Abstract
Locus heterogeneity characterizes a variety of skeletal dysplasias often due to interacting or overlapping signaling pathways. Robinow syndrome is a skeletal disorder historically refractory to molecular diagnosis, potentially stemming from substantial genetic heterogeneity. All current known pathogenic variants reside in genes within the noncanonical Wnt signaling pathway including ROR2, WNT5A, and more recently, DVL1 and DVL3. However, ∼70% of autosomal-dominant Robinow syndrome cases remain molecularly unsolved. To investigate this missing heritability, we recruited 21 families with at least one family member clinically diagnosed with Robinow or Robinow-like phenotypes and performed genetic and genomic studies. In total, four families with variants in FZD2 were identified as well as three individuals from two families with biallelic variants in NXN that co-segregate with the phenotype. Importantly, both FZD2 and NXN are relevant protein partners in the WNT5A interactome, supporting their role in skeletal development. In addition to confirming that clustered –1 frameshifting variants in DVL1 and DVL3 are the main contributors to dominant Robinow syndrome, we also found likely pathogenic variants in candidate genes GPC4 and RAC3, both linked to the Wnt signaling pathway. These data support an initial hypothesis that Robinow syndrome results from perturbation of the Wnt/PCP pathway, suggest specific relevant domains of the proteins involved, and reveal key contributors in this signaling cascade during human embryonic development. Contrary to the view that non-allelic genetic heterogeneity hampers gene discovery, this study demonstrates the utility of rare disease genomic studies to parse gene function in human developmental pathways.
Keywords: Frizzled, human embryonic development, skeletal dysplasia, dual molecular diagnosis
Introduction
The study of rare disease informs human biology. Bridging the gap between disease and gene can often provide the initial insights into gene function and disease mechanism, drive experimental clinical or laboratory questions, and provide immediate practical application in clinical genomic dignostics.1 Robinow syndrome (RS) is a congenital skeletal dysplasia, with autosomal-dominant (DRS) and -recessive (RRS) inheritance and evidence for genetic heterogeneity. Originally described by Meinhard Robinow in 1969, it is characterized by short stature, mesomelic limb shortening, broad thumbs and toes, genital hypoplasia, and distinctive and recognizable craniofacial features with frontal bossing, high, broad forehead, depressed nasal bridge, and prominent eyes; the tall forehead relative to the face leads to facial disproportion often termed the “fetal face.”2 The clinically more severe recessive Robinow syndrome (RRS [MIM: 268310]) has been associated with biallelic variants in the tyrosine kinase-like orphan receptor, ROR2 (MIM: 602337), in an estimated 80% of the case subjects described.3, 4 ROR2 is essential for noncanonical Wnt (β-catenin-independent) signaling that establishes cellular orientation via the Wnt/planar cell polarity pathway (PCP).5 Ensuing discovery of heterozygous hypomorphic missense alleles in WNT5A (MIM: 164975), the gene that encodes the signal transducer and the putative ligand for ROR2, resulting in dominant Robinow syndrome (DRS1 [MIM: 180700]),6 supports the hypothesis that a specific pathway could be involved in the disease etiology. More recently, de novo indels resulting in clustered protein-truncating variants affecting two out of the three human orthologs of the Drosophila dishevelled (dsh) gene, DVL1 and DVL3 (MIM: 601365, 601368), were shown to be another cause of dominant Robinow syndrome (DRS2 [MIM: 616331], DRS3 [MIM: 616894]).7, 8, 9 This finding is consistent with the observation that human genes with two or more paralogs for the Drosophila ortholog are eight times more likely to be disease relevant.10 DVL is a key downstream mediator of the Wnt pathway, including the Wnt/noncanonical PCP pathway via interaction with ROR2,11, 12 which further strengthens the link of the disease with a perturbed PCP signaling cascade during human development.
The mutational mechanism observed in DVL1- and DVL3-associated RS is exclusively clustered mutations in the penultimate or last exon resulting in −1 frameshifting variants that result in a premature stop codon within the last exon. Those variants generate a stable mRNA that is predicted to translate into a mutant protein with a similarly truncated C terminus. This is remarkably specific and recurs independently in DRS-affected families.7, 8, 9 Whether the pathogenicity of these truncated proteins results from a gain-of-function or dominant-negative effect is actively being investigated, but preliminary experimental data suggest that it is not driven by the loss of the C terminus alone.8 Collectively, the pathogenesis of RS appears to be a result of perturbation of the WNT5A/ROR2 signaling cascade required for efficient PCP.
The Wnt signaling pathways control many critical early developmental and post-natal physiological processes. One fundamentally important function of noncanonical Wnt signaling is to provide directional information by regulating the evolutionarily conserved PCP pathway. The PCP pathway is essential for vertebrate embryogenesis, including convergent-extension movements and limb bud outgrowth and patterning.13 The core system of Wnt/PCP signaling is composed of a set of highly conserved proteins that interact with each other to form opposing complexes at opposite sides of the cell, thus allowing cellular orientation.14 Mutation in genes crucial for PCP have been linked to human diseases, including neural tube defects15, 16 and nearly identical mouse phenotypes,17, 18 most notably RS.3, 6, 7, 9 Moreover, despite discovery of some of the genes and variants underlying DRS, it is estimated that 70% to 80% of individuals with DRS have an unknown underlying molecular etiology.7, 9 Based on the fact that all the Robinow syndrome-associated proteins play a vital role in noncanonical Wnt/PCP signaling, we hypothesized that studying subjects with Robinow syndrome would reveal other causative variants within genes important to the establishment of PCP and human development.
Herein we report the analyses of a Robinow syndrome-affected cohort where we applied a combination of targeted Sanger and whole-exome sequencing (WES) studies to 21 unrelated families diagnosed clinically with RS, some of whom have more than one individual affected. This study revealed de novo and/or private variants affecting four genes for which translated proteins directly interact with DVL1 and DVL3 or have a role in the Wnt/PCP pathway: FZD2 (MIM: 600667), NXN (MIM: 612895), RAC3 (MIM: 602050), and GPC4 (MIM: 300168). Importantly, mutations of the Frizzled receptor, FZD2, alone accounts for 3/21 (14%) of the clinically ascertained RS-affected case subjects, two of which are due to substitution of an identical amino acid that maps adjacent to the domain that directly interacts with DVL. In addition, we report that NXN, which encodes a regulator of the Wnt pathway, is also associated with RRS.19, 20 In total, WNT5A, DVL1, DVL3, and FZD2 explain 12/21 (57%) of our DRS-affected case subjects. In summary, these data strongly support the notion that RS results from perturbation of the Wnt/PCP pathway during human development and reveals specific gene paralogs, key proteins, and potential domains involved in human limb and craniofacial formation.
Subjects and Methods
Study Participants
We recruited 21 unrelated individuals with a clinical diagnosis of RS or presenting clinical features consistent with a Robinow-like syndrome. Of the 21 probands, 10 were ascertained during the 13th Biennial Robinow Syndrome Conference in Minneapolis, MN. All individuals were screened for pathogenic variants in ROR2 before inclusion into this study, and therefore the cohort was biased toward individuals with the dominant form of RS. Inclusion was based on core clinical phenotypes including characteristic facial features, short stature, and limb abnormalities but not altogether meeting a strict criterion for DRS clinical diagnosis (see Supplemental Note for detailed clinical synopsis). RS was of sporadic occurrence, except for two families with an affected mother and child. DNA was obtained after all relevant family members provided written informed consent. Two additional families, which were not included in the original cohort, were subsequently ascertained via GeneMatcher. This study was approved by the institutional review board at Baylor College of Medicine (protocol no. H-29697).
Sanger Sequencing
From our previous studies, we estimate that nearly 20% of DRS results from truncating alleles in either DVL1 or DVL3.7, 9 Therefore, before performing WES, we first Sanger sequenced the penultimate and final exons of DVL1 and DVL3 from the 21 probands, since DVL-mediated RS is the unique result of clustered −1 frameshifting indels in either DVL1 or DVL3, the mRNAs being predicted to escape nonsense-mediated decay.7, 9 All PCR products containing candidate frameshift alleles detected by Sanger sequencing were further confirmed using allele-specific sequencing via manually cloning both alleles into a standard TOPO TA cloning vector (Life Technologies) and then transforming the recombinant clones into chemically competent Escherichia coli to be grown overnight. Individual clonal colonies were then sequenced by a standard Sanger dideoxy capillary sequencing.
