high expression in lymphoid cells,85 and its domain structure suggests a role in chromatin-mediated regulation of gene expression.86 Several GWASs identified SP140 SNPs that are significantly associated with chronic lymphocytic leukemia,87 multiple sclerosis,88 Crohn disease,89 and inflammatory bowel disease.90 However, the causal mechanism underlying these GWAS signals remained unknown. On the basis of sQTL analysis of RNA-seq data of LCLs from a European population, a significant sQTL signal was found for exon 7 of SP140, and the peak SNP was a C-to-T exonic SNP, rs28445040 (Figures 4A and 4B).57 Although this SNP does not alter the encoded protein sequence of SP140, minigene splicing reporter assays demonstrated its role in regulating the splicing level of SP140 exon 7, such that the T allele is associated with significantly reduced exon inclusion.57 Because the exon is 78 bp in length, skipping of this exon would remove an in-frame 26 amino acid peptide from the protein product without affecting the downstream reading frame. Strikingly, this SNP is in high LD with GWAS SNPs of all four diseases (Figure 4C), suggesting that this is the causal variant underlying the association between SP140 and these diseases. Furthermore, the association between this sQTL and multiple sclerosis was replicated in a recent case-control study.92 In another example, several studies identified an sQTL in exon 10 of ERAP2,57, 93, 94 a gene encoding a protease that processes antigenic epitopes for MHC class I antigen presentation.95 An A-to-G intronic SNP (rs2248374) within the 5′ splice site of ERAP2 deactivates the canonical 5′ splice site and activates a downstream cryptic 5′ splice site. This change leads to the production of an aberrant transcript that contains a premature termination codon subject to nonsense-mediated mRNA decay. RNA-seq data of LCLs indicate a significant switch in splicing among different genotypes of rs2248374, along with a significant change in steady-state mRNA levels due to alternative-splicing-coupled mRNA decay (Figures 4D and 4E). The G allele is associated with lower levels of MHC class I molecules at the surface of B cells94 and is in LD with GWAS signals for several diseases, such as Crohn disease89 and inflammatory bowel disease90 (Figure 4F). These two examples are just the tip of the iceberg for many sQTLs identified across various studies, and they illustrate that sQTLs can influence complex traits and diseases by altering protein activity and function (SP140) or mRNA stability and steady-state mRNA levels (ERAP2). It is also worth noting that the causal variants for these two GWAS-associated sQTLs are silent exonic (SP140) or intronic (ERAP2) and would therefore be missed by many commonly used tools for variant annotation.96
Figure 4.
Two Examples of sQTLs Associated with GWAS Signals for Complex Diseases
(A–C) Alternative splicing of SP140 exon 7 is associated with chronic lymphocytic leukemia, Crohn disease, inflammatory bowel disease, and multiple sclerosis. The alternative splicing event is an exon-skipping event. The C allele is associated with a higher level of exon inclusion, whereas the T allele is associated with a higher level of exon skipping. (A) Boxplot showing the significant association between SNP rs28445040 and the splicing level (PSI value) of SP140 exon 7 within the Geuvadis CEU (Utah residents with ancestry from northern and western Europe) population. Each dot represents the PSI value from a particular individual, and the size of each dot is proportional to the RNA-seq read coverage for the alternative splicing event in that individual. (B) Sashimi plot indicating the average RNA-seq read density and splice junction counts for each genotype. Exons and introns are not drawn to scale, and the relative width of exons is increased for clarity. (C) LD plot showing multiple GWAS SNPs (green boxes) linked with the sQTL SNP (purple box).
(D–F) Alternative splicing of ERAP2 exon 10 is associated with Crohn disease, ulcerative colitis, inflammatory bowel disease, and birdshot chorioretinopathy. The alternative splicing event is an alternative 5′ splice site event. The A allele is associated with a higher level of the upstream canonical 5′ splice site, whereas the G allele is associated with a higher level of the downstream cryptic 5′ splice site. Usage of the downstream cryptic 5′ splice site introduces a premature stop codon and results in nonsense-mediated mRNA decay. (D) Boxplot showing the significant association between SNP rs2248374 and the splicing level (PSI value) of ERAP2 exon 10 (i.e., usage of the downstream cryptic 5′ splice site) within the Geuvadis CEU population. Each dot represents the PSI value from a particular individual, and the size of each dot is proportional to the RNA-seq read coverage for the alternative splicing event in that individual. (E) Sashimi plot indicating the average RNA-seq read density and splice junction counts for each genotype. Exons and introns are not drawn to scale, and the relative width of exons is increased for clarity. (F) LD plot showing multiple GWAS SNPs (green boxes) linked with the sQTL SNP (purple box).
RNA-seq data of 89 CEU individuals are from the Geuvadis project.64 Sashimi plots were drawn with rmats2sashimiplot (see Web Resources). LD plots were drawn with Haploview 4.291 and include CEU individuals from the 1000 Genomes Project (phase 3). For each boxplot, the top and bottom of the box represent the third and first quartiles, respectively. The band in the middle of the box represents the median. The whiskers of each boxplot extend to the most extreme data points within 1.5 times the interquartile range from each box.
Characterizing Causal Variants of Alternative Splicing via Massively Parallel Reporter Assays
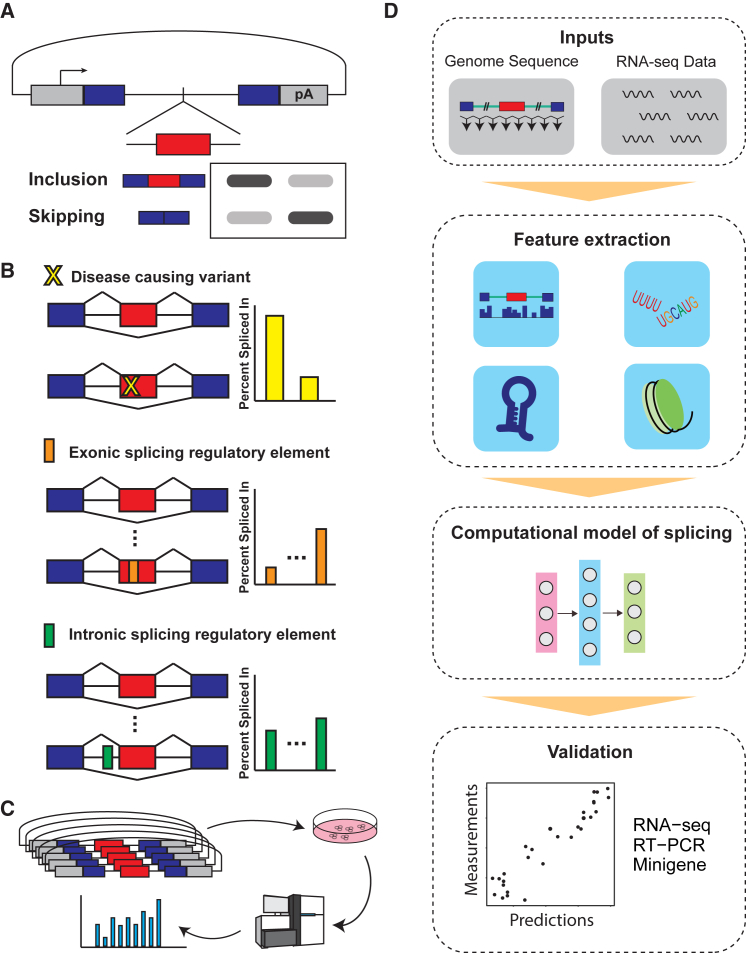
Although RNA-seq can reveal associations between genetic variants and alternative splicing, identifying the causal variants underlying the detected associations remains a challenging task. In an sQTL analysis, multiple variants within a haplotype block can be significantly associated with alternative splicing, but we do not know which variant(s) causally affect(s) splicing regulation. A widely used molecular biology approach to the study of splicing regulation is the minigene splicing reporter assay.97 A minigene splicing reporter is constructed via the insertion of a piece of genomic DNA that contains the exon of interest and its flanking intronic sequences into a position where it is flanked either by exons from another gene (i.e., heterologous minigene reporter) or by the upstream and downstream constitutive exons from the same gene (Figure 5A). Site-directed mutagenesis within a minigene splicing reporter can be used for assessing the impact of specific genomic variants or splicing regulatory elements (Figure 5B). Coupled with high-throughput screens, minigene splicing reporters can be used for identifying splicing enhancer or silencer elements and discovering trans-acting factors or small-molecule compounds that regulate the splicing of specific exons.
Figure 5.
Experimental and Computational Tools for Characterizing the Causal Impacts of Genomic Variants on Alternative Splicing
(A) Schematic diagram of a minigene splicing reporter. An exon of interest, along with its flanking intronic sequences, is inserted into a splicing reporter construct, where it is flanked by upstream and downstream exons containing a promoter and a polyA site. The splicing profile of the minigene splicing reporter can be determined by RT-PCR or RNA-seq.
(B) Use of minigene splicing reporters for characterizing the effects of disease-causing variants or exonic and intronic splicing regulatory elements on splicing.
(C) Minigene splicing reporters can be used in massively parallel reporter assays (MPRAs) for determining the consequences of many sequence variants on splicing in a high-throughput manner. A library of minigenes is transfected into a cell line, and splicing levels are measured for all variants simultaneously by RNA-seq.
(D) Deep learning framework for analyzing alternative splicing. Starting with input data, including the genome sequence and RNA-seq data, the framework extracts genomic and RNA features. These features include diverse types of quantitative or qualitative features, such as conservation score, sequence motifs, secondary structure, and epigenetic marks. A computational model is trained to predict splicing patterns and levels by using the extracted features. The predictions can be evaluated with experimental validation (e.g., by RNA-seq, RT-PCR, or minigene).