Exome Sequencing
The DNA samples from “unsolved” individuals lacking pathogenic variants in either DVL1 or DVL3 were subjected to WES, through the Baylor-Hopkins Center for Mendelian Genomics initiative.21 WES was performed at the Baylor College of Medicine-Human Genome Sequencing Center (BCM-HGSC), and pre-captured libraries were pooled and then hybridized in solution using the BCM-HGSC in-house VCRome 2.1 design22 according to the manufacturer’s protocol NimbleGen SeqCap EZ Exome Library SR User’s Guide with minor revisions. All samples achieved 96% of the targeted exome bases covered to a depth of 20× or greater with an average depth of coverage of 95× . The choice of variant annotation software can influence the filtering and parsing of rare variants, thus affecting one’s ability to identify potential disease-relevant variants. Differing annotation software tools often provide distinct interpretations and varying levels of false-positive and false-negative findings; this appears particularly true for indels.23 To maximize our variant discovery from the Illumina sequencing data, we used two variant discovery methods in parallel starting with the BCM-HGSC Mercury analysis pipeline,24 which moves data from the initial sequence generation on the instrument to annotated variant call files (VCF) via various analysis tools. In addition, we used the Genome Analysis Toolkit (GATK) HaplotypeCaller to produce joint called files with indel realignment and base recalibration in all families that underwent WES. We identified de novo mutations in silico by using read-depth information extracted from the BAM files of either parent and proband using the in-house developed software DNM-Finder.25 Candidate variants were filtered against exome data in publicly available databases, including the 1000 Genomes Project, the NHLBI Exome Sequencing Project (ESP) Exome Variant Server, the Atherosclerosis Risk in Communities Study (ARIC) database, and our internal Baylor-Hopkins Centers for Mendelian Genomics variant analyzer database of approximately 6,400 exomes. In addition, all affected individuals were screened for copy-number variants (CNV) with exome data, utilizing XHMM26 and our in-house developed algorithm for detecting homozygous exonic deletions, HMZDelFinder.27 We also used WES data to calculate a B-allele frequency and delineate genomic intervals for absence of heterozygosity consistent with identity by descent.27 We verified and evaluated potential disease-associated variants identified via WES for co-segregation with the phenotype by using standard PCR amplification. PCR products were purified with ExoSAP-IT (Affymetrix) and sequenced with dideoxy nucleotide Sanger sequencing at the DNA Sequencing and Gene Vector Core at Baylor College of Medicine.
Results
Contribution of Genes Associated with Robinow Syndrome
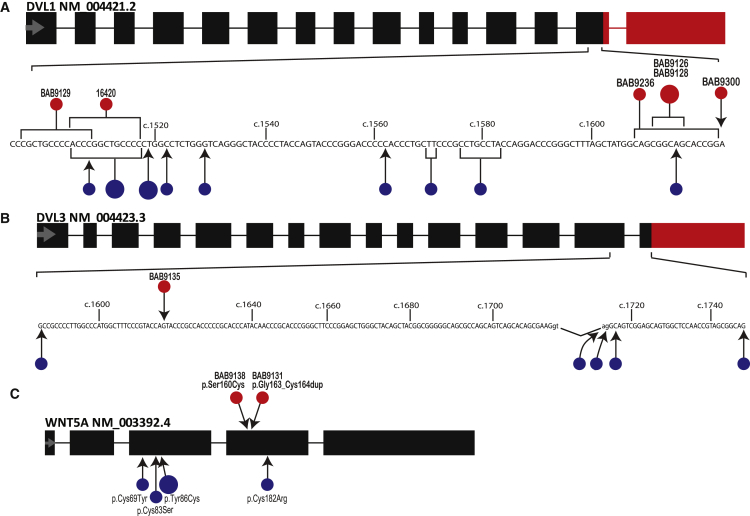
We initially set out to confirm and assess the contribution of DVL1 and DVL3 to DRS by targeted Sanger sequencing of the penultimate and final exons in DVL1 and DVL3 from our in-house database of clinically diagnosed RS-affected subjects (n = 21). Seven individuals (33%) presenting with DRS harbored likely pathogenic variants in either DVL1 or DVL3, offering strong support to our assertion that at least 20% of DRS are caused by indels in DVL1 or DVL3. The identified variants in DVL1 and DVL3 are shown in Figures 1A, 1B, S1, and Table S1. Five individuals had truncating alleles distributed within a highly GC-rich, 116-nucleotide stretch of the penultimate exon of DVL1 according to RefSeq transcript GenBank: NM_004421.2. Two unrelated individuals contained an identical 4-base-pair duplication (c.1612_1616dup [p.Ser539Argfs∗112]), another individual harbored a single base-pair deletion (c.1623del [p.Ser542Valfs∗107]), and one individual had a 16-base-pair deletion (c.1608_1623del [p.Ser537Valfs∗107]). Lastly, two unrelated individuals contained distinct 13-base-pair deletions (c.1496_1508del [p.Pro499Argfs∗146], c.1505_1517del [p.His502Profs∗143]) located in a region where half (9/17) of all reported variants in DVL1 are tightly clustered, likely the result of sequence complexity of this region. The repeated sequence “GCTGCCC” is potentially mediating the recurrently observed 13-bp deletion variants via secondary structure mutagenesis.28 In total, the four newly identified pathogenic variants in DVL1 are consistent with previous reports wherein variants are tightly clustered in the penultimate exon and result in a −1 frameshift.
Figure 1.
Location of Identified Variants in DVL1, DVL3, and WNT5A Resulting in Dominant Robinow Syndrome
The variants in DVL1 and DVL3 are mostly small insertions or deletions, except for two splicing variants in DVL3; all of them are predicted to lead to −1 frameshifting. Black rectangles represent transcript segments identical to the reference DVL1 (A), DVL3 (B), or WNT5A (C) mRNAs. Red rectangles indicate the shared transcript regions affected by the −1 frameshift in the predicted protein structure in all subjects. Part of exon 14 (DVL1) or exons 14 and 15 (DVL3) transcript sequence is shown in detail. Previously described variants are displayed by blue circles, whereas variants identified in this study are displayed by red circles. Larger circles represent identical variants in unrelated individuals. For complete description of all variants, see Tables S1 and S2.
In addition to the six subjects with variants in DVL1, one individual had a likely pathogenic variant in DVL3. According to the DVL3 Refseq transcript GenBank: NM_004423.3, this variant is located in the penultimate exon (c.1617del [p.Gln539Hisfs∗129]). Collectively, and in agreement with previous reports, the variants in DVL1 and DVL3 indicate that DVL-mediated RS is the unique result of clustered −1 frameshifting indels supporting the hypothesis that translation from this mutant reading frame is necessary for the pathogenic effect, due to either a gain-of-function or dominant-negative effect.7, 8, 9
The first gene to be associated with DRS was WNT5A, but hypomorphic missense alleles of WNT5A in DRS-affected individuals has only been reported in a small number of subjects with DRS (<10 individuals). All individuals without pathogenic variants in DVL1 or DVL3 were studied by WES and two likely pathogenic variants in WNT5A were found in two unrelated probands (Figure 1C). In addition to the annotated WES results, to ensure complete sequence read coverage, we manually visualized the coding regions of WNT5A in the Integrative Genomic Viewer (IGV) for all affected individuals studied. One individual, BAB9138, contained a private missense in WNT5A (GenBank: NM_003392.4; c.479C>G [p.Ser160Cys]), but this individual was adopted and parental studies to segregate this allele are unavailable. Another individual, BAB9131, was found to carry a de novo non-frameshift 6-base-pair insertion affecting WNT5A (GenBank: NM_003392.4; c.487_492dup [p.Gly163_Cys164dup]). The variant was observed in 16/70 (23%) mapped reads, which is consistent with a mosaic state of the mutant allele. The identification of only two individuals (∼9.5%) in our cohort of 21 RS-affected probands indicates that WNT5A is an infrequent cause for this disorder. All publicly available pathogenic WNT5A variants are demonstrated in Figure 1C.6, 29
The clinical phenotype of BAB9131 and BAB9138 were generally more severe than previous reports of individuals with WNT5A-mediated RS as both individuals presented with hemivertebrae and significant mesomelic limb shortening indicating that these variants, including one that appears to be mosaic in blood DNA, may result in a more severe RS phenotype (see Supplemental Note for the detailed clinical findings).
Differential Diagnosis of Robinow Syndrome
Approximately 57% of our cohort (12 out of 21) did not yield a pathogenic variant in DVL1, DVL3, or WNT5A. The remaining “unsolved” individuals were then studied by WES, thus allowing for gene discovery and a comprehensive and agnostic screening of all the genes within the broader differential diagnosis of clinical observations consistent with RS.
Clinical findings in subjects diagnosed with RS can elicit a range of potential differential diagnoses, including other skeletal disorders, most notably Aarskog-Scott syndrome (MIM: 305400), Opitz G/BBB syndrome (MIM: 145410), and omodysplasia type 2 (MIM: 164745). In our cohort, four individuals were found to carry likely pathogenic single-nucleotide variants (SNVs) in genes associated with diseases considered within the context of the differential clinical diagnoses of DRS. In addition, one individual, BAB8836, has two large copy-number variants that were revealed by XHMM and/or HMZDelFinder and further confirmed by aCGH (Figure S2). The CNVs are constituted by an ∼8.3 Mb deletion of the short arm of chromosome X and a ∼13 Mb duplication spanning the long arm of chromosome 6. Array analysis in parental samples indicates a de novo event but chromosome analysis did not reveal any visible cytogenetic abnormalities in the proband, which supports the hypothesis that BAB8836 carries an unbalanced chromosomal translocation; karyotype 46,XY,der(X)t(X;6)(p22.31;q25.3).arr[GRCh37]6q25.3q27(157,870,814_170,881,475)x3, Xp22.33p22.31(409,876_8,199,541)x0. Parental samples were not available for further testing so we could not rule out the possibility that BAB8836 has inherited an unbalanced chromosome translocation that resulted from the transmission of a derivative chromosome involved in a balanced event in the mother. The Xpter deletion encompasses the pseudoautosomal gene SHOX (MIM: 312865), deletion of which causes Leri-Weill dyschondrosteosis (MIM: 127300). The partial duplication of the long arm of chromosome 6 and the hemizygous deletion of Xpter also encompasses several known contiguous gene deletion syndromes including chondrodysplasia punctata (MIM: 302950) and ichthyosis (MIM: 308100). Collectively, the clinical phenotype of BAB8836 includes short stature, developmental delay, distal phalangeal hypoplasia, dryness of the skin, scoliosis, hemivertebrae, and an increase of sclerosis in bones, representing a unique amalgamation resulting from these large copy-number variants (see Supplemental Note).
Two male subjects, BAB8747 and BAB8751, were found to have hemizygous frameshift mutations in FGD1 (GenBank: NM_004463.2; c.892dup [p.Cys298Leufs∗5] and c.527dup [Leu177Thrfs∗40]) (Table S1). Truncating alleles in FGD1 (MIM: 300546) are an established cause of Aarskog-Scott syndrome, which has a wide phenotypic variability that largely overlaps with DRS. One individual, BAB8743, was found to have a previously reported pathogenic variant in PTPN11 (MIM: 176876; GenBank: NM_002834.4; c.836A>G [p.Tyr279Cys]).30 The resulting disorder, Noonan syndrome (MIM: 163950), often includes facial features similar to those of DRS.
Lastly, in BAB8759, who was the offspring of consanguineous parents, homozygous pathogenic variants were identified in two genes mapping to distinct segments of absence of heterozygosity (AOH) on heterologous chromosomes. A homozygous frameshift deletion was detected in SH3PXD2B (MIM: 613293; GenBank: NM_001017995.2; c.969del [p.Arg324Glyfs∗19]). Recessive loss-of-function mutations in SH3PXD2B cause Frank-Ter Haar syndrome (MIM: 249420)31 and multiple skeletal anomalies and heart defects which likely explain these clinical observations in subject BAB8759. Additionally, the homozygous missense variant in INPPL1 (MIM: 600829; GenBank: NM_001567.3; c.1636G>A [p.Val546Ile]) likely contributes to the additional clinical features present in BAB8759, including nuchal edema and recurrent respiratory infections, which are observed in opsismodysplasia (MIM: 258480).32 Therefore, the resulting clinical features of BAB8759 are likely a blended phenotype33 resulting from a dual molecular diagnoses in which homozygous single gene defects in both INPPL1 and SH3PXD2B contributed. From our cohort of 21 families, identification of one individual with a likely dual molecular diagnosis (4.8%) is consistent with previous estimates of the rate of dual molecular diagnosis in clinical WES.33, 34, 35 In total, five individuals from our cohort of 21 families who were ascertained due to a clinical impression of RS were found to contain likely pathogenic variants in other relevant genes that represent a broad range of overlapping features. This demonstrates the utility of exome sequencing and molecular diagnosis as an adjuvant approach to achieve characterization of affected individuals as well as detecting disease-gene associations in a complex clinical landscape.35, 36
Recurrent Missense and Truncating Variants in FZD2 Cause Robinow Syndrome Features
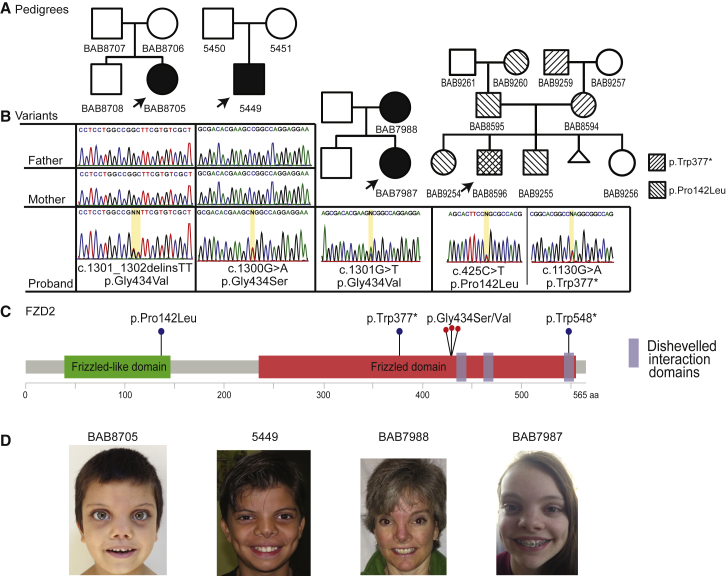
Similar to the “solve-rate” for RS, after analyzing our cohort for known genes with a disease association, seven individuals from our cohort (7/21) remained without a molecular diagnosis. We then performed a personal genome analysis to identify rare and potentially damaging variants that might represent additional genes underlying DRS. Most notably, five affected individuals from three distinct families with DRS or Robinow-like traits had variants affecting FZD2 either de novo or co-segregating with the disease. We then performed a GeneMatcher37 search which led to the identification of a fourth family with a single simplex case of Robinow-like features and a de novo variant in FZD2. The four families, identified FZD2 variants, and selected subjects are shown as Figure 2 and described in Table S1.
Figure 2.
Location of Identified Variants in FZD2 Segregating with the Associated Phenotypic Features
(A) Pedigrees of the four probands with Robinow syndrome features, carrying variants in FZD2.
(B) Sanger sequencing traces for index case subjects, demonstrating the detected variants at the nucleotide level. Family of BAB8596, far right, contains two variants in FZD2: p.Trp377∗ and p.Pro142Leu. The stop gain was inherited from an affected mother and the missense represents a variant of uncertain significance inherited from father.
(C) Representation of the known functional domains of FZD2 (green and red rectangles).86 Location of protein-coding variants identified in our cohort: one stop gain and three recurrent variants all affecting glycine 434 located within the third intracellular loop (red dots). One additional variant (p.Trp548∗) from the literature, and reported in association with omodysplasia, is also included.46
(D) Photographs of consenting subjects with FZD2 variants demonstrating shared facial characteristics consisting of a high, broad forehead, prominent eyes, broad and low nasal bridge, low-set ears, broad nasal tip, and anteverted nares.
Of note, the same amino acid residue, glycine 434, is altered in three out of four families. One family with an affected mother and child co-segregate a missense variant (GenBank: NM_001466.3; c.1301G>T [p.Gly434Val]) and one subject has a dinucleotide substitution (c.1301_1302delinsTT [p.Gly434Val]), but the same modification at the protein level (Figures 2A–2C). Lastly, in one affected individual, a de novo variant (c.1300G>A [p.Gly434Ser]), which also changes the same codon, altering residue Gly434 was observed. Residue Gly434 is conserved throughout all vertebrates and is located at the edge of a transmembrane domain; the adjacent intracellular loop is one of the three motifs that were shown to be required for binding and stabilizing the interaction with Dvl1.38 Therefore, alterations in Gly434 potentially interfere or enhance the affinity or stability of the FZD2-DVL interaction, which has been shown to occur via the C terminus of DVL1 including the DEP domain.39
In contrast to the above individuals with FZD2 variants, one subject (BAB8596) carries a nonsense truncating allele (c.1130G>A [p.Trp377∗]) in addition to a variant of uncertain significance (VUS) (c.425C>T [p.Pro142Leu]) affecting FZD2 in trans (Figure 2). The nonsense single-nucleotide variant is located upstream of the recurrent variant present in all other families and is predicted to result in truncation of nearly 200 aa at the C terminus. FZD2 is a single-exon gene not subject to nonsense-mediated decay, thus indicating that this truncating allele may produce a stable mRNA product. BAB8596 inherited this allele from his affected mother, BAB8594, who has a milder form of DRS; her phenotype includes short stature, a high and broad forehead, and mesomelia. The proband presents with short stature, RS facial characteristics (more prominent in him than in his mother), and mesomelia early in life that improved with age as assessed both clinically and by radiographic measurement (Figure S3 and Supplemental Note). The VUS was inherited from his father, BAB8595, and is present in two siblings, BAB9254 and BAB9255, all of whom have short stature but no other features consistent with RS (Table 1). Therefore, we are unable to determine the potential genetic contribution of this VUS to the phenotypes observed in this family. An alternative interpretation of the FZD2 variant data and segregation in this family is that the VUS may be benign and the short stature observed in this pedigree, a result of assortative mating, as both maternal and paternal lineages are significant for short stature with the proband’s expected mid-parental height of 162 cm (<2nd centile).
Table 1.