With recent advances in oligonucleotide synthesis technologies and high-throughput sequencing, massively parallel reporter assays (MPRAs) have become an increasingly popular approach to the study of gene regulation, including alternative splicing.98 MPRAs test the functional impacts of many sequence variants in parallel. These sequences are inserted into a reporter construct and transfected into cells or combined with cellular extracts for determining the functional impacts of sequence variants (Figure 5C). Two recent studies conducted MPRAs with minigene splicing reporters to determine the effects of cis sequence variants on splicing.99, 100 Rosenberg et al. tested over two million synthetic minigenes in a high-throughput fashion.99 Specifically, they created two separate libraries to study alternative 5′ or 3′ splice sites and analyzed the ability of random sequences to influence splice site selection. The authors split a single-gene sequence (Citrine, a derivative of YFP) into two exons as the backbone of the reporter and inserted introns with degenerate sequences between the two exons. For the alternative 5′ splice site library, each intron was designed to have two competing alternative 5′ splice sites, and two random 25 bp sequences were inserted into positions between the two competing 5′ splice sites or downstream of the distal 5′ splice site. The library for alternative 3′ splice site analysis was designed in the same manner. The resulting libraries were transfected into cells, and the splicing profiles of all sequences were measured in parallel by RNA-seq. Leveraging the abundant synthetic reporter data, the authors were able to use machine learning to model splicing patterns and predict the effects of human SNPs on splicing. Interestingly, the models learned from alternative 5′ and 3′ splice sites can also predict exon skipping in vivo. In another study, Soemedi et al. developed a massively parallel splicing assay (MaPSy) to interrogate the effects of 4,964 exonic disease-causing mutations on alternative splicing.100 The authors synthesized a 170 bp genomic sequence library for all mutant and wild-type exon pairs. Disease-mutation-containing exons that were less than or equal to 100 bp in length were selected and synthesized to include at least 55 bp of the upstream intron and at least 15 bp of the downstream intron. Two parallel assays were performed. The first assay tested the impact of the mutation on the exon's inclusion or skipping in vivo when the reporter was transfected into cells, and the second tested whether the mutation influenced the splicing of the upstream intron in vitro when the sequence was incubated with nuclear extracts. Even though they used distinct experimental systems, the two assays reached general agreement. Approximately 10% of the tested disease-causing mutations perturbed splicing in both assays. By contrast, only 3% of common SNPs perturbed splicing in both assays. This 10% is most likely a lower-bound estimate for the percentage of pathogenic exonic mutations that disrupt splicing, considering the cell-type-specific nature of splicing regulation and that only a single cell type (HEK293) was used for the in vivo assay.
MPRAs provide a powerful tool for characterizing the causal genetic variants of alternative splicing. A major advantage of MPRAs is that these experiments generate a massive amount of data. As demonstrated by Rosenberg et al.,99 these data-rich experiments can be coupled with computational modeling for learning important features of splicing regulation and predicting the impact of cis variants on splicing. Additionally, although both studies performed MPRA experiments in the HEK293 cell line, these reporters can be transfected into other cell lines for determining the splicing effects of cis variants in other cell types. Moreover, MPRAs can be coupled with sQTL analyses for identifying causal variants underlying sQTL signals, or they can be utilized in clinical exome or genome sequencing studies for identifying splicing-altering variants in disease-affected individuals. One inherent limitation of MPRAs is that the reporter system might not completely recapitulate the exact cellular environment that allows splicing to occur. For example, factors such as chromatin states, DNA methylation, and histone marks are known to influence alternative splicing.101 CRISPR-Cas9-based genome editing could address these issues and has been used in recent work for characterizing splicing regulatory elements in endogenous genes.102 MPRAs are also limited by the ability to generate libraries; thus, not all exons or variants are assessable by current systems. Future improvements in oligonucleotide synthesis technologies could address this limitation and allow a broader set of exons and deep intronic variants to be examined.
Alternative Splicing Meets Machine Learning
There has been a long-standing interest in developing in silico methods of predicting alternative splicing. The basic scientific premise is that there exists a “splicing code,” a set of genomic and RNA features and associated rules that determine the splicing pattern of any primary transcript in a given cell type.12 Machine learning serves the general purpose of learning underlying patterns from data to allow pattern recognition, classification, and prediction. In computational biology, machine learning has been extensively employed in genomics, transcriptomics, proteomics, and other domains.103 For example, algorithms have been developed to predict regulatory elements such as promoters, enhancers, and splice sites.103
Shortly after the EST-based discovery of widespread alternative splicing, several studies applied machine learning methods to predict a binary classification of alternative versus constitutive exons.104, 105, 106, 107 Alternative exons have distinct sequence features such as exon and intron length, splice site strength, divisibility by three, sequence conservation within exonic and flanking intronic regions, and composition of oligonucleotides reflecting splicing regulatory elements.107 Machine learning methods can leverage these features to predict whether an exon undergoes alternative splicing.104, 105, 106, 107
In a landmark study, Barash et al. used quantitative splicing microarray data across 27 mouse tissues to predict tissue-specific patterns of alternative splicing.108 They grouped the 27 tissues into four broad categories and converted the PSI value of each exon for each tissue category into three probabilities representing an increase, a decrease, or no change in exon inclusion in that tissue category. Then, the authors collected 1,014 features representing RNA sequence motifs and transcript features. They applied a single-layer logistic Bayesian network that models how individual features cooperate or compete to influence splicing in each tissue type. Importantly, the resulting splicing code can reveal novel regulatory features and predict mutation-induced changes in splicing patterns. This work represents a breakthrough in the field because it was the first demonstration that in silico models can successfully predict tissue-regulated alternative splicing. After this work, Xiong et al. added hidden layers to the Bayesian network to construct a Bayesian neural network (BNN).109 These hidden layers helped the authors model non-linear relationships between features, leading to an improved prediction accuracy. Based on the BNN framework, the web tool AVISPA was constructed for splicing prediction and analysis and was trained with more data and an expanded feature set.110