Phenotypic Features of Individuals with FZD2 (GenBank: NM_001466.3) Variants
| Individual ID | 5449 | BAB8705 | BAB7987 | BAB7988 | BAB8596 | BAB8594 | BAB9254∗ | BAB9255∗ |
|---|---|---|---|---|---|---|---|---|
| Zygosity | het | het | het | het | comp het | het | het | het |
| Variant | c.1300G>A | c.1301_1302delinsTT | c.1301G>T | c.1301G>T | c.1130G>A, c.425C>T | c.1130G>A | c.425C>T | c.425C>T |
| Effect | p.Gly434Ser | p.Gly434Val | p.Gly434Val | p.Gly434Val | p.Trp377∗, p.Pro142Leu | p.Trp377∗ | p.Pro142Leu | p.Pro142Leu |
| Age last examination | 10 y 3 mo | 5 y 8 mo | 15 y | 47 y | 6 y 7 mo | 30 y | 8 y 4 mo | 4 y 3 mo |
| Inheritance | de novo | de novo | inherited | unknown; affected mother of BAB7987 | inherited | inherited; affected mother of BAB8596 | inherited; sister of BAB8596 | inherited; brother of BAB8596 |
| Gender | M | F | F | F | M | F | F | M |
| Growth | ||||||||
| Height SD | 45th centile | −2.9 SD | −2.25 SD | −1.7 SD | −3.5 SD | −2.1 SD | −4.5 SD | −2.6 SD |
| Facial Features | ||||||||
| Macrocephaly | − | relative | − | + | − | − | − | − |
| Broad forehead | + | + | + | + | + | + | − | − |
| High forehead | + | + | − | − | + | + | − | − |
| Midface hypoplasia | + | + | + | + | − | − | − | − |
| Hypertelorism | + | + | + | + | − | − | − | − |
| Long eyelashes | mild + | + | + | + | + | − | − | − |
| Prominent eyes | + | + | + | + | − | − | − | − |
| Anteverted nares | + | + | + | + | + | + | − | − |
| Wide nasal bridge | + | + | − | ND | − | − | − | − |
| Thin vermillion border | + | + | + | + | − | − | − | − |
| Gingival hyperplasia | + | + | + | + | ND | ND | − | − |
| Bilobed tongue | − | + | + | + | − | − | − | − |
| Dental anomalies | + | + | + | + | − | − | − | − |
| Low set ears | + | + | + | + | + | + | − | − |
| Skeletal | ||||||||
| Hand length | 14.5 cm (−1 SD) | ND | ND | ND | 11.5 cm | 15.5L 15.7R | 12 cm | 10 cm |
| Limbs | mesomelia | micromelia | mesomelia | mesomelia | mesomelia, improved with age | mesomelia | micromelia | micromelia |
| Brachydactyly | + | + | + | + | − | − | − | − |
| Clinodactyly | 5th fingers + | 5th fingers + | + | + | − | − | − | − |
| Camptodactyly | short low implanted thumbs | 4th fingers + | − | − | − | − | − | − |
| Broad thumb | − | + | − | − | − | − | − | − |
| Broad 1st toe | − | + | + | + | − | − | − | − |
| Other Features | ||||||||
| Genital hypoplasia | − | + | + | + | − | ND | − | − |
| Renal anomalies | − | − | − | + | − | − | − | − |
| Cardiac anomalies | − | − | − | − | − | − | − | − |
Abbreviations: ND, no data; het, heterozygous. Asterisk (∗) indicates subjects not diagnosed with Robinow syndrome.
Pertinent clinical information in all individuals with variants in FZD2 is shown in Table 1 (see Supplemental Note for detailed clinical case reports). The clinical presentation of affected individuals could be considered a constellation of features representing a distinct disorder that includes short stature, limb defects, and shared facial characteristics that includes a high, broad forehead, flat face, low-set ears, broad nasal tip, and anteverted nares (Figure 2D). In addition, all affected individuals (6/6) have mesomelia or micromelia, 4/6 have brachydactyly, and 5/6 have significant short stature. Alternatively, as a result of the broad variable expressivity of associated RS features, individuals with FZD2 variants could be considered as atypical RS case subjects. In fact, FZD2 has been proposed in a single family as a candidate gene for omodysplasia40 (MIM: 258315), a disorder that significantly overlaps with RS. Indeed, the clinical similarities between omodysplasia and DRS are striking. Given the observations that FZD2, WNT5A, and DVL1 and DVL3 interact molecularly and the shared phenotype between omodysplasia and RS, these data are consistent with the hypothesis that omodysplasia represents a subset of RS with predominant short humeri and radial head dislocation collectively representing a point on the spectrum of RS, rather than a distinct clinical syndrome. The characteristic “fetal face” is present in all affected individuals with additional skeletal and limb defects of varying severity but this would not meet a strict clinical diagnostic criterion for DRS, except for BAB7987, BAB7988, and BAB8596. Although there have been previous reports of RS-affected individuals without limb defects, it is unclear whether this cohort of affected individuals would constitute necessary phenotypic overlap to clinically describe this as RS or a distinct disorder of variable severity.41
Biallelic Variants in NXN Cause Recessive Robinow Syndrome
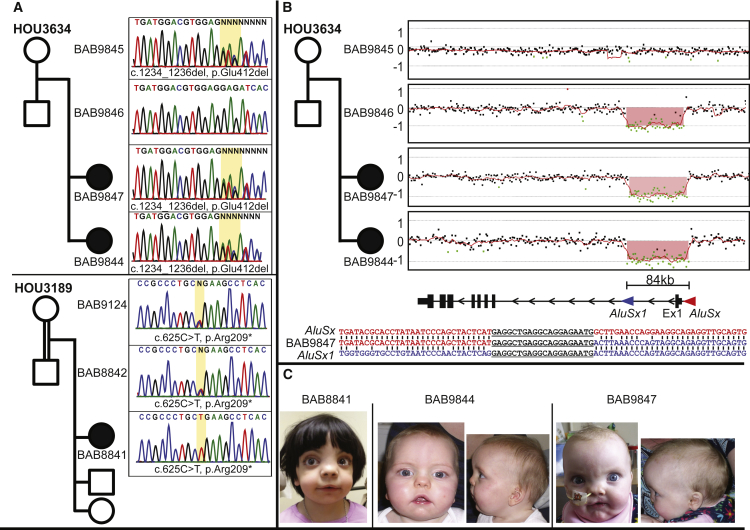
One individual clinically diagnosed with RS, BAB8841, carries a homozygous stopgain variant in NXN (GenBank: NM_022463.4; c.625C>T [p.Arg209∗]) inherited from consanguineous parents (Figure 3A). GeneMatcher37 identified a second family with two affected siblings (BAB9844, BAB9847) who shared compound heterozygous biallelic variants in NXN: a maternally inherited in-frame 3-bp deletion (GenBank: NM_022463.4; c.1234_1236del [p.Glu412del]) and a paternally inherited intragenic 84-kb deletion that encompasses the entire first exon: seq[GRCh37]del (17)(p13.3) chr17:g.805043::GAGG…..AATG::889090) (Figures 3A, 3B, and S4). Deletion CNVs can often account for disease-associated pathogenic alleles at both AD and AR disease trait loci.42, 43, 44 This specific NXN exon 1 deletion CNV is mediated by two directly orientated Alu elements (AluSx-AluSx1) sharing 81% nucleotide identity (Figure 3B); such Alu-Alu-mediated exonic deletions often involve different Alu family members located within flanking introns. Remarkably, NXN maps to 17p13.3, which has a high Alu density and likely elevated rates of Alu-Alu-mediated rearrangements,45 indicating that future diagnostic efforts should include copy-number analyses for this locus. The first and last exons of a multi-exon gene may be particularly prone to Alu-Alu-mediated deletion CNVs related to the abundance of Alu elements flanking the 5′ and 3′ ends of the gene which can be used as template switch substrates during microhomology-mediated break-induced replicative repair.46, 47
Figure 3.
Identified Biallelic Variants in NXN
(A) Pedigrees and Sanger sequencing traces for the two families with variants in NXN; a homozygous stop gain in BAB8841 (family HOU3189) and a heterozygous 3-bp in-frame deletion in BAB9847 and BAB9844 (family HOU3634).
(B) High-density arrays and breakpoint mapping showing the second mutant allele in BAB9847 and BAB9844. The deletion is mediated by two highly similar Alu elements in direct orientation that flank the first coding exon.
(C) Facial pictures demonstrating shared facial features including high forehead, prominent eyes, broad and low nasal bridge, broad nasal tip, anteverted nares, and micrognathia.
NXN encodes an oxidative stress response protein, nucleoredoxin, which is highly expressed in the developing limb bud of mice.48 Abnormal activation of the Wnt/β-catenin signaling in Nxn−/− knockout mice is hypothesized to lead to the observed craniofacial defects, a phenotype perhaps relevant to the subjects with RRS reported herein due to biallelic NXN variants that are likely loss-of-function alleles. Moreover, studies in Xenopus laevis have collectively shown that Nxn acts as a negative regulator of the Wnt/PCP pathway potentially by blocking the ubiquitination and degradation of Dvl as well as inhibiting Dvl-induced c-jun phosphorylation via Rac, the crucial biochemical mechanism underlying the Wnt/PCP pathway.19, 20 Two independent Nxn−/− mouse models show abnormal craniofacial morphology with a shortened snout and cleft palate partially recapitulating the RS subjects’ phenotype.49, 50 The individuals with biallelic variants in NXN present with a phenotype consistent with a diagnosis of RS, wherein all three identified individuals have typical facial characteristics, mesomelia, brachydactyly, and broad thumbs/toes (Figure 3C and Supplemental Note for detailed clinical case reports).51
Identification of Additional Robinow Syndrome Candidate Disease-Associated Genes
In two affected individuals we identified likely pathogenic rare variants in association with the rare RS phenotype in two distinct candidate genes whose function has been demonstrated to play an important role in Wnt/PCP signaling: RAC3 and GPC4. These include one sporadic Robinow-like individual (BAB8740) with a de novo missense variant in RAC3 (GenBank: NM_005052.2; c.176C>G [p.Ala59Gly]). Another individual (BAB8295) has a hemizygous nonsynonymous missense variant in GPC4 (GenBank: NM_001448.2; c.1235G>A [p.Arg412Lys]) inherited from a heterozygous unaffected mother, potentially responsible for an X-linked form of RS. We propose RAC3 and GPC4 to be candidate genes underlying Robinow-like features due to the shared interactome and signaling network of known (WNT5A, ROR2, DVL1, DVL3) and currently described (FZD2, NXN) RS disease-relevant genes. In addition, both affected subjects share the same rare phenotype and the exceedingly rare nature of the mutant alleles. RAC3 is a member of the Rac subfamily of the Rho family of GTPases, which specifically interacts with DVL in response to Wnt ligand to activate downstream effects including JNK/c-jun phosphorylation essential for cytoskeletal reorganization.52 GPC4 is one of the six members of the heparin sulfate proteoglycans, which are key players in endochondral ossification53 with defects in GPC6 (MIM: 604404) and GPC3 (MIM: 300037) resulting in the skeletal disorders omodysplasia54 and Simpson-Golabi-Behmel syndrome (MIM: 312870),55 respectively. Specifically, GPC4 has been shown to be a positive regulator of Wnt/PCP signaling by promoting the accumulation of dishevelled at the cell membrane56 and can rescue Drosophila Dfz2 aberrant Wnt signaling phenotypes.57 Indeed, duplications of GPC4 have been associated with Simpson-Golabi-Behmel syndrome;58 however, loss of gpc4 in Danio rerio results in dwarfism phenotypes, which may represent an unspecific finding.59 The clinical phenotype of individuals harboring candidate variants is consistent with earlier clinical descriptions of DRS with varying severity of associated traits;51 shared phenotypes are shown in Table 2 (see Supplemental Note for detailed clinical descriptions).
Table 2.
Phenotypic Features of Individuals with Variants in RS Candidate Genes NXN (Recessive), RAC3 (Dominant), and GPC4 (X-Linked)
| Individual ID | BAB8841 | BAB9847 | BAB9844 | BAB8740 | BAB8295 |
|---|---|---|---|---|---|
| Gene | NXN | NXN | NXN | RAC3 | GPC4 |
| GenBank | NM_022463.4 | NM_022463.4 | NM_022463.4 | NM_005052.2 | NM_001448.2 |
| Zygosity | hom | comp het | comp het | het | hemizygous |
| Variant(s) | c.625C>T | c.1234_1236del | c.1234_1236del | c.176C>G | c.1235G>A |
| chr17:g.805043::GAGG….AATG::889090 | chr17:g.805043::GAGG….AATG::889090 | ||||
| Effect | p.Arg209∗ | p.Glu412del, ? | p.Glu412del, ? | p.Ala59Gly | p.Arg412Lys |
| Consanguinity | + | − | − | − | − |
| Age last examination | 5 y | 2 y 5 mo | 4 wk | 13 y 2 mo | 8 y |
| Inheritance | inherited | inherited | inherited | de novo | inherited |
| Gender | F | F | F | F | M |
| Facial Features | |||||
| Macrocephaly | + | relative | relative | − | + |
| Frontal bossing | − | + | + | − | + |
| High forehead | + | + | + | − | + |
| Midface hypoplasia | − | + | + | + | + |
| Hypertelorism | + | + | + | + | + |
| Long eyelashes | + | − | − | + | − |
| Prominent eyes | + | + | + | + | − |
| Anteverted nares | + | + | + | + | + |
| Wide nasal bridge | − | + | + | + | + |
| Short nose | − | + | + | + | − |
| Long philtrum | + | + | + | + | − |
| Triangular mouth | + | + | + | − | + |
| Gingival hyperplasia | + | + | + | − | + |
| Absent uvula | + | − | + | − | − |
| Cleft soft palate | − | − | + | − | + |
| Dental anomalies | + | − | ND | + | delayed dental eruption |
| Micrognathia | + | + | + | + | − |
| Skeletal | |||||
| Mesomelia | + | + improved with age | + | − | + |
| Brachydactyly | + | + | + | − | + |
| Clinodactyly | + | + | + | + | + |
| Camptodactyly | − | + | + | − | + |
| Broad thumb | + | + | + | − | + |
| Fetal finger/toe pads | − | + | + | + | − |
| Broad 1st toe | + | + | + | − | + |
| Other Features | |||||
| Genital hypoplasia | ND | − | − | + | − |
| Renal anomalies | + | − | − | − | − |
| Cardiac anomalies | + | − | − | − | PDA |
Abbreviations: ND, no data; hom, homozygous; het, heterozygous; PDA, patent ductus arteriosus.
Discussion
The concept that mutation in distinct genes could lead to the same phenotype was first suggested almost a century ago by William Allan when he observed multiple modes of inheritance (AD, AR, XL) for Charcot-Marie-Tooth disease consistent with at least two genetic loci (autosomal and X-linked);60 this was further demonstrated by N.E. Morton more than 60 years ago, wherein elliptocytosis was mathematically linked to two distinct human loci in different families.61 Although genetic heterogeneity has been known for some time, only recently with comprehensive clinical phenotyping, syndrome characterization, and cataloguing of Mendelian disease traits through the efforts of OMIM can we begin to appreciate the frequency of genetic heterogeneity.62 As of April 2017, an estimated 12.5% (395/4,931) of Mendelian phenotypes have had disease-causing mutations identified in two or more genes represented as a “phenotypic series.”63 Genetic heterogeneity can be viewed as an obstacle to identifying disease-gene associations, although it is also recognized that overlapping phenotypes arising from distinct loci might reflect perturbed protein interactions or shared biological relationships.64 Accordingly, genetically heterogeneous disorders provide an opportunity for insight into the pathways and biological underpinnings of disease, wherein a single syndrome provides evidence for the aberrant interactome, network, or biological signaling cascade.
Noncanonical Wnt signaling regulates, via the PCP pathway, convergent extension movements, an essential cell migration process during vertebrate gastrulation.65 The aforementioned developmental processes in different organisms are largely controlled by the same set of core PCP proteins, which were originally identified in Drosophila. However, compared to Drosophila, the vertebrate PCP pathway has evolved and diversified by specification and emergence of multiple paralogs from a single fly gene, throughout nearly all of the core components of this pathway. Not surprisingly, recent studies have demonstrated that genes that are essential in flies and have multiple human orthologs are more likely to be associated with human disease than are fly genes with a single human ortholog,10 potentially indicating duplication and further paralog specialization during evolution from flies to human. Indeed, in vertebrates, Wnt/PCP signaling partially drives the patterning and formation of the limb-bud outgrowth and growth plate in skeletal formation.66, 67 Of particular interest is the organized cellular division and planar alignment of proliferating chondrocytes at the growth plate, as this process is dependent on Wnt/PCP activation and defects result in abnormal growth plate chondrocyte localization and morphology with skeletal defects analogous to RS.66, 68, 69
Genetic screening in Drosophila identified many of the components of the PCP pathway including both frizzled70 and dishevelled.71 We provide evidence that variants in multiple WNT/PCP signaling genes can be associated with RS including FZD2 and NXN in autosomal-dominant and -recessive inheritance patterns, respectively. FZD2 encodes the highly conserved seven-pass transmembrane protein of the Frizzled family of membrane receptors. Frizzled was originally identified by Vinson and Adler in Drosophila melanogaster with “clinical findings” of misoriented cuticle hairs, which they named frizzled.70 Interestingly, similar to all other Robinow-associated proteins, FZD2 is implicated in the Wnt/PCP pathway, in which studies have demonstrated Frizzled proteins functioning as Wnt receptors and co-receptors72, 73 and is required for the WNT5A-mediated recruitment of DVL2 (MIM: 602151) to the membrane.74 WNT5A binding, in the presence of ROR1 (MIM: 602336), ROR2, and DVL2, induces clathrin-mediated internalization of FZD2, which is necessary for RAC1 activation in HEK293 cells.74
NXN demonstrates a specific redox-dependent interaction with dishevelled via the PDZ domain; in fact, pull-down assays of mouse fibroblasts indicate that NXN is a substantial interacting partner of DVL1, particularly under oxidative conditions,20 often stimulated by growth factors.75 In vitro association with DVL2 and DVL3 has also been demonstrated.76 The interaction of NXN and DVL may be a key regulatory mechanism to maintain a spatial or temporal balance between canonical and noncanonical Wnt pathways during development. Evidence for this includes the fact that NXN specifically inhibits DVL function in vitro whereas it also blocks ubiquitination and degradation of DVL via KLHL12 in a tissue-specific manner; the latter helps to regulate transmission of Wnt/β-catenin signal upon Wnt stimulation.20, 49 In addition, tissue-specific dysregulation of Wnt/β-catenin signaling was experimentally demonstrated in Nxn−/− mice in which osteoblastic cells show excessive differentiation associated with strong Wnt/β-catenin signaling activation compared to osteoblasts of Nxn+/+ mice whereas mouse embryonic fibroblast cells show impaired Wnt/β-catenin signaling.49 Collectively, there is ample evidence in the literature that disruption of Frizzled and noncanonical Wnt signaling leads to delay or block in chondrocyte maturation and shortening of skeletal elements,67, 76 which could be the basis of short stature and mesomelia in individuals with RS. Pathogenic variants found in individuals with RS provide insights into the underlying biology of the disease, e.g., loss-of-function mutations affecting the WNT5A receptor ROR2 in ∼80% of individuals with RRS3, 4 and hypomorphic mutations affecting WNT5A in ∼9.5% of individuals with DRS6 (Figure 4A), suggesting that the Wnt/PCP pathway is affected in those subjects. This hypothesis is strengthened with the discovery that recurrent truncating variants in both DVL1 and DVL3, leading to C-terminal replacement of those proteins with a highly basic tail, is currently the major cause (33%) of DRS. Furthermore, it has been demonstrated that a subset of DVL1-mediated DRS-affected subjects present with osteosclerosis, a phenotype that is associated with perturbed Wnt signaling.8 Such mutational specificity in both DVL1 and DVL3 is intriguing and suggest a specific role in the noncanonical pathway. Despite DVL being an important contributor in diverse signaling pathways,11 its C terminus is characterized by (1) having a conserved DEP domain that works in the PCP signaling, (2) being required for membrane recruitment via Wnt-mediated signaling,77 (3) carrying relevant phosphorylation sites that specify DVL functions and subcellular localization,78, 79 and (4) being necessary to establish and to stabilize protein-protein interactions with ROR212 as well as FZD2.38 In fact, our current study indicates that variants affecting FZD2 specifically lead to DRS or Robinow-like traits40 when all the DVL interaction domains either are removed (p.Trp377∗) or are adjacent to them (p.Gly434Ser/Val) (Figure 2C). Moreover, NXN, a gene that encodes a redox-dependent regulator of DVL, was identified in 5% (one family) of this clinically ascertained RS cohort and appears to cause a new recessive form of RS. Compound heterozygous variants within this gene co-segregating with the disease were further observed in another family, confirming our initial observation. NXN is a potent regulator of DVL stability, particularly in conditions of high oxidative stress,49 circumstances intriguingly similar to the high levels of reactive oxygen species during chondrocyte hypertrophy at the growth plate.80 The relevance of NXN mutations for RRS requires further studies in other unsolved RS cases.