Recently, deep learning, a state-of-the-art machine learning technology, has been applied to predicting alternative splicing111, 112, 113 (Figure 5D). Deep learning refers to methods that map raw input feature data to increasingly abstract feature representations, where higher layers contain more abstract representations.114 Compared with canonical machine learning methods, deep learning is capable of automatically learning complex functions without a need for handcrafted features or rules, and it scales well to large and high-dimensional datasets.114, 115 Deep learning has been successfully applied in a variety of fields, including image classification and speech recognition114 and more recently in computational biology.115 In two studies, Frey and colleagues used RNA-seq data from mouse and human tissues to construct deep learning models that predict the splicing levels of individual exons across different tissues111 and the effects of cis genetic variants on splicing.112 Unlike their previous work that treated tissue-specific splicing patterns as categorical data,108 these new methods attempted to predict the numerical PSI values for each exon in each tissue.111, 112 Evaluations using independent RNA-seq datasets showed good agreement (R2 = 0.65) between predicted and empirical PSI values.112 The authors then applied the deep learning model to predict the effects of cis genetic variants on RNA splicing. Their predictions on clinical variants of selected exons matched well with data from minigene splicing reporters. Furthermore, they applied their model to genome sequencing data of people with autism spectrum disorder (ASD) and control individuals and predicted misregulated splicing in 19 candidate genes with ASD-related neuronal functions. This study demonstrates that deep-learning-based modeling of splicing provides a powerful tool for annotating clinical variants and elucidating the genetic determinants of complex diseases.112 In another interesting application, Huang et al. developed a method called BRIE, which learns prior information from RNA sequence features to augment splicing quantification by using single-cell RNA-seq data.116
With the rapid accumulation of RNA-seq data and RBP-RNA interaction maps in the public domain,25, 117 future work should take advantage of more comprehensive training data and feature space coupled with more advanced machine learning frameworks to improve in silico prediction of alternative splicing. As a step in this direction, Jha et al. recently developed a new deep learning framework to integrate additional RNA genomics data, such as CLIP-seq data of RBP-RNA interactions, and RNA-seq data after the knockdown or overexpression of RBPs.113 The integrative model generalizes well for RBP perturbation data and improves the accuracy of alternative splicing prediction.113 Another interesting direction for future work is to incorporate chromatin states, epigenetic marks, and 3D genome organization in a predictive model, given that splicing is a co-transcriptional process and these features influence splicing via a variety of molecular mechanisms.101
In addition to using machine learning techniques to directly predict splicing patterns and PSI values, other studies have adopted an alternative strategy of predicting splicing-altering genomic variants by using prior variant annotations as training data.118, 119, 120, 121 The basic idea is to collect variants known to affect splicing and/or cause human diseases along with common “splicing-neutral” variants that are likely to have no effect on splicing and then build classifiers to distinguish these two categories of variants. The potential shortcomings of these approaches are that the classification of positive versus negative training data might not be accurate and that the results might suffer from selection bias or overfitting. Nonetheless, these tools offer a complementary strategy for evaluating the potential effects of genomic variants on splicing. An interesting method called ExonImpact was recently developed to prioritize disease-associated splicing-altering variants on the basis of the predicted effects of alternative splicing at the protein level.121 The rationale behind this work is that not all aberrant splicing events are equally detrimental at the protein level, and pathogenic splicing mutations have distinct protein features that can be incorporated into the predictive model.121
Alternative Splicing for Disease Diagnosis
Given the importance of splicing in disease pathogenesis and progression, several therapeutic strategies have been pursued for correcting splicing defects in disease.17 A notable success is the recent FDA approval of nusinersen, an antisense oligonucleotide drug for correcting splicing in spinal muscular atrophy.122
New data are emerging that alternative splicing might provide diagnostic biomarkers for disease status or outcome.26 An example of the predictive power of alternative splicing for disease prognosis was demonstrated in two recent studies showing that alternative splicing profiles can predict cancer patients’ survival time at a comparable and often better accuracy than gene expression levels.54, 123 One possible explanation for these observations is the intrinsic feature of alternative splicing data. Given that alternative splicing is quantified as the relative ratio of multiple isoforms from a single gene, alternative splicing data are self-normalized on a per-gene basis and can be viewed as having an “internal control” that could provide a more robust molecular signature than gene expression levels, especially for large clinical RNA-seq datasets that are prone to technical biases and confounding issues.54 Consistent with these observations, a new study reported that alternative-splicing-based classifiers generally outperform gene-expression-based classifiers for a wide range of biological classification problems.124
In a major advance with broad implications, Cummings et al. demonstrated the potential of RNA-seq and alternative splicing analysis for diagnosing rare diseases.55 The authors analyzed the muscle transcriptomes of 63 individuals with muscle disorders and compared their RNA-seq data with GTEx RNA-seq data of 184 control muscle samples. Of the 63 individuals with muscle disorders, 50 were genetically undiagnosed. Strikingly, through RNA-seq analysis, the authors obtained a genetic diagnosis for 35% of the previously undiagnosed individuals by identifying novel disease-associated aberrant splicing events in known disease-associated genes. In four individuals, a recurrent aberrant splicing event was discovered in COL6A1, in which a GC-to-GT genetic variant created a novel 5′ splice site, leading to the exonization of a 72 bp intronic segment that disrupted the COL6A1 protein product. This variant would not be easily identifiable by exome or genome sequencing alone, given that exome sequencing would miss this deep intronic variant, and genome sequencing would identify too many variants, making it difficult to determine their pathogenicity in the absence of RNA-seq information. Thus, this study offers an important proof of concept that alternative splicing analysis via the integration of RNA-seq with exome or genome sequencing improves disease diagnosis.