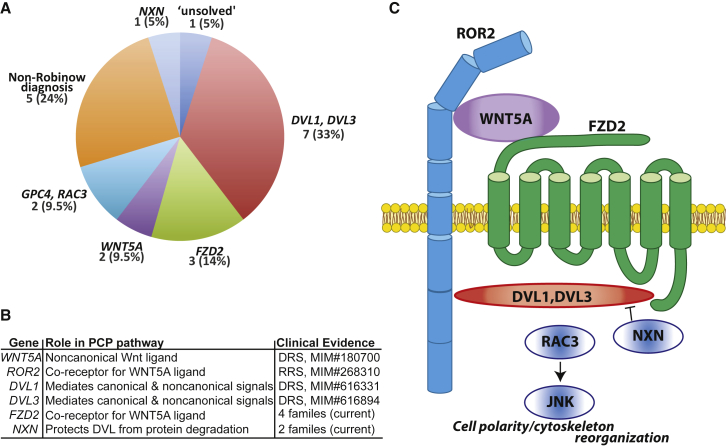
Figure 4.
Robinow Syndrome-Associated Genes in the Wnt/PCP Pathway Identified in Human Subjects
(A) Molecular diagnosis pie chart from the cohort of 21 Robinow syndrome-affected individuals who had a combination of direct Sanger screening and whole-exome sequencing to identify the molecular cause of their disorder highlighting the contribution of DVL1, DVL3, and FZD2 variants to RS.
(B and C) The establishment of planar cell polarity is vital for vertebrate development; in humans, variants affecting genes in that pathway are linked to skeletal defects, including Robinow syndrome. In vitro studies from several model organisms have demonstrated that ROR2 binds to WNT5A and acts as a co-receptor with FZD2.5 The downstream effect is routed by the dishevelled homologs, which are stabilized by NXN in a context-dependent manner.49 The downstream readouts, which ultimately involves cytoskeletal reorganization, are a combination of small GTPases including RAC to activate JNK signaling.52 Pathogenic variants in all of the aforementioned genes have been identified in individuals with Robinow syndrome, underscoring the notion that this disorder results from aberrant Wnt/PCP signaling, likely due to disturbance of the organized development of chondrocytes at the growth plate.
In aggregate, our data provide supportive experimental evidence that the clinical phenotype in RS results from subtle dysregulation of the Wnt/PCP pathway, contextually affecting the organized proliferation of chondrocytes at the growth plate. A delicate balance between canonical and PCP signaling requires precise regulation of DVL, so it appears that the pathogenesis of Robinow is not simply loss of Wnt/PCP signaling, but a context-specific perturbation in the Wnt signaling. These new findings also support the hypothesis that additional proteins underlying RS or overlapping genetic syndromes will also have a relevant role in this pathway (Figure 4B). Our data initially indicated that 3 of the 21 families from the RS-affected case subjects remained “unsolved” or without a plausible molecular diagnosis (Figure 4A). However, two of the three unsolved probands in our RS cohort have variants affecting genes biologically relevant in the context of Wnt signaling: a de novo mutation in an autosomal gene (RAC3) and an X-linked variant in a male from a heterozygous, unaffected mother (GPC4). Both genes RAC3 and GPC4 are established components of the Wnt/PCP pathway and therefore represent candidate genes that need to be confirmed by the ascertainment of additional affected individuals. In aggregate, personal genome analyses provide a plausible molecular diagnosis that parsimoniously explains the observed clinical phenotype in 95% of individuals clinically diagnosed with Robinow; i.e., 20/21 subjects investigated.
In summary, we demonstrate that the study of rare human syndromes and phenotypes can provide insight into human biology, a Garrodian principle stemming from the early pioneers of human genetics.1, 81 Figure 4C depicts a representative model for the physical interactions of all identified Robinow-associated proteins with a role in the Wnt/PCP pathway that have been identified in human mutational studies thus far. Of note, in this cohort all variant types—single-nucleotide variants (SNVs), indels, and copy-number variants (CNVs)—were found to be pathogenic and a multitude of mutational processes, including Alu-Alu-mediated rearrangement resulting in exonic deletion82, 83 and secondary structure mutagenesis for recurrent indel formation,84 were shown to be operative. In dissecting the etiology of non-allelic genetic heterogeneity in RS, we observe strong evidence that disturbed levels of a complex interplay of noncanonical and canonical Wnt signaling underlies the core phenotype exhibited by affected individuals. The relevance of this observation becomes clear when considering that canonical and noncanonical pathways use common intracellular components and that specificity of distinct Wnt signaling pathways is regulated by the combination of ligands and receptors in a particular cellular context. In this way, the same proteins in distinct cellular environments may inhibit or trigger a given signal branch. For example, FZD2 can either inhibit or trigger canonical and non-canonical β-catenin pathways depending on the presence of additional transmembrane receptors.74 Ligation of WNT5A to FZD2 was shown to inhibit canonical β-catenin in addition to activate the noncanonical pathway in HEK293 cells, as measured by downstream RAC1 activation, the latter depending on the presence of ROR1, ROR2, and DVL2. The discovery of the main gene contributors in RS will likely further clarify underlying molecular mechanism in other skeletal dysplasias with an overlapping phenotype, as for example, in omodysplasia.
As we progress in our molecular and genetic understanding of many Mendelian disease traits, it is important to understand and appreciate genetic heterogeneity, clinical variability, and overlap of clinical features (i.e., phenotypic signs/symptoms) with other disorders. Here we have approached gene discovery in a phenotype-driven manner, using an unbiased genomics rather than locus-specific approach, i.e., a Mendelian genomics versus Mendelian genetics single-gene/locus approach. As shown here, Mendelian genomics can readily investigate the underpinnings of genetic heterogeneity of disease traits as well as explore mutational burden and multi-locus variation models for disease.33, 85 This study has enabled the identification of pathway-specific candidate disease-associated genes in RS, the specific paralogs involved, assessed the contribution of known genes, and provided insights into underlying mechanisms of a disease marked by genetic heterogeneity, which ultimately revealed the contribution of key proteins to limb formation and human development.
Conflicts of Interest
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc., and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the Baylor-Genetics Laboratories.
Acknowledgments
The authors would like to thank the individuals and their families who contributed to this study and the Robinow Syndrome Foundation for facilitating collaboration. Supported by the US National Human Genome Research Institute/National Heart Blood Lung Institute jointly funded Baylor Hopkins Center for Mendelian Genomics (UM1 HG006542). J.J.W. was funded in part by the Smith-Magenis Syndrome Research Foundation (SMSRF). A.M.V.-M. is funded by FAPESP (CEPID 2013/08028-1). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHGRI/NHBLI, NIH.
Published: December 21, 2017
Footnotes
Supplemental Data include four figures, two tables, and Supplemental Notes of case reports and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.10.002.
Accession Numbers
The accession numbers for identified variants were deposited into ClinVar with the following identifiers: SCV000583563–SCV000583582.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Atherosclerosis Risk in Communities Study (ARIC) Database, http://www2.cscc.unc.edu/aric/
Baylor Genetics Laboratory, http://bmgl.com/
BCM HGSC Mercury, https://www.hgsc.bcm.edu/software/mercury
ExAC Browser, http://exac.broadinstitute.org/
OMIM, http://www.omim.org/
Robinow Syndrome Foundation, http://www.Robinow.org
Supplemental Data
References
- 1.Garrod A. The lessons of rare maladies: annual oration before the medical society of London by Sir Archibald Garrod. BMJ. 1928;1:914–915. [PMC free article] [PubMed] [Google Scholar]
- 2.Robinow M., Silverman F.N., Smith H.D. A newly recognized dwarfing syndrome. Am. J. Dis. Child. 1969;117:645–651. doi: 10.1001/archpedi.1969.02100030647005. [DOI] [PubMed] [Google Scholar]
- 3.van Bokhoven H., Celli J., Kayserili H., van Beusekom E., Balci S., Brussel W., Skovby F., Kerr B., Percin E.F., Akarsu N., Brunner H.G. Mutation of the gene encoding the ROR2 tyrosine kinase causes autosomal recessive Robinow syndrome. Nat. Genet. 2000;25:423–426. doi: 10.1038/78113. [DOI] [PubMed] [Google Scholar]
- 4.Afzal A.R., Rajab A., Fenske C.D., Oldridge M., Elanko N., Ternes-Pereira E., Tüysüz B., Murday V.A., Patton M.A., Wilkie A.O., Jeffery S. Recessive Robinow syndrome, allelic to dominant brachydactyly type B, is caused by mutation of ROR2. Nat. Genet. 2000;25:419–422. doi: 10.1038/78107. [DOI] [PubMed] [Google Scholar]
- 5.Oishi I., Suzuki H., Onishi N., Takada R., Kani S., Ohkawara B., Koshida I., Suzuki K., Yamada G., Schwabe G.C. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8:645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- 6.Person A.D., Beiraghi S., Sieben C.M., Hermanson S., Neumann A.N., Robu M.E., Schleiffarth J.R., Billington C.J., Jr., van Bokhoven H., Hoogeboom J.M. WNT5A mutations in patients with autosomal dominant Robinow syndrome. Dev. Dyn. 2010;239:327–337. doi: 10.1002/dvdy.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White J., Mazzeu J.F., Hoischen A., Jhangiani S.N., Gambin T., Alcino M.C., Penney S., Saraiva J.M., Hove H., Skovby F., Baylor-Hopkins Center for Mendelian Genomics DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am. J. Hum. Genet. 2015;96:612–622. doi: 10.1016/j.ajhg.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunn K.J., Daniel P., Rösken H.S., O’Neill A.C., Cameron-Christie S.R., Morgan T., Brunner H.G., Lai A., Kunst H.P., Markie D.M., Robertson S.P. Mutations in DVL1 cause an osteosclerotic form of Robinow syndrome. Am. J. Hum. Genet. 2015;96:623–630. doi: 10.1016/j.ajhg.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White J.J., Mazzeu J.F., Hoischen A., Bayram Y., Withers M., Gezdirici A., Kimonis V., Steehouwer M., Jhangiani S.N., Muzny D.M., Baylor-Hopkins Center for Mendelian Genomics DVL3 alleles resulting in a -1 frameshift of the last exon mediate autosomal-dominant Robinow syndrome. Am. J. Hum. Genet. 2016;98:553–561. doi: 10.1016/j.ajhg.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto S., Jaiswal M., Charng W.-L., Gambin T., Karaca E., Mirzaa G., Wiszniewski W., Sandoval H., Haelterman N.A., Xiong B. A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao C., Chen Y.G. Dishevelled: The hub of Wnt signaling. Cell. Signal. 2010;22:717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Witte F., Bernatik O., Kirchner K., Masek J., Mahl A., Krejci P., Mundlos S., Schambony A., Bryja V., Stricker S. Negative regulation of Wnt signaling mediated by CK1-phosphorylated Dishevelled via Ror2. FASEB J. 2010;24:2417–2426. doi: 10.1096/fj.09-150615. [DOI] [PubMed] [Google Scholar]
- 13.Mlodzik M. Planar cell polarization: do the same mechanisms regulate Drosophila tissue polarity and vertebrate gastrulation? Trends Genet. 2002;18:564–571. doi: 10.1016/s0168-9525(02)02770-1. [DOI] [PubMed] [Google Scholar]
- 14.Strutt H., Warrington S.J., Strutt D. Dynamics of core planar polarity protein turnover and stable assembly into discrete membrane subdomains. Dev. Cell. 2011;20:511–525. doi: 10.1016/j.devcel.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei Y.-P., Zhang T., Li H., Wu B.-L., Jin L., Wang H.-Y. VANGL2 mutations in human cranial neural-tube defects. N. Engl. J. Med. 2010;362:2232–2235. doi: 10.1056/NEJMc0910820. [DOI] [PubMed] [Google Scholar]
- 16.Allache R., De Marco P., Merello E., Capra V., Kibar Z. Role of the planar cell polarity gene CELSR1 in neural tube defects and caudal agenesis. Birth Defects Res. Part A - Clin. Mol. Teratol. 2012;94:176–181. doi: 10.1002/bdra.23002. [DOI] [PubMed] [Google Scholar]
- 17.Torban E., Patenaude A.-M., Leclerc S., Rakowiecki S., Gauthier S., Andelfinger G., Epstein D.J., Gros P. Genetic interaction between members of the Vangl family causes neural tube defects in mice. Proc. Natl. Acad. Sci. USA. 2008;105:3449–3454. doi: 10.1073/pnas.0712126105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson A., Escuin S., Doudney K., Vekemans M., Stevenson R.E., Greene N.D., Copp A.J., Stanier P. Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum. Mutat. 2012;33:440–447. doi: 10.1002/humu.21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funato Y., Michiue T., Terabayashi T., Yukita A., Danno H., Asashima M., Miki H. Nucleoredoxin regulates the Wnt/planar cell polarity pathway in Xenopus. Genes Cells. 2008;13:965–975. doi: 10.1111/j.1365-2443.2008.01220.x. [DOI] [PubMed] [Google Scholar]
- 20.Funato Y., Michiue T., Asashima M., Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat. Cell Biol. 2006;8:501–508. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- 21.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy D.J., Humburg P., Kanapin A., Rivas M.A., Gaulton K., Cazier J.B., Donnelly P. Choice of transcripts and software has a large effect on variant annotation. Genome Med. 2014;6:26. doi: 10.1186/gm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eldomery M.K., Coban-Akdemir Z., Harel T., Rosenfeld J.A., Gambin T., Stray-Pedersen A., Küry S., Mercier S., Lessel D., Denecke J. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26. doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fromer M., Moran J.L., Chambert K., Banks E., Bergen S.E., Ruderfer D.M., Handsaker R.E., McCarroll S.A., O’Donovan M.C., Owen M.J. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gambin T., Akdemir Z.C., Yuan B., Gu S., Chiang T., Carvalho C.M.B., Shaw C., Jhangiani S., Boone P.M., Eldomery M.K. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45:1633–1648. doi: 10.1093/nar/gkw1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krawczak M., Cooper D.N. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum. Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 29.Roifman M., Marcelis C.L.M., Paton T., Marshall C., Silver R., Lohr J.L., Yntema H.G., Venselaar H., Kayserili H., van Bon B., FORGE Canada Consortium De novo WNT5A-associated autosomal dominant Robinow syndrome suggests specificity of genotype and phenotype. Clin. Genet. 2015;87:34–41. doi: 10.1111/cge.12401. [DOI] [PubMed] [Google Scholar]
- 30.Digilio M.C., Conti E., Sarkozy A., Mingarelli R., Dottorini T., Marino B., Pizzuti A., Dallapiccola B. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am. J. Hum. Genet. 2002;71:389–394. doi: 10.1086/341528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iqbal Z., Cejudo-Martin P., de Brouwer A., van der Zwaag B., Ruiz-Lozano P., Scimia M.C., Lindsey J.D., Weinreb R., Albrecht B., Megarbane A. Disruption of the podosome adaptor protein TKS4 (SH3PXD2B) causes the skeletal dysplasia, eye, and cardiac abnormalities of Frank-Ter Haar Syndrome. Am. J. Hum. Genet. 2010;86:254–261. doi: 10.1016/j.ajhg.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Below J.E., Earl D.L., Shively K.M., McMillin M.J., Smith J.D., Turner E.H., Stephan M.J., Al-Gazali L.I., Hertecant J.L., Chitayat D., University of Washington Center for Mendelian Genomics Whole-genome analysis reveals that mutations in inositol polyphosphate phosphatase-like 1 cause opsismodysplasia. Am. J. Hum. Genet. 2013;92:137–143. doi: 10.1016/j.ajhg.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posey J.E., Harel T., Liu P., Rosenfeld J.A., James R.A., Coban Akdemir Z.H., Walkiewicz M., Bi W., Xiao R., Ding Y. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tauriello D.V.F., Jordens I., Kirchner K., Slootstra J.W., Kruitwagen T., Bouwman B.A.M., Noutsou M., Rüdiger S.G.D., Schwamborn K., Schambony A., Maurice M.M. Wnt/β-catenin signaling requires interaction of the Dishevelled DEP domain and C terminus with a discontinuous motif in Frizzled. Proc. Natl. Acad. Sci. USA. 2012;109:E812–E820. doi: 10.1073/pnas.1114802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punchihewa C., Ferreira A.M., Cassell R., Rodrigues P., Fujii N. Sequence requirement and subtype specificity in the high-affinity interaction between human frizzled and dishevelled proteins. Protein Sci. 2009;18:994–1002. doi: 10.1002/pro.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saal H.M., Prows C.A., Guerreiro I., Donlin M., Knudson L., Sund K.L., Chang C.F., Brugmann S.A., Stottmann R.W. A mutation in FRIZZLED2 impairs Wnt signaling and causes autosomal dominant omodysplasia. Hum. Mol. Genet. 2015;24:3399–3409. doi: 10.1093/hmg/ddv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKusick V.A. On lumpers and splitters, or the nosology of genetic disease. Perspect. Biol. Med. 1969;12:298–312. doi: 10.1353/pbm.1969.0039. [DOI] [PubMed] [Google Scholar]