Conclusions
The past decade since the advent of RNA-seq has seen tremendous growth in the amount of human transcriptome data. Advances in RNA-seq technologies and computational methods have transformed the study of alternative splicing in health and disease. Population-scale RNA-seq studies have discovered many naturally occurring genomic variants that modulate alternative splicing. Many of these variants are associated with GWAS signals, suggesting a ubiquitous contribution of alternative splicing to phenotypic variability and disease susceptibility in human populations. These genetically regulated, GWAS-associated mRNA isoforms are prime candidates for functional studies of alternative splicing. Future work using isoform-specific gain-of-function or loss-of-function assays should elucidate how genetic variation of alternative splicing affects gene functions and consequently cellular and organismal phenotypes.
The prevalent role of alternative splicing in Mendelian and complex diseases suggests that evaluating the impact of genomic variants on splicing needs to be an integral part of clinical variant prioritization. Many computational tools and online resources exist for prioritizing and annotating variants discovered by exome or genome sequencing.96 Most tools are designed to predict the pathogenic effects of missense variants on protein products. However, there is overwhelming evidence that missense, nonsense, and silent variants within exons, as well as intronic variants, can disrupt splicing and cause disease.14 Currently, it is challenging to predict the pathogenic effects of splicing variants within exonic and intronic regions, except for variants affecting the conserved splice site signals, and they are thus ignored by many commonly used pipelines for variant assessment.96 Recent advances in experimental (e.g., MPRAs) and computational (e.g., deep learning) tools will allow researchers and clinicians to screen a large number of variants for their effects on splicing in a systematic and unbiased manner. Beyond SNPs, other non-SNP variants such as indels or short tandem repeats can modify cis splicing regulatory elements and affect alternative splicing.125, 126 The genetic associations between these non-SNP variants and alternative splicing can also be discovered and characterized by the computational and experimental approaches described in this review. A comprehensive catalog of alternative splicing variation in human populations, along with the ability to discover and characterize splicing-altering variants in specific individuals, holds great value for improving disease diagnoses and ultimately patient care in the era of sequencing and precision medicine.
Acknowledgments
We thank Maggie Lam, John Phillips, Douglas Black, Andrey Damianov, Yang Pan, Levon Demirdjian, and Rocky Cheung for their helpful comments and Shihao Shen for technical assistance. This work was supported by National Institutes of Health grants R01GM088342 and R01GM117624 to Y.X. Y.X. is supported by an Alfred Sloan Research Fellowship (BR2013-117). E.P. is supported by National Institutes of Health postdoctoral training grant T32AR059033. Y.X. is a scientific cofounder of Trimontia Genomics Inc. and IsoTex Biotechnology Inc.
Web Resources
rmats2sashimiplot, https://github.com/Xinglab/rmats2sashimiplot
References
- 1.Sharp P.A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaquero-Garcia J., Barrera A., Gazzara M.R., González-Vallinas J., Lahens N.F., Hogenesch J.B., Lynch K.W., Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife. 2016;5:e11752. doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmucker D., Clemens J.C., Shu H., Worby C.A., Xiao J., Muda M., Dixon J.E., Zipursky S.L. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 9.Braunschweig U., Gueroussov S., Plocik A.M., Graveley B.R., Blencowe B.J. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelemen O., Convertini P., Zhang Z., Wen Y., Shen M., Falaleeva M., Stamm S. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalsotra A., Cooper T.A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Burge C.B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu X.D., Ares M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 15.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 16.Osborne R.J., Thornton C.A. RNA-dominant diseases. Hum. Mol. Genet. 2006;15:R162–R169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 17.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Percifield R., Murphy D., Stoilov P. Medium throughput analysis of alternative splicing by fluorescently labeled RT-PCR. Methods Mol. Biol. 2014;1126:299–313. doi: 10.1007/978-1-62703-980-2_22. [DOI] [PubMed] [Google Scholar]