- 42.Boone P.M., Campbell I.M., Baggett B.C., Soens Z.T., Rao M.M., Hixson P.M., Patel A., Bi W., Cheung S.W., Lalani S.R. Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles. Genome Res. 2013;23:1383–1394. doi: 10.1101/gr.156075.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lalani S.R., Liu P., Rosenfeld J.A., Watkin L.B., Chiang T., Leduc M.S., Zhu W., Ding Y., Pan S., Vetrini F. Recurrent muscle weakness with rhabdomyolysis, metabolic crises, and cardiac arrhythmia due to bi-allelic TANGO2 mutations. Am. J. Hum. Genet. 2016;98:347–357. doi: 10.1016/j.ajhg.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harel T., Yoon W.H., Garone C., Gu S., Coban-Akdemir Z., Eldomery M.K., Posey J.E., Jhangiani S.N., Rosenfeld J.A., Cho M.T., Baylor-Hopkins Center for Mendelian Genomics. University of Washington Center for Mendelian Genomics Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 2016;99:831–845. doi: 10.1016/j.ajhg.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu S., Yuan B., Campbell I.M., Beck C.R., Carvalho C.M.B., Nagamani S.C.S., Erez A., Patel A., Bacino C.A., Shaw C.A. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum. Mol. Genet. 2015;24:4061–4077. doi: 10.1093/hmg/ddv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayle R., Campbell I.M., Beck C.R., Yu Y., Wilson M., Shaw C.A., Bjergbaek L., Lupski J.R., Ira G. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho C.M.B., Lupski J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurooka H., Kato K., Minoguchi S., Takahashi Y., Ikeda J., Habu S., Osawa N., Buchberg A.M., Moriwaki K., Shisa H., Honjo T. Cloning and characterization of the nucleoredoxin gene that encodes a novel nuclear protein related to thioredoxin. Genomics. 1997;39:331–339. doi: 10.1006/geno.1996.4493. [DOI] [PubMed] [Google Scholar]
- 49.Funato Y., Terabayashi T., Sakamoto R., Okuzaki D., Ichise H., Nojima H., Yoshida N., Miki H. Nucleoredoxin sustains Wnt/β-catenin signaling by retaining a pool of inactive dishevelled protein. Curr. Biol. 2010;20:1945–1952. doi: 10.1016/j.cub.2010.09.065. [DOI] [PubMed] [Google Scholar]
- 50.Boles M.K., Wilkinson B.M., Wilming L.G., Liu B., Probst F.J., Harrow J., Grafham D., Hentges K.E., Woodward L.P., Maxwell A. Discovery of candidate disease genes in ENU-induced mouse mutants by large-scale sequencing, including a splice-site mutation in nucleoredoxin. PLoS Genet. 2009;5:e1000759. doi: 10.1371/journal.pgen.1000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazzeu J.F., Pardono E., Vianna-Morgante A.M., Richieri-Costa A., Ae Kim C., Brunoni D., Martelli L., de Andrade C.E., Colin G., Otto P.A. Clinical characterization of autosomal dominant and recessive variants of Robinow syndrome. Am. J. Med. Genet. A. 2007;143:320–325. doi: 10.1002/ajmg.a.31592. [DOI] [PubMed] [Google Scholar]
- 52.Habas R., Dawid I.B., He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bishop J.R., Schuksz M., Esko J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 54.Campos-Xavier A.B., Martinet D., Bateman J., Belluoccio D., Rowley L., Tan T.Y., Baxová A., Gustavson K.H., Borochowitz Z.U., Innes A.M. Mutations in the heparan-sulfate proteoglycan glypican 6 impair endochondral ossification and cause recessive omodysplasia. Am. J. Hum. Genet. 2009;84:760–770. doi: 10.1016/j.ajhg.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pilia G., Hughes-Benzie R.M., MacKenzie A., Baybayan P., Chen E.Y., Huber R., Neri G., Cao A., Forabosco A., Schlessinger D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat. Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 56.Ohkawara B., Yamamoto T.S., Tada M., Ueno N. Role of glypican 4 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development. 2003;130:2129–2138. doi: 10.1242/dev.00435. [DOI] [PubMed] [Google Scholar]
- 57.Tsuda M., Kamimura K., Nakato H., Archer M., Staatz W., Fox B., Humphrey M., Olson S., Futch T., Kaluza V. The cell-surface proteoglycan Dally regulates Wingless signalling in Drosophila. Nature. 1999;400:276–280. doi: 10.1038/22336. [DOI] [PubMed] [Google Scholar]
- 58.Waterson J., Stockley T.L., Segal S., Golabi M. Novel duplication in glypican-4 as an apparent cause of Simpson-Golabi-Behmel syndrome. Am. J. Med. Genet. A. 2010;152A:3179–3181. doi: 10.1002/ajmg.a.33450. [DOI] [PubMed] [Google Scholar]
- 59.Wiweger M.I., Avramut C.M., de Andrea C.E., Prins F.A., Koster A.J., Ravelli R.B.G., Hogendoorn P.C.W. Cartilage ultrastructure in proteoglycan-deficient zebrafish mutants brings to light new candidate genes for human skeletal disorders. J. Pathol. 2011;223:531–542. doi: 10.1002/path.2824. [DOI] [PubMed] [Google Scholar]
- 60.Allan W. Relation of hereditary pattern to clinical severity as illustrated by peroneal atrophy. Arch. Intern. Med. (Chic.) 1939;63:1123–1131. [Google Scholar]
- 61.Morton N.E. The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. Am. J. Hum. Genet. 1956;8:80–96. [PMC free article] [PubMed] [Google Scholar]
- 62.Sobreira N.L., Valle D. Lessons learned from the search for genes responsible for rare Mendelian disorders. Mol. Genet. Genomic Med. 2016;4:371–375. doi: 10.1002/mgg3.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brunner H.G., van Driel M.A. From syndrome families to functional genomics. Nat. Rev. Genet. 2004;5:545–551. doi: 10.1038/nrg1383. [DOI] [PubMed] [Google Scholar]
- 65.Qian D., Jones C., Rzadzinska A., Mark S., Zhang X., Steel K.P., Dai X., Chen P. Wnt5a functions in planar cell polarity regulation in mice. Dev. Biol. 2007;306:121–133. doi: 10.1016/j.ydbio.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamaguchi T.P., Bradley A., McMahon A.P., Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- 67.Li Y., Dudley A.T. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development. 2009;136:1083–1092. doi: 10.1242/dev.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwabe G.C., Trepczik B., Süring K., Brieske N., Tucker A.S., Sharpe P.T., Minami Y., Mundlos S. Ror2 knockout mouse as a model for the developmental pathology of autosomal recessive Robinow syndrome. Dev. Dyn. 2004;229:400–410. doi: 10.1002/dvdy.10466. [DOI] [PubMed] [Google Scholar]
- 69.Kuss P., Kraft K., Stumm J., Ibrahim D., Vallecillo-Garcia P., Mundlos S., Stricker S. Regulation of cell polarity in the cartilage growth plate and perichondrium of metacarpal elements by HOXD13 and WNT5A. Dev. Biol. 2014;385:83–93. doi: 10.1016/j.ydbio.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 70.Vinson C.R., Adler P.N. Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature. 1987;329:549–551. doi: 10.1038/329549a0. [DOI] [PubMed] [Google Scholar]
- 71.Klingensmith J., Nusse R., Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- 72.Mikels A.J., Nusse R. Purified Wnt5a protein activates or inhibits β-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhanot P., Brink M., Samos C.H., Hsieh J.C., Wang Y., Macke J.P., Andrew D., Nathans J., Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 74.Sato A., Yamamoto H., Sakane H., Koyama H., Kikuchi A. Wnt5a regulates distinct signalling pathways by binding to Frizzled2. EMBO J. 2010;29:41–54. doi: 10.1038/emboj.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sundaresan M., Yu Z.X., Ferrans V.J., Irani K., Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 76.Hartmann C., Tabin C.J. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;127:3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- 77.Pan W.J., Pang S.Z., Huang T., Guo H.Y., Wu D., Li L. Characterization of function of three domains in dishevelled-1: DEP domain is responsible for membrane translocation of dishevelled-1. Cell Res. 2004;14:324–330. doi: 10.1038/sj.cr.7290232. [DOI] [PubMed] [Google Scholar]
- 78.Bernatík O., Šedová K., Schille C., Ganji R.S., Červenka I., Trantírek L., Schambony A., Zdráhal Z., Bryja V. Functional analysis of dishevelled-3 phosphorylation identifies distinct mechanisms driven by casein kinase 1ϵ and frizzled5. J. Biol. Chem. 2014;289:23520–23533. doi: 10.1074/jbc.M114.590638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho H.-Y.H., Susman M.W., Bikoff J.B., Ryu Y.K., Jonas A.M., Hu L., Kuruvilla R., Greenberg M.E. Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc. Natl. Acad. Sci. USA. 2012;109:4044–4051. doi: 10.1073/pnas.1200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morita K., Miyamoto T., Fujita N., Kubota Y., Ito K., Takubo K., Miyamoto K., Ninomiya K., Suzuki T., Iwasaki R. Reactive oxygen species induce chondrocyte hypertrophy in endochondral ossification. J. Exp. Med. 2007;204:1613–1623. doi: 10.1084/jem.20062525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boone P.M., Yuan B., Campbell I.M., Scull J.C., Withers M.A., Baggett B.C., Beck C.R., Shaw C.J., Stankiewicz P., Moretti P. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am. J. Hum. Genet. 2014;95:143–161. doi: 10.1016/j.ajhg.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boone P.M., Liu P., Zhang F., Carvalho C.M.B., Towne C.F., Batish S.D., Lupski J.R. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet. Med. 2011;13:582–592. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trinh T.Q., Sinden R.R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 85.Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L.B., Francescatto L., Ozes B., Karaca E., Jhangiani S.N., Bainbridge M.N., Baylor-Hopkins Center for Mendelian Genomics Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 2015;12:1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y., Macke J.P., Abella B.S., Andreasson K., Worley P., Gilbert D.J., Copeland N.G., Jenkins N.A., Nathans J. A large family of putative transmembrane receptors homologous to the product of the Drosophila tissue polarity gene frizzled. J. Biol. Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.