- 19.Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 20.Lee C., Roy M. Analysis of alternative splicing with microarrays: successes and challenges. Genome Biol. 2004;5:231. doi: 10.1186/gb-2004-5-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Z.X., Jiang P., Xing Y. Genetic variation of pre-mRNA alternative splicing in human populations. Wiley Interdiscip. Rev. RNA. 2012;3:581–592. doi: 10.1002/wrna.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 25.Collado-Torres L., Nellore A., Kammers K., Ellis S.E., Taub M.A., Hansen K.D., Jaffe A.E., Langmead B., Leek J.T. Reproducible RNA-seq analysis using recount2. Nat. Biotechnol. 2017;35:319–321. doi: 10.1038/nbt.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byron S.A., Van Keuren-Jensen K.R., Engelthaler D.M., Carpten J.D., Craig D.W. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet. 2016;17:257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilgner H., Knowles D.G., Johnson R., Davis C.A., Chakrabortty S., Djebali S., Curado J., Snyder M., Gingeras T.R., Guigó R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang E.T., Cody N.A., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhatt D.M., Pandya-Jones A., Tong A.J., Barozzi I., Lissner M.M., Natoli G., Black D.L., Smale S.T. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 33.Song Y., Botvinnik O.B., Lovci M.T., Kakaradov B., Liu P., Xu J.L., Yeo G.W. Single-Cell Alternative Splicing Analysis with Expedition Reveals Splicing Dynamics during Neuron Differentiation. Mol. Cell. 2017;67:148–161.e5. doi: 10.1016/j.molcel.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilgner H., Jahanbani F., Blauwkamp T., Moshrefi A., Jaeger E., Chen F., Harel I., Bustamante C.D., Rasmussen M., Snyder M.P. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat. Biotechnol. 2015;33:736–742. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaisson M.J., Wilson R.K., Eichler E.E. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 2015;16:627–640. doi: 10.1038/nrg3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhoads A., Au K.F. PacBio Sequencing and Its Applications. Genomics Proteomics Bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y., Zhang Y., Ying C., Wang D., Du C. Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics. 2015;13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Au K.F., Sebastiano V., Afshar P.T., Durruthy J.D., Lee L., Williams B.A., van Bakel H., Schadt E.E., Reijo-Pera R.A., Underwood J.G., Wong W.H. Characterization of the human ESC transcriptome by hybrid sequencing. Proc. Natl. Acad. Sci. USA. 2013;110:E4821–E4830. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharon D., Tilgner H., Grubert F., Snyder M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolisetty M.T., Rajadinakaran G., Graveley B.R. Determining exon connectivity in complex mRNAs by nanopore sequencing. Genome Biol. 2015;16:204. doi: 10.1186/s13059-015-0777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T., DuBois R.M., Forsberg E.C., Akeson M., Vollmers C. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alamancos G.P., Agirre E., Eyras E. Methods to study splicing from high-throughput RNA sequencing data. Methods Mol. Biol. 2014;1126:357–397. doi: 10.1007/978-1-62703-980-2_26. [DOI] [PubMed] [Google Scholar]
- 45.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X., Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xing Y., Yu T., Wu Y.N., Roy M., Kim J., Lee C. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 2006;34:3150–3160. doi: 10.1093/nar/gkl396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 49.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J., Akerman M., Sun S., McCombie W.R., Krainer A.R., Zhang M.Q. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics. 2011;27:3010–3016. doi: 10.1093/bioinformatics/btr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alamancos G.P., Pagès A., Trincado J.L., Bellora N., Eyras E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA. 2015;21:1521–1531. doi: 10.1261/rna.051557.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen S., Wang Y., Wang C., Wu Y.N., Xing Y. SURVIV for survival analysis of mRNA isoform variation. Nat. Commun. 2016;7:11548. doi: 10.1038/ncomms11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cummings B.B., Marshall J.L., Tukiainen T., Lek M., Donkervoort S., Foley A.R., Bolduc V., Waddell L.B., Sandaradura S.A., O’Grady G.L., Genotype-Tissue Expression Consortium Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9:eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stein S., Lu Z.X., Bahrami-Samani E., Park J.W., Xing Y. Discover hidden splicing variations by mapping personal transcriptomes to personal genomes. Nucleic Acids Res. 2015;43:10612–10622. doi: 10.1093/nar/gkv1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao K., Lu Z.X., Park J.W., Zhou Q., Xing Y. GLiMMPS: robust statistical model for regulatory variation of alternative splicing using RNA-seq data. Genome Biol. 2013;14:R74. doi: 10.1186/gb-2013-14-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ongen H., Dermitzakis E.T. Alternative Splicing QTLs in European and African Populations. Am. J. Hum. Genet. 2015;97:567–575. doi: 10.1016/j.ajhg.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monlong J., Calvo M., Ferreira P.G., Guigó R. Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat. Commun. 2014;5:4698. doi: 10.1038/ncomms5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia C., Hu Y., Liu Y., Li M. Mapping Splicing Quantitative Trait Loci in RNA-Seq. Cancer Inform. 2015;14(Suppl 1):45–53. doi: 10.4137/CIN.S24832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Q., Hu Y., Li J., Zhang X. ulfasQTL: an ultra-fast method of composite splicing QTL analysis. BMC Genomics. 2017;18(Suppl 1):963. doi: 10.1186/s12864-016-3258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Battle A., Mostafavi S., Zhu X., Potash J.B., Weissman M.M., McCormick C., Haudenschild C.D., Beckman K.B., Shi J., Mei R. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li G., Bahn J.H., Lee J.H., Peng G., Chen Z., Nelson S.F., Xiao X. Identification of allele-specific alternative mRNA processing via transcriptome sequencing. Nucleic Acids Res. 2012;40:e104. doi: 10.1093/nar/gks280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park E., Guo J., Shen S., Demirdjian L., Wu Y.N., Lin L., Xing Y. Population and allelic variation of A-to-I RNA editing in human transcriptomes. Genome Biol. 2017;18:143. doi: 10.1186/s13059-017-1270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tilgner H., Grubert F., Sharon D., Snyder M.P. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. USA. 2014;111:9869–9874. doi: 10.1073/pnas.1400447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsiao Y.H., Bahn J.H., Lin X., Chan T.M., Wang R., Xiao X. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res. 2016;26:440–450. doi: 10.1101/gr.193359.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montgomery S.B., Sammeth M., Gutierrez-Arcelus M., Lach R.P., Ingle C., Nisbett J., Guigo R., Dermitzakis E.T. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fadista J., Vikman P., Laakso E.O., Mollet I.G., Esguerra J.L., Taneera J., Storm P., Osmark P., Ladenvall C., Prasad R.B. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X., Battle A., Karczewski K.J., Zappala Z., Knowles D.A., Smith K.S., Kukurba K.R., Wu E., Simon N., Montgomery S.B. Transcriptome sequencing of a large human family identifies the impact of rare noncoding variants. Am. J. Hum. Genet. 2014;95:245–256. doi: 10.1016/j.ajhg.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pala M., Zappala Z., Marongiu M., Li X., Davis J.R., Cusano R., Crobu F., Kukurba K.R., Gloudemans M.J., Reinier F. Population- and individual-specific regulatory variation in Sardinia. Nat. Genet. 2017;49:700–707. doi: 10.1038/ng.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takata A., Matsumoto N., Kato T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat. Commun. 2017;8:14519. doi: 10.1038/ncomms14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lalonde E., Ha K.C., Wang Z., Bemmo A., Kleinman C.L., Kwan T., Pastinen T., Majewski J. RNA sequencing reveals the role of splicing polymorphisms in regulating human gene expression. Genome Res. 2011;21:545–554. doi: 10.1101/gr.111211.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum. Genet. 1986;73:320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- 78.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ward M.C., Gilad Y. Human genomics: Cracking the regulatory code. Nature. 2017;550:190–191. doi: 10.1038/550190a. [DOI] [PubMed] [Google Scholar]
- 82.Warren C.R., Jaquish C.E., Cowan C.A. The NextGen Genetic Association Studies Consortium: A Foray into In Vitro Population Genetics. Cell Stem Cell. 2017;20:431–433. doi: 10.1016/j.stem.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 83.Zhang X., Joehanes R., Chen B.H., Huan T., Ying S., Munson P.J., Johnson A.D., Levy D., O’Donnell C.J. Identification of common genetic variants controlling transcript isoform variation in human whole blood. Nat. Genet. 2015;47:345–352. doi: 10.1038/ng.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bloch D.B., de la Monte S.M., Guigaouri P., Filippov A., Bloch K.D. Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem. 1996;271:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- 86.Zucchelli C., Tamburri S., Quilici G., Palagano E., Berardi A., Saare M., Peterson P., Bachi A., Musco G. Structure of human Sp140 PHD finger: an atypical fold interacting with Pin1. FEBS J. 2014;281:216–231. doi: 10.1111/febs.12588. [DOI] [PubMed] [Google Scholar]
- 87.Di Bernardo M.C., Crowther-Swanepoel D., Broderick P., Webb E., Sellick G., Wild R., Sullivan K., Vijayakrishnan J., Wang Y., Pittman A.M. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 88.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., International Multiple Sclerosis Genetics Consortium. International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 92.Matesanz F., Potenciano V., Fedetz M., Ramos-Mozo P., Abad-Grau Mdel.M., Karaky M., Barrionuevo C., Izquierdo G., Ruiz-Peña J.L., García-Sánchez M.I. A functional variant that affects exon-skipping and protein expression of SP140 as genetic mechanism predisposing to multiple sclerosis. Hum. Mol. Genet. 2015;24:5619–5627. doi: 10.1093/hmg/ddv256. [DOI] [PubMed] [Google Scholar]
- 93.Coulombe-Huntington J., Lam K.C., Dias C., Majewski J. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet. 2009;5:e1000766. doi: 10.1371/journal.pgen.1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andrés A.M., Dennis M.Y., Kretzschmar W.W., Cannons J.L., Lee-Lin S.Q., Hurle B., Schwartzberg P.L., Williamson S.H., Bustamante C.D., Nielsen R., NISC Comparative Sequencing Program Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tanioka T., Hattori A., Masuda S., Nomura Y., Nakayama H., Mizutani S., Tsujimoto M. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J. Biol. Chem. 2003;278:32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 96.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat. Rev. Genet. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh G., Cooper T.A. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques. 2006;41:177–181. doi: 10.2144/000112208. [DOI] [PubMed] [Google Scholar]
- 98.Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant Interpretation: Functional Assays to the Rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rosenberg A.B., Patwardhan R.P., Shendure J., Seelig G. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell. 2015;163:698–711. doi: 10.1016/j.cell.2015.09.054. [DOI] [PubMed] [Google Scholar]
- 100.Soemedi R., Cygan K.J., Rhine C.L., Wang J., Bulacan C., Yang J., Bayrak-Toydemir P., McDonald J., Fairbrother W.G. Pathogenic variants that alter protein code often disrupt splicing. Nat. Genet. 2017;49:848–855. doi: 10.1038/ng.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown S.J., Stoilov P., Xing Y. Chromatin and epigenetic regulation of pre-mRNA processing. Hum. Mol. Genet. 2012;21(R1):R90–R96. doi: 10.1093/hmg/dds353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Linares A.J., Lin C.H., Damianov A., Adams K.L., Novitch B.G., Black D.L. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife. 2015;4:e09268. doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Libbrecht M.W., Noble W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015;16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sorek R., Shemesh R., Cohen Y., Basechess O., Ast G., Shamir R. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dror G., Sorek R., Shamir R. Accurate identification of alternatively spliced exons using support vector machine. Bioinformatics. 2005;21:897–901. doi: 10.1093/bioinformatics/bti132. [DOI] [PubMed] [Google Scholar]
- 106.Rätsch G., Sonnenburg S., Schölkopf B. RASE: recognition of alternatively spliced exons in C.elegans. Bioinformatics. 2005;21(Suppl 1):i369–i377. doi: 10.1093/bioinformatics/bti1053. [DOI] [PubMed] [Google Scholar]
- 107.Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 109.Xiong H.Y., Barash Y., Frey B.J. Bayesian prediction of tissue-regulated splicing using RNA sequence and cellular context. Bioinformatics. 2011;27:2554–2562. doi: 10.1093/bioinformatics/btr444. [DOI] [PubMed] [Google Scholar]
- 110.Barash Y., Vaquero-Garcia J., González-Vallinas J., Xiong H.Y., Gao W., Lee L.J., Frey B.J. AVISPA: a web tool for the prediction and analysis of alternative splicing. Genome Biol. 2013;14:R114. doi: 10.1186/gb-2013-14-10-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leung M.K., Xiong H.Y., Lee L.J., Frey B.J. Deep learning of the tissue-regulated splicing code. Bioinformatics. 2014;30:i121–i129. doi: 10.1093/bioinformatics/btu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jha A., Gazzara M.R., Barash Y. Integrative deep models for alternative splicing. Bioinformatics. 2017;33:i274–i282. doi: 10.1093/bioinformatics/btx268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 115.Angermueller C., Pärnamaa T., Parts L., Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 2016;12:878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang Y., Sanguinetti G. BRIE: transcriptome-wide splicing quantification in single cells. Genome Biol. 2017;18:123. doi: 10.1186/s13059-017-1248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Van Nostrand, E.L., Freese, P., Pratt, G.A., Wang, X., Wei, X., Blue, S.M., Dominguez, D., Cody, N.A.L., Olson, S., Sundararaman, B., et al. (2017). A Large-Scale Binding and Functional Map of Human RNA Binding Proteins. bioRxiv doi: https://doi.org/10.1101/179648. [DOI] [PMC free article] [PubMed]
- 118.Lim K.H., Ferraris L., Filloux M.E., Raphael B.J., Fairbrother W.G. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. USA. 2011;108:11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Woolfe A., Mullikin J.C., Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome Biol. 2010;11:R20. doi: 10.1186/gb-2010-11-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mort M., Sterne-Weiler T., Li B., Ball E.V., Cooper D.N., Radivojac P., Sanford J.R., Mooney S.D. MutPred Splice: machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol. 2014;15:R19. doi: 10.1186/gb-2014-15-1-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li M., Feng W., Zhang X., Yang Y., Wang K., Mort M., Cooper D.N., Wang Y., Zhou Y., Liu Y. ExonImpact: Prioritizing Pathogenic Alternative Splicing Events. Hum. Mutat. 2017;38:16–24. doi: 10.1002/humu.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Corey D.R. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 2017;20:497–499. doi: 10.1038/nn.4508. [DOI] [PubMed] [Google Scholar]
- 123.Trincado J.L., Sebestyén E., Pagés A., Eyras E. The prognostic potential of alternative transcript isoforms across human tumors. Genome Med. 2016;8:85. doi: 10.1186/s13073-016-0339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnson, N.T., Dhroso, A., Hughes, K.J., and Korkin, D. (2017). Biological classification with RNA-Seq data: Can alternative splicing enhance machine learning classifier? bioRxiv, https://doi.org/10.1101/146340. [DOI] [PMC free article] [PubMed]
- 125.Gymrek M., Willems T., Guilmatre A., Zeng H., Markus B., Georgiev S., Daly M.J., Price A.L., Pritchard J.K., Sharp A.J., Erlich Y. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 2016;48:22–29. doi: 10.1038/ng.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang X., Lin H., Zhao H., Hao Y., Mort M., Cooper D.N., Zhou Y., Liu Y. Impact of human pathogenic micro-insertions and micro-deletions on post-transcriptional regulation. Hum. Mol. Genet. 2014;23:3024–3034. doi: 10.1093/hmg/ddu019. [DOI] [PMC free article] [PubMed] [Google Scholar